Abstract
Peptide nucleic acids (PNAs) are the only class of nucleic acid mimics developed to date that has been shown to be capable of invading double helical B-form DNA. Recently, we showed that sequence limitation associated with PNA recognition can be relaxed by utilizing conformationally-preorganized γ-peptide nucleic acids (γPNAs). However, like all the previous studies, with the exception of triplex-binding, DNA strand invasion was performed in relatively low salt concentrations. When subjected to physiological ionic strengths, little to no binding was observed. On the basis of this finding it was not clear whether the lack of binding is due to the lack of base-pair opening, or to the lack of binding free energy—either of which would result in no productive binding. In this Article, we show that it is the latter. Under simulated physiological conditions DNA double helix is sufficiently dynamic to permit strand invasion by the designer oligonucleotide molecules provided that the required binding free energy can be met. This finding has important implication for the design oligonucleotides for recognition of B-DNA via direct Watson-Crick base-pairing.
Double helical DNA has traditionally been thought to be a relatively static molecular entity, specifically designed for the storage and safeguard of genetic information (1). Early theoretical estimations (2) and NMR imino-proton exchange studies (3, 4) placed the probability of internal base-pair opening right around 10−5, open base-pair lifetime ~ 10−7s, and base-pair activation energy in the range of 43–65 kJ/mol at ambient temperature. These initial estimates cast a grim outlook on the prospect of being able to access the Watson-Crick (W-C) hydrogen-bond donors and acceptors of nucleobases for recognition. As a consequence, efforts to establish sequence-specific interactions with double-stranded DNA (dsDNA) in the past have generally been focused on chemical groups that reside in the minor (5) and major (6, 7) grooves, in large part, because of the ease of accessibility and the precedence for their recognition in nature. However, over the past two decades, peptide nucleic acid (PNAs) (8), a particular class of nucleic acid mimics comprising a pseudopeptide backbone (Chart 1a, top), have been shown to be capable of invading dsDNA (8–10). This finding was significant because, contrary to the traditional belief, it demonstrates that W-C base-pairing interactions can be established with intact dsDNA at physiological temperature. Though they are promising as antigene reagents, because of the generality and specificity of recognition, the originally designed achiral PNAs can only recognize homopurine (11) and homopyrimidine targets (12). Mixed-sequence PNAs can invade topologically constrained supercoiled plasmid DNA (13, 14) and structurally perturbed regions of genomic DNA (15); however, they are unable to invade linear, double helical B-form DNA (B-DNA).
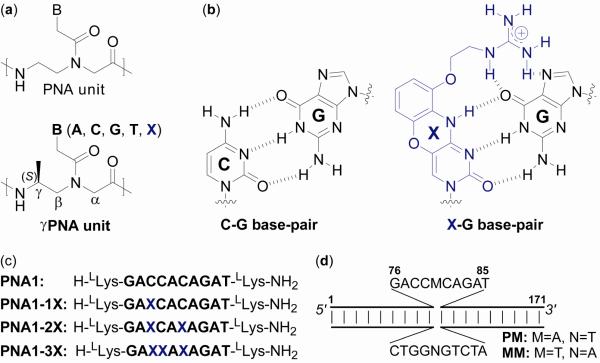
Chart 1.
Chemical structure of (a) PNA and γPNA building blocks and (b) C-G and X-G base-pairs, (c) sequence of γPNA oligomers, and (d) illustration of the DNA targets.
Recently, two approaches, tail-clamp (16, 17) and double-duplex invasion (18), have been developed, enabling mixed-sequence PNAs to invade B-DNA. However, they are not without limitations. The first approach still requires a stretch of homopyrimidine tract for anchoring the triplex binding, while the second approach, although more relaxed in sequence selection, is complicated by the need to use two separate strands of pseudocomplementary PNA to invade dsDNA. More recently, we showed that sequence restriction can be relaxed by utilizing conformationally preorganized γPNAs as molecular reagents (Chart 1a, bottom) (19). However, like all the other studies, with the exception of triplex-binding by homopyrimidine PNAs (20), strand invasion of linear double helical B-DNA was performed in buffers containing relatively low salt concentrations (11, 18, 19, 21, 22). When subjected to physiological ionic strengths, little to no binding was observed (23). On the basis of this result, it was not clear whether the lack of binding is due to the lack of base-pair opening (accessibility) or to the lack of binding free energy, since the kinetic of base-pair breathing and thermodynamic stability of DNA duplex are tightly coupled. Knowing the answer to this question is important because it will tell us whether W-C recognition is a viable option for targeting mixed-sequence B-DNA under physiological conditions. If it the former, there is little that can be done from a design standpoint. However, if it is the latter, this issue could be resolved through molecular design. Herein we show that it is the latter. Under simulated physiological conditions, DNA double helix is sufficiently dynamic to permit strand invasion by the designer oligonucleotide molecules provided that the required binding free energy can be met.
