Abstract
In animal cells, the primary cilium transduces extracellular signals through signaling receptors localized in the ciliary membrane, but how these ciliary membrane proteins are retained in the cilium is unknown. We found that ciliary membrane proteins were highly mobile, but their diffusion was impeded at the base of the cilium by a diffusion barrier. Septin 2 (SEPT2), a member of the septin family of guanosine triphosphatases that form a diffusion barrier in budding yeast, localized at the base of the ciliary membrane. SEPT2 depletion resulted in loss of ciliary membrane protein localization and Sonic hedgehog signal transduction, and inhibited ciliogenesis. Thus, SEPT2 is part of a diffusion barrier at the base of the ciliary membrane and is essential for retaining receptor-signaling pathways in the primary cilium.
The primary cilium is an antenna-like organelle protruding from the apical surface of almost every cell in a wide variety of organisms. The ciliary membrane is contiguous with the apical plasma membrane but has a unique set of proteins that sense and transduce a variety of extracellular signals, such as Sonic hedgehog (Shh) (1). These signaling pathways regulate gene expression during development and in adult life, and mutations in ciliary proteins give rise to a range of developmental defects (2, 3).
Signal transduction by the primary cilium depends on the enrichment of specific proteins in the ciliary membrane. Protein trafficking and intraflagellar transport from the cytoplasm into the cilium establish the characteristic intracellular distributions of ciliary membrane proteins (4, 5), but it is unknown how these proteins are subsequently retained in the ciliary membrane: For example, they might be immobilized in the cilium or impeded by a diffusion barrier (6-10).
To investigate these possible mechanisms, we used fluorescence recovery after photobleaching (FRAP) (11) to measure the diffusional mobility of four ciliary membrane proteins tagged with green fluorescent protein (GFP): two G protein–coupled receptors [serotonin receptor 6 (FlHtr6SEP) and somatostatin receptor (Sstr3GFP) (12)], and the membrane-anchored and ciliary-targeted cytoplasmic tail (amino acids 1 to 193) of fibrocystin (CTSPKHD1GFP) (13) in primary kidney inner medullary collecting duct (IMCD3) cells, and the Shh signaling pathway transducer smoothened (SmoYFP) (14) in mouse embryonic fibroblasts (MEF). All of these proteins localized to the cilium, but none recovered fluorescence when the whole cilium was photobleached (Fig. 1, A and B; fig. S1, A and B; and movies S1 to S6) (15), even though their mobility was high in the periciliary membrane, as shown for Htr6SEP (fig. S1C, table S1, and movie S7). In contrast, fluorescence recovery of the intraflagellar transport complex protein IFT88 (IFT88YFP) (16, 17) was high (Fig. 1, A and B, table S1, and movie S8).
Fig. 1.
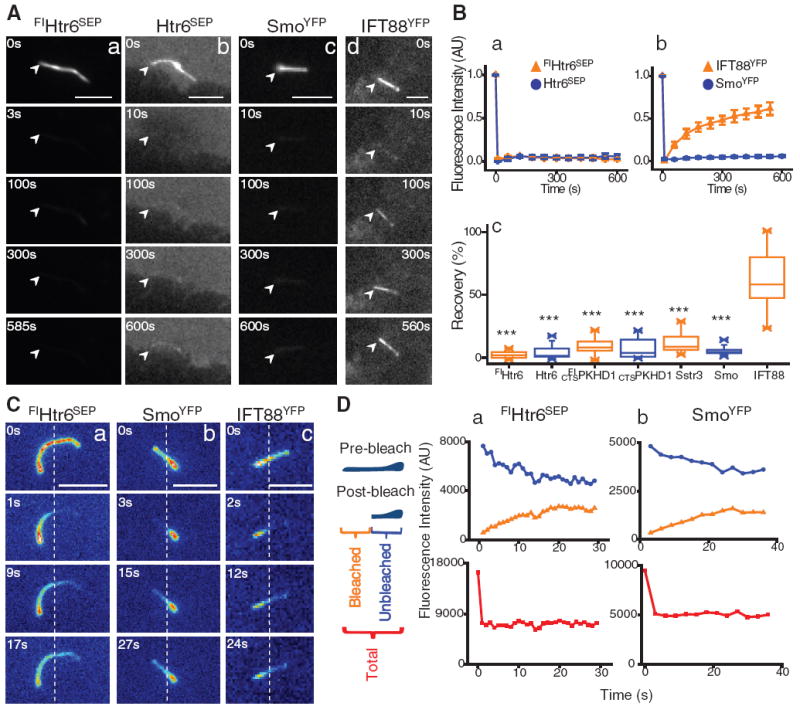
(A) FRAP of proteins in the whole cilium 24 hours after serum starvation. IMCD3 cells stably (a) or transiently (b) expressing Htr6SEP or IFT88YFP (d), and SmoYFP stably expressed in MEFs treated with 100 nM SAG for 24 hours (c); arrowheads mark one end of the cilium. Scale bars, 5 μm. (B) Kinetics of average (±SEM) fluorescence recovery of proteins photobleached in the whole cilium (a, b; n = 8 to 12). Summary of fluorescence recovery of ciliary membrane proteins as a percentage of the initial unbleached fluorescence level compared with IFT88YFP distribution (c; ***P < 0.0001). (C) FRAP of Htr6SEP (a), SmoYFP (b), and IFT88YFP (c) represented as heat-map images after photobleaching part of the cilium in IMCD3 cells; dotted lines mark the photobleached/unbleached boundary. Scale bars, 5 µm. (D) Representative example of kinetics of fluorescence recovery of photobleached region (orange curve), unbleached region (blue curve), and the two regions combined (red curve) of a primary cilium (n = 12 to 13).