Materials and Methods
Monomer and oligomer synthesis
Boc-protected PNA monomers were obtained from ASM Research Chemicals. Methyl-containing γPNA monomers were prepared according to published procedures (24). G-clamp nucleobase and corresponding γPNA monomer were prepared according to the protocol reported by Chenna and coworkers (23). The oligomers were synthesized on MBHA (4-methylbenzhydrylamine) resin according to standard procedures (25). The oligomers were cleaved from the resin using m-cresol:thioanisole:TFMSA:TFA (1:1:2:6) cocktail, and the resulting mixtures were precipitated with ethyl ether, purified by RP-HPLC and characterized by MALDI-TOF mass spectrometer.
Cloning
Two DNA inserts, containing a perfectly-matched (PM) and single-base mismatched (MM) binding site, were cloned into the BamHI-EcoRI restriction site of pSuper plasmid vector (26). The recombinant vectors were transformed into E. coli (X-Gold 10). Following selection, clones were expanded and the plasmids were extracted using Qiagen plasmid extraction kit. The identities of the inserts were verified by sequencing. Insert 1 (PM): p-gatcgatatctatgagaccacagatctaagctagc-3'/p-aattgctagcttagatctgtggtctcatagatatc-3', Insert 2 (MM): p-gatcgatatctatgagacctcagatctaagctagc-3'/p-aattgctagcttagatctgaggtctcatagatatc-3'.
Polymerase Chain Reaction (PCR)
The 171bp-DNA fragments containing PM and MM binding sites were PCR-amplified from the corresponding plasmid vectors using the following primer set: 5'-gctactcaagctttcactatagggcgaattgga-3'/5'-gtgttcccgcctagtgaca-3'. The PCR reaction was performed as followed: (i) 95 °C for 2 min, (ii) 95 °C for 30 sec, (iii) 55 °C for 30 sec, (iv) 72 °C for 1 min, (v) repeated steps ii–iv for 32 cycles. The reaction mixtures were pooled and quenched with 0.2× vol of 10 mM EDTA. The mixtures were extracted twice with 1× vol phenol:chloroform:IAA (24:5:1), and the aqueous portions were combined and precipitated by adding 1 μL of 5 mg/mL glycogen, 0.1× vol of 3 M sodium acetate and 3× vol of cold ethanol, and placed in dried ice for 30 min. Following centrifugation and removal of the liquid layers, the remaining DNA pellets were washed 3× with 70% ethanol, air-dried, and then reconstituted with Nanopure water. The concentration of the PCR products (171bp-linear DNA fragments) were estimated based on the following parameters: 1 OD260nm = 50 μg/mL and MW = 649 Dalton/bp.
Gel-shift assay
Following incubation of the DNA targets with the appropriate oligomers at the indicated concentrations, time-points and temperatures, the samples were separated on precasted, 10% nondenaturing polyacrylamide gels using 0.5× TBE buffer. The gels were run at 10 V/cm for 3 hr in a water-bath at 37 °C. After electrophoresis, the gels were stained with 1× SYBR-Gold for 5 min, washed 2× with 0.5× TBE buffer, and then imaged using a gel-documentation system (BioDoc-It System). The images were then inverted using Adobe Photoshop 6.0.
DNase-I footprinting assay
The primer set: 5'-gctactcaagctttcactatagggcgaattgga-3'/5'-gtgttcccgcctagtgaca-3' was used to PCR-amplify the DNA fragments containing PM and MM binding sites from the corresponding vectors. The PCR products were digested with HindIII, heat-inactivated and then 3'-labeled with P-32 using the Klenow fragment of E. coli DNA polymerase. Notice that in this case the 3-end of the target DNA strand was labeled with P-32. The DNA targets were gel-purified and ethanol-precipitated. The radioactively-labeled targets were then redissolved in water and divided into aliquots containing approximately 10,000 cpm each. The invasion complex was initiated by incubating 10,000 cpm of the labeled and 0.2 μM of the cold (unlabeled) DNA target with the indicated oligomers and concentrations in a simulated physiological buffer (10 mM sodium phosphate, 2 mM MgCl2, 150 mM KCl; pH 7.4) at 37 °C. Footprinting of the invasion complex was performed by incubating the mixtures with 1 unit of DNase-I for 30 sec. The mixtures were then immediately quenched by adding 20 μL of the Stop Buffer (1.5 M sodium acetate, 10 mM EDTA, 1 M β-mercaptoethanol, 250 μg/mL calf thymus) and 400 μL of chilled ethanol. The samples were briefly vortexed and placed in dried-ice for 30 min, followed by centrifugation and washing 3× with 70% ethanol. The samples were air-dried and then separated on 10% urea-denaturing polyacrylamide gels. The cleavage patterns were visualized by autoradiography.
DEPC chemical probing assay
The PCR primer set: 5'-tcactatagggcgaattgga-3'/5'-gctactcaagcttgtgttcccgcctagtgaca-3' was used to PCR-amplify the DNA targets containing PM and MM binding sites from the corresponding plasmid vectors. The PCR products were digested with HindIII and 3'-labeled with P-32 under identical conditions as described above. The invasion complex was prepared the same way as that for DNase-I footprinting. DEPC treatment was initiated by adding 10% vol of DEPC into the invasion samples. The mixtures were incubated at room temperature for 5 min, followed by precipitation and piperidine treatment. The samples were then separated on 10% urea-denaturing polyacrylamide gels and the cleavage patterns were visualized by autoradiography.
Results and Discussion
To address the question of accessibility versus thermodynamic stability, we synthesized a series of decameric γPNA oligomers containing the same nucleobase sequence content but with a different number of cytosine (C) to G-clamp (X) nucleobase substitutions (Chart 1b and c). We then assessed their DNA strand invasion capability using a combination of gel-shift, enzymatic footprinting, and chemical probing assays. The C→X nucleobase substitutions were made in an attempt to systematically improve the binding free energy of γPNAs without resorting to changing the nucleobase sequence or chain length. This particular cytosine analogue was chosen because prior studies showed that C→X substitution can significantly enhance the thermodynamic stability of the PNA-DNA duplex (27). Given the already strong binding affinity of γPNAs (22), making such nucleobase substitutions should significantly further enhance their thermodynamic stability. Implementing such a design would allow us to survey the invasion capability of γPNAs over a range of binding free energy. Our working hypothesis is that if DNA strand invasion under simulated physiological conditions is predominated by thermodynamics, we would expect a gradual increase in the invasion efficiency with binding free energy, or in this case the number of C→X nucleobase substitutions. On the other hand, if base-pair accessibility is the determining factor in DNA strand invasion, we do not expect the above correlation but instead expect a steep dependence on temperature, which has a direct effect on the rate of base-pair opening.