The low diffusional mobility of these ciliary membrane proteins might be due to their immobilization within the primary cilium or to the presence of a diffusion barrier at the base of the ciliary membrane. To distinguish between these possibilities, we photobleached part of the cilium and measured the fluorescence recovery of ciliary membrane proteins (Fig. 1, C and D; fig. S1, D and E; and movies S9 to S13). Strikingly, all proteins were highly mobile within the ciliary membrane (fig. S1F and table S2), although fluorescence recovery was still confined within the cilium (Fig. 1, C and D, and fig. S1, D and E). Note that constituent plasma membrane proteins (GPIYFP and VSV-GGFP) were also excluded from the ciliary membrane (fig. S2). Therefore, the diffusion of membrane proteins between the ciliary and periciliary membrane, unlike the movement of IFT88 and cytoplasmic GFP (18), is impeded at the base of the cilium by a diffusion barrier.
To identify molecular components of this ciliary membrane diffusion barrier, we focused on septins. Septins are oligomeric guanosine triphosphatases (GTPases) that form a diffusion barrier at the mother-bud neck of budding yeast (10, 19, 20) and localize at the annulus that may form a diffusion barrier between the middle and principal piece of the mammalian sperm flagellum (21, 22). Septin 2 (SEPT2) (23) localized to the base of the cilium at the boundary between the ciliary and periciliary membrane in IMCD3 cells (Fig. 2A and fig. S3A) and in MEFs stably expressing SmoYFP (Fig. 2A). In some optical sections SEPT2 staining appeared as a ringlike structure of ~500 nm diameter (Fig. 2A), reminiscent of structures formed by recombinant septins in vitro (24), and as two fused dots (Fig. 2A and movies S14 and S15), similar to septin staining at the sperm annulus (21). In ~10% of cells, punctate SEPT2 also localized along and at the tip of the axoneme (fig. S3B).
Fig. 2.
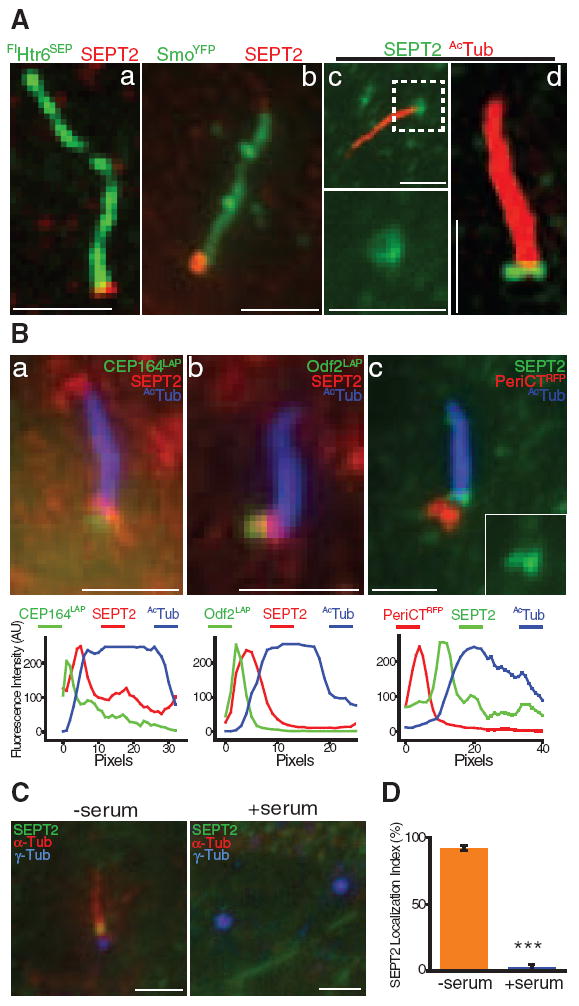
(A) SEPT2 localization at the cilium of IMCD3 cells stably expressing FlHtr6SEP (a), MEFs stably expressing SmoYFP treated with 100 nM SAG for 24 hours (b), and IMCD3 cells stained for acetylated α-tubulin (AcTub) (c; higher magnification of boxed region shown at the bottom), and imaged with a super-resolution Optical Microscope Experimental (OMX) system using three-dimensional structured illumination (d). Scale bars, 2 µm. (B) SEPT2 and AcTub localization at the cilium of IMCD3 cells transiently expressing CEP164LAP (a), Odf2LAP (b) or PeriCTRFP (c). Scale bars, 2 µm. (Lower panels) Fluorescence intensity profiles of protein staining from the basal body to the tip of the cilium. (C) IMCD3 cells grown for 24 hours without (–) or with (+) serum and stained for SEPT2, α-tubulin (α-tub), and γ-tubulin (γ-tub). Scale bars, 2 µm. (D) Percentage of cells with SEPT2 localized at the base of primary cilia (–serum) or pericentriolar region (+serum). Error bars represent SD of three independent experiments (n = 60 to 178 each; ***P < 0.0001).