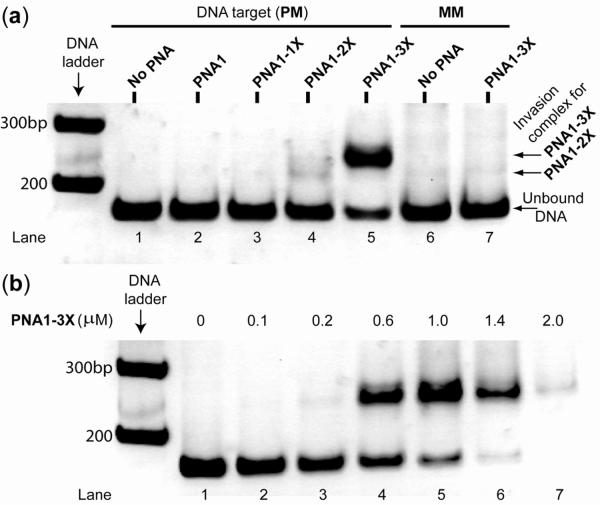
A gel-shift assay was performed by incubating a 171bp, linear doubled-stranded B-DNA containing an internal, perfectly-matched (PM) binding site (Chart 1d) with various γPNA oligomers in a buffer containing physiological salt concentrations (28) (10 mM sodium phosphate, 2 mM MgCl2, 150 mM KCl; pH 7.4) at 37 °C. The mixtures were separated on nondenaturing polyacrylamide gels and stained with SYBR-Gold for visualization. Inspection of Figure 1a reveals no binding for PNA1 or PNA1-1X, but a small amount for PNA1-2X, as evidenced from formation of a shifted band following 16 hr incubation. This result is consistent with an earlier finding (23). Upon further increase in the number of C→X nucleobase substitutions from 2 to 3, we noticed a dramatic increase in the intensity of the shifted band, from ~ 3 % with PNA1-2X (lane 4) to ~ 80% with PNA1-3X (lane 5), indicating that more of the bound complex was formed. Formation of this complex was sequence-specific, since no trace of the shifted band was observed with DNA containing an inverted, single-base mismatched (MM) binding site (compare lane 7 to lane 6). Nucleobase inversion is expected to have minimal impact on the thermodynamic stability (or base-pair breathing rate) of the DNA double helix. Thus, the difference in binding efficiency is predominantly due to sequence mismatch discrimination. We attribute the high level of sequence specificity to the competitive binding of the native complementary DNA strand. Our attempt to drive the reaction to completion by increasing the concentration of PNA1-3X was unsuccessful because beyond a 5:1 (PNA1-3X:DNA) ratio, the DNA bands gradually disappeared from the gel (Figure 1b, compare lanes 6 & 7 to lane 5). This is probably due to aggregation and nonspecific binding of PNA1-3X to DNA as the result of the charge-neutral polyamide backbone and hydrophobic character of the methyl group at the γ-backbone. In addition to the simulated physiological buffer, we have examined a number of other biologically relevant buffers including those employed in restriction digestion, transcription, and replication to assess the generality of PNA1-3X binding. In all of these buffers, some of which contained concentrations of MgCl2 as high as 10 mM, binding efficiencies greater than 60% were achieved within a 16 hr incubation period (Figure 2). Longer incubation times did not produce additional binding, indicating that equilibrium had been reached. Taken together, these results show that PNA1-3X can invade dsDNA under physiologically relevant conditions. Strand invasion is unique to γPNA, since no evidence of binding was observed for the unmodified PNA containing the same nucleobase sequence as PNA1-3X.
Figure 1.
Result of a gel-shift assay following incubation of 0.2 μM of a 171bp-DNA fragment containing (a) a perfectly-matched (PM, lanes 1–5) and single-base mismatched (MM, lanes 6 and 7) binding site with 1.0 μM of the indicated γPNA oligomers, and (b) a PM binding site with the indicated concentrations of PNA1-3X oligomer in a simulated physiological buffer at 37 °C for 16 hr, followed by separation on nondenaturing PAGE and staining with SYBR-Gold.
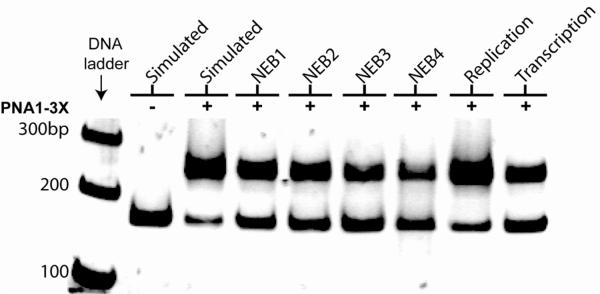
Figure 2.
Effects of biological buffers on DNA strand invasion by PNA1-3X. Gel-shift assay was performed by incubating 0.2 μM of DNA containing a PM binding site with 1.0 μM of PNA1-3X in the indicated buffers at 37 °C for 16 hr. Simulated: 10 mM NaPi, 2 mM MgCl2, 150 mM KCl; NEB1 (New England Biolabs buffer 1, 1×): 10 mM Bis tris propane·HCl, 10 mM MgCl2, 1 mM dithiothreitol; NEB2: 10 mM Tris·HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol; NEB3: 10 mM Tris·HCl, 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol; NEB4: 20 mM Tris·OAc, 50 mM KOAc, 10 mM MgCl2, 1 mM dithiothreitol; Replication: 10 mM Tris·HCl, 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100; and Transcription: 40 mM Tris·HCl, 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine.
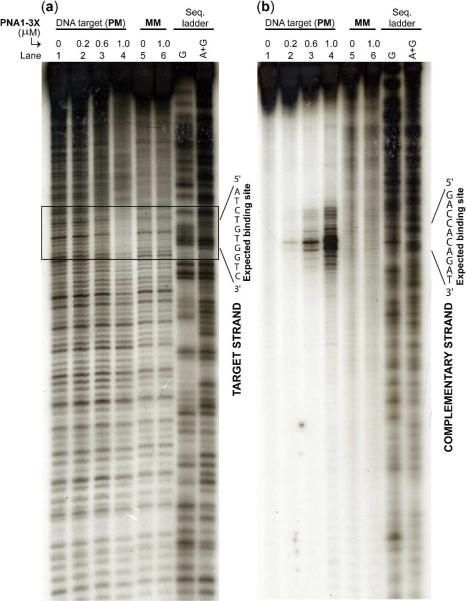
To confirm that binding of PNA1-3X to DNA occurred at the expected site and through a strand invasion mechanism, we performed DNase footprinting and DEPC (diethyl pyrocarbonate) chemical probing assays. DNase is an endonuclease enzyme that indiscriminately cleaves dsDNA. Selective binding of PNA1-3X to dsDNA is expected to block DNase digestion, resulting in a footprinting pattern following electrophoretic separation and detection of the DNA targets. DEPC, on the other hand, is a chemical reagent known to react selectively with adenine and to a smaller extent with the guanine nucleobase of single-stranded or perturbed regions of double-stranded DNA (29). Strand invasion of DNA by PNA1-3X is expected to result in local displacement of the homologous DNA strand. The looped-out strand can be revealed in the form of strand cleavage following DEPC and subsequent piperidine treatments. Consistent with these predictions, we observed a distinct footprinting pattern on the target strand, localized at the expected binding site following incubation of DNA with PNA1-3X (Figure 3a). The extent of the footprint became more pronounced as PNA1-3X concentrations increased, and it was only observed with DNA containing a PM binding site (lanes 1–4). Likewise, we observed cleavage of the complementary DNA strand directly across from the binding site (Figure 3b). Strand cleavage occurred only with DNA containing a PM binding site (lanes 1–4). These results are consistent with PNA1-3X binding selectively to its target site through a strand invasion mechanism.
Figure 3.
(a) DNase-I footprinting, and (b) DEPC-chemical probing assay following incubation 0.2 μM of the cold and 10,000 cpm of the 3'-labeled DNA target containing a PM (lanes 1–4) and MM (lanes 5 and 6) binding site with the indicated concentrations of PNA1-3X in a simulated physiological buffer at 37 °C for 16 hr. In panel (a) the 3'-end of the target strand was labeled with P-32, and in panel (b) the homologous strand was labeled with P-32.