SEPT2 localization at the base of the axoneme is similar to basal body distal appendage proteins. SEPT2, however, clearly localized between the ciliary axoneme [marked by acetylated α-tubulin (AcTub)] and structures marked by the distal appendage protein CEP164 (25), the subdistal/distal appendage protein outer dense fiber 2 (Odf2) (26), and the pericentriolar protein pericentrin (PeriCT) (27) (Fig. 2B and fig. S3, C to E). SEPT2, but not the ciliary axoneme or these basal body proteins, was solubilized in buffer containing 0.5% TritonX-100 (fig. S3F), indicating that SEPT2, like other septins (28, 29), is associated with membrane. In addition, SEPT2 did not localize to the centrosome in nonciliated cells (Fig. 2, C and D), further indicating that it is not a constituent centrosomal protein.
To investigate whether SEPT2 is required for cilium formation and ciliary membrane barrier function, we used transient transfection with small interfering RNA (siRNA) oligonucleotides to deplete ~ 69% SEPT2 from serum-starved IMCD3 cells (Fig. 3, A and B). In general, cells that were completely depleted of SEPT2 lacked a cilium (Fig. 3C and fig. S4, A and B), whereas cells that were partially depleted (~54.3% depletion at the base of primary cilia) had a cilium that was significantly shorter (3.46 ± 0.20 µm) than controls (4.78 ± 0.31 µm). We also established stable IMCD3 cell lines depleted of SEPT2 using three different short hairpin RNAs (shRNAs), which reduced SEPT2 levels by 80 to 90% (Fig. 3, D and E) and resulted in a more complete defect in ciliogenesis across the cell population (Fig. 3E and fig. S4, C and D).
Fig. 3.
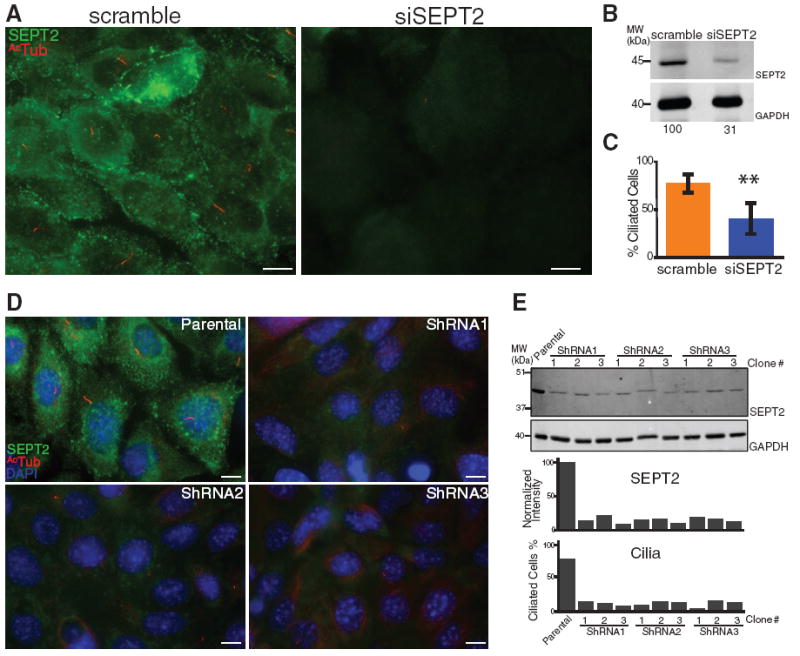
(A) IMCD3 cells transfected with scrambled (scramble) or SEPT2 siRNA (siSEPT2), serum-starved to induce ciliogenesis, and stained for SEPT2 and AcTub. Scale bars, 10 µm. (B) Whole-cell lysates of scramble and siSEPT2 cells immunoblotted for SEPT2 and glyceraldehyde phosphate dehydrogenase (GAPDH); lower panel, SEPT2 band intensities normalized to GAPDH. (C) Percentage of scramble and siSEPT2 cells with a cilium >1 μm long (±SD of three independent experiments; n = 101 to 205 each; **P = 0.0098). (D) IMCD3 cells (Parental), and IMCD3 cells stably depleted of SEPT2 using shRNA1, shRNA2, or shRNA3, stained for SEPT2, AcTub, and 4′,6′-diamidino-2-phenylindole (DAPI). Scale bars, 10 µm. (E) Whole-cell lysates of parental IMCD3 cells and three arbitrarily chosen clones of shRNA1, 2, or 3 cells immunoblotted for SEPT2 and GAPDH; lower panel, SEPT2 band intensities normalized to GAPDH, and percentage of parental and shRNA1, 2, and 3 IMCD3 cells with a cilium (n = 115 to 282).