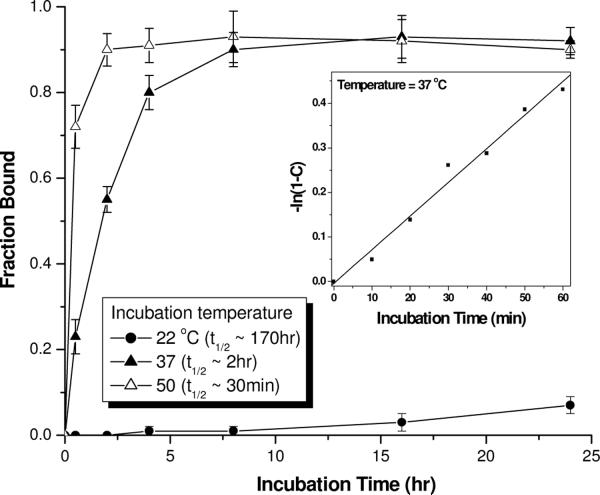
Next, we assessed the effects of temperature and incubation-time on the efficiency of DNA strand invasion. Our result shows that the efficiency of strand invasion is strongly dependent on temperature in the 22–37 °C regimes (Figure 4). This is not surprising since temperature has a direct effect on the rate of base-pair opening. Strand invasion was extremely inefficient at ambient temperature, in which case less than 10% invasion took place after 24 hr incubation. The rate of strand invasion dramatically increased at 37 °C, reaching equilibrium within ~ 6 hr incubation. Further increase in temperature resulted in further increase in the rate of strand invasion, but the difference in the rate at 37 and 50 °C is not as dramatic as compared to that at 22 and 37 °C (t1/2, 22°C ~ 170 hr; t1/2, 37°C ~ 2 hr; t1/2, 45°C ~ 30 min). There is only a 4-fold difference in the t1/2 in the higher temperature regimes as compared to 135-fold in the lower temperature regimes. Regardless of whether the samples were incubated at 37 or 50 °C, the invasion profiles reached a plateau corresponding to ~ 90% binding efficiency. This result indicates that at 37 °C, DNA double helix is sufficiently dynamic to permit strand invasion by γPNAs. However, whether successful strand invasion takes place depends on whether γPNA has the required binding free energy to displace the native complementary DNA strand.
Figure 4.
Effects of temperature and incubation-time on DNA strand invasion by PNA1-3X. The fraction bound was determined by gel-shift assays following incubation of 0.2 μM of DNA containing a PM binding site with 1.0 μM of PNA1-3X in a simulated physiological buffer at the indicated temperatures and time-points. t1/2 is defined as the time it took to reach 50% binding. Inset: A plot of −ln(1−C) vs. incubation time; the data shows that strand of invasion of B-DNA by PNA1-3X at 37 °C obeys pseudofirst-order kinetics: C = 1 − exp(−kpst), where C is the fraction bound at time t and kps is the pseudofirst-order rate constant (kps, 37 °C = 0.0075 min−1).
Conclusion
In summary, we have shown that PNA1-3X can invade double helical B-DNA under physiologically relevant conditions. Improvements in thermodynamic stability significantly enhance the efficiency of DNA strand invasion. We have already demonstrated how additional binding free energy may be achieved—through backbone preorganization and nucleobase substitution in this study, covalent attachment of a DNA-intercalating agent, such as acridine, as demonstrated in prior studies (20, 22), and extending the length of oligonucleotides (19). Besides cytosine, other nucleobases, including thymine, could also be modified to improve their hydrogen-bonding and base-stacking capabilities. Important work in this area has already begun in earnest by Nielsen and coworkers (30, 31). Replacement of natural nucleobases with synthetic analogues capable of forming additional hydrogen-bonding interactions and larger and geometrically aligned hydrophobic cores would further expand the recognition repertoire of PNA and γPNA. Issues with aggregation and nonspecific binding, as observed at high PNA and γPNA concentrations, could be alleviated by installing a hydrophilic group at the γ-backbone (24). We have already embarked on this front, the result of which will be reported in due course.
γPNAs are attractive, as compared to other classes of antigene reagents developed to date (5, 32–36), because they are relatively easy to synthesize and they hybridize to their targets (via strand invasion) in a highly sequence-specific and predictable manner in accordance with the Watson-Crick base-pairing rules. Molecules that can recognize and bind double-stranded DNA in a sequence-specific manner are of considerable interest in biology, biotechnology and medicine. They could be used as molecular tools to probe sequence information, manipulate genome's structure and function, and regulate gene expression at the transcriptional level, which may be more effective than the antisense approach because it shuts down gene expression at the initial step—as well as potential therapeutic and diagnostic reagents for the treatment and detection of genetic diseases.
Acknowledgement
We thank Prof. B. A. Armitage, M. Ly, and J. Deems for reading the manuscript and for providing insightful comments and suggestions.
This work was supported by the National Institutes of Health (GM076251) and the National Science Foundation (CHE-1012467).
Abbreviations and Textual Footnotes
- PNA
peptide nucleic acid
- γPNA
γ-backbone modified peptide nucleic acid
- A
adenine
- C
cytosine
- G
guanine
- T
thymine
- X
G-clamp
- TFA
trifluoroacetic acid
- TFMSA
trifluoromethanesulfonic acid
- DEPC
diethyl pyrocarbonate
References
- 1.Frank-Kamenetskii MD. How the double helix breathes. Nature. 1987;328:17–18. doi: 10.1038/328017a0. [DOI] [PubMed] [Google Scholar]
- 2.Frank-Kamenetskii MD. Structure & Motion: Membranes, Nucleic Acids & Proteins. In: Clementi E, Corongiu G, Sarma MH, Sarma RH, editors. Proc. Int. Symp. Struct. Dyn. Membranes, Nucleic Acids & Proteins. Adenine Press; Rome, Italy: 1985. pp. 417–432. [Google Scholar]
- 3.Gueron M, Kochoyan M, Leroy J-L. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987;328:89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- 4.Leroy JL, Kochoyan M, Huynh-Dinh T, Gueron M. Characterization of base-pair opening in deoxynucleotide duplexes using catalyzed exchange of the imino proton. J. Mol. Biol. 1988;200:223–238. doi: 10.1016/0022-2836(88)90236-7. [DOI] [PubMed] [Google Scholar]
- 5.Dervan PB. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 6.Thuong NT, Helene C. Sequence-specific recognition and modification of double-helical DNA by oligonucleotides. Angew. Chem.-Int. Edit. Engl. 1993;32:666–690. [Google Scholar]
- 7.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen PE. Peptide nucleic acid. A molecule with two identities. Acc. Chem. Res. 1999;32:624–630. [Google Scholar]
- 10.Ishihara T, Corey DR. Rules for strand invasion by chemically modified oligonucleotides. J. Am. Chem. Soc. 1999;121:2012–2020. [Google Scholar]
- 11.Nielsen PE, Christensen L. Strand displacement binding of a duplex-forming homopurine PNA to a homopyrimidine duplex DNA target. J. Am. Chem. Soc. 1996;118:2287–2288. [Google Scholar]
- 12.Egholm M, Nielsen PE, Buchardt O, Berg RH. Recognition of guanine and adenine in DNA by cytosine and thymine containing peptide nucleic acids (PNA) J. Am. Chem. Soc. 1992;114:9677–9678. [Google Scholar]
- 13.Bentin T, Nielsen PE. Enhanced peptide nucleic acid binding to supercoiled DNA: Possible implications for DNA “breathing” dynamics. Biochemistry. 1996;35:8863–8869. doi: 10.1021/bi960436k. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Ishihara T, Corey DR. Strand invasion by mixed base PNAs and a PNA-peptide chimera. Nucleic Acids Res. 2000;28:3332–3338. doi: 10.1093/nar/28.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, Mendelson CR, Corey DR. Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nat. Chem. Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- 16.Bentin T, Larsen HJ, Nielsen PE. Combined triplex/duplex invasion of double-stranded DNA by “tail-clamp” peptide nucleic acid. Biochemistry. 2003;42:13987–13995. doi: 10.1021/bi0351918. [DOI] [PubMed] [Google Scholar]
- 17.Kaihatsu K, Shah RH, Zhao X, Corey DR. Extending recognition by peptide nucleic acids (PNAs): binding to duplex DNA and inhibition of transcription by tail-clamp PNA-peptide conjugate. Biochemistry. 2003;42:13996–14003. doi: 10.1021/bi035194k. [DOI] [PubMed] [Google Scholar]
- 18.Lohse J, Dahl O, Nielsen PE. Double duplex invasion by peptide nucleic acid: A general principle for sequence-specific targeting of double-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11804–11808. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He G, Rapireddy S, Bahal R, Sahu B, Ly DH. Strand invasion of extended, mixed sequence B-DNA by gammaPNAs. J. Am. Chem. Soc. 2009;131:12088–12090. doi: 10.1021/ja900228j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentin T, Nielsen PE. Superior duplex DNA strand invasion by acridine conjugated peptide nucleic acids. J. Am. Chem. Soc. 2003;125:6378–6379. doi: 10.1021/ja029936t. [DOI] [PubMed] [Google Scholar]