To investigate whether SEPT2 is part of a diffusion barrier at the base of the ciliary membrane, we measured ciliary membrane protein mobility in the 10 to 15% of the stable SEPT2-depleted IMCD3 cells (shRNA1) that had a short cilium but did not have significant SEPT2 staining near the basal body (fig. S4C). We detected a significant increase in the diffusional mobility of four ciliary membrane proteins when the whole cilium was photobleached in SEPT2-depleted cells, compared with controls (Fig. 4, A to C; fig. S5, A and B; table S3; and movies S16 to S23). Furthermore, the barrier index, defined as the ratio of mean fluorescence intensity of these proteins in the ciliary membrane to surrounding periciliary membrane, was reduced significantly in SEPT2-depleted cells compared with controls (Fig. 4D and fig. S5C). Thus, SEPT2 depletion removed the ciliary membrane diffusion barrier in cells that could still assemble a cilium. Because the cilium was shorter in these cells, and absent in most SEPT2-depleted cells, it is possible that loss of the diffusion barrier contributed to the overall defect in ciliogenesis.
Fig. 4.
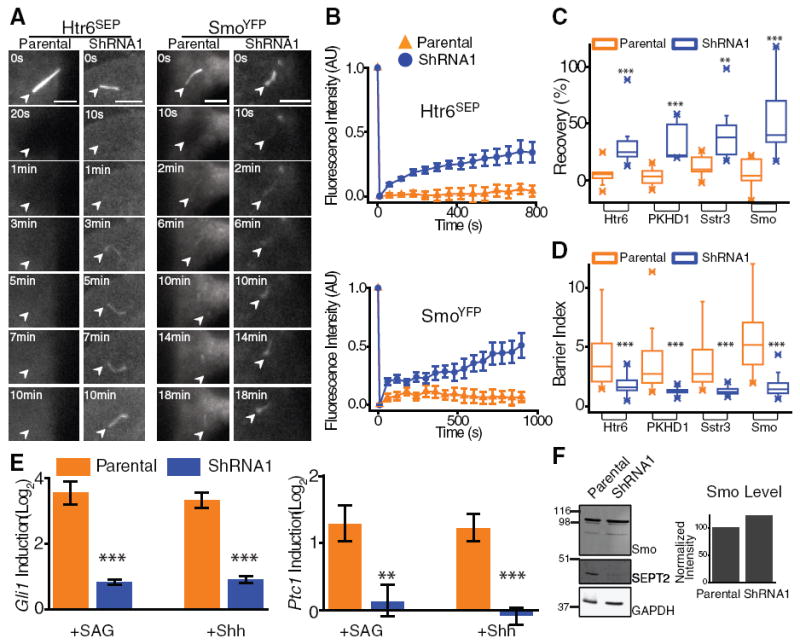
(A and B) FRAP of the whole cilium of parental IMCD3 (Parental) and shRNA1 IMCD3 cells transiently expressing Htr6SEP or SmoYFP and treated with 100 nM SAG for 24 hours. Scale bars, 5 μm. (B) Kinetics of average (±SEM) fluorescence recovery of photobleached proteins (n = 9 to 12). (C) Summary of percentage of fluorescence recovery of photobleached ciliary membrane proteins in parental and shRNA1 cells (**P < 0.002; ***P <0.0005). (D) Summary of barrier index in parental and shRNA1 cells (n = 31 to 59; ***P < 0.0001). (E) Gli1 (left) and Ptc1 (right) mRNA induction in parental and shRNA1 cells after 100 nM SAG or Shh treatment for 24 hours (average of three independent experiments ±SD; **P = 0.005; ***P <0.0007). (F) Whole-cell lysates of parental and shRNA1 cells immunoblotted for Smo, SEPT2, and GAPDH; normalized Smo levels in parental and shRNA1 cells (right).
We investigated whether loss of the ciliary membrane diffusion barrier after SEPT2 depletion affected