- 21.Smolina IV, Demidov VV, Soldatenkov VA, Chasovskikh SG, Frank-Kameneteskii MD. End invasion of peptide nucleic acids (PNAs) with mixed-sequence composition into linear DNA duplexes. Nucleic Acid Res. 2005;33:e146. doi: 10.1093/nar/gni151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapireddy S, He G, Roy S, Armitage BA, Ly DH. Strand invasion of mixed-sequence B-DNA by acridine-linked, gamma-peptide nucleic acid (gamma-PNA) J. Am. Chem. Soc. 2007;129:15596–15600. doi: 10.1021/ja074886j. [DOI] [PubMed] [Google Scholar]
- 23.Chenna V, Rapireddy S, Sahu B, Ausin C, Pedroso E, Ly DH. A simple cytosine-to-G-clamp nucleobase substitution enables chiral gamma-PNAs to invade mixed-sequence double helical B-form DNA. ChemBioChem. 2008;9:2388–2391. doi: 10.1002/cbic.200800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dragulescu-Andrasi A, Rapireddy S, Frezza BM, Gayathri C, Gil RR, Ly DH. A simple gamma-backbone modification preorganizes peptide nucleic acid into a helical structure. J. Am. Chem. Soc. 2006;128:10258–10267. doi: 10.1021/ja0625576. [DOI] [PubMed] [Google Scholar]
- 25.Christensen L, Fitzpatrick R, Gildea B, Petersen KH, Hansen HF, Koch T, Egholm M, Buchardt O, Nielsen PE, Coull J, Berg RH. Solid-phase synthesis of peptide nucleic acids. J. Pept. Sci. 1995;1:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 26.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 27.Ortega J-A, Blas JR, Orozco M, Grandas A, Pedroso E, Robles J. Binding affinities of oligonucleotides and PNAs containing phenoxazine and G-clamp cytosine analogues are unusually sequence dependent. Org. Lett. 2007;9:4503–4506. doi: 10.1021/ol701826x. [DOI] [PubMed] [Google Scholar]
- 28.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Nat. Acad. Sci. U.S.A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furlong JC, Lilley DMJ. Highly selective chemical modification of cruciform loops by diethyl pyrocarbonate. Nucleic Acids Res. 1986;14:3995–4007. doi: 10.1093/nar/14.10.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eldrup AB, Nielsen BB, Haaima G, Rasmussen H, Kastrup JS, Christensen C, Nielsen PE. 1,8-Naphtharidin-2(1H)-ones--novel bicyclic and tricyclic analogues of thymine in peptide nucleic acids (PNAs) Eur. J. Org. Chem. 2001:1781–1790. [Google Scholar]
- 31.Eldrup AB, Christensen C, Haaima G, Nielsen PE. Substituted 1,8-naphthyridin-2(1H)-ones are superior to thymine in the recognition of adenine in duplex as well as triplex structure. J. Am. Chem. Soc. 2002;124:3254–3262. doi: 10.1021/ja0117027. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen PE. Sequence-selective DNA recognition by synthetic ligands. Bioconj. Chem. 1991;2:1–12. doi: 10.1021/bc00007a001. [DOI] [PubMed] [Google Scholar]
- 33.Sun JS, Helene C. Oligonucleotide-directed triple-helix formation. Curr. Opin. Struct. Biol. 1993;3:345–356. doi: 10.1016/s0959-440x(96)80051-0. [DOI] [PubMed] [Google Scholar]
- 34.Beerli RR, Barbas CFI. Engineering polydactyl zinc-finger transcription factors. Nat. Biotech. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 35.Jantz D, Amann BT, Gatto GJ, Berg JM. The design of functional DNA-binding proteins based on zinc finger domains. Chem. Rev. 2004;104:789–800. doi: 10.1021/cr020603o. [DOI] [PubMed] [Google Scholar]
- 36.Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat. Rev. Drug Discovery. 2003;2:361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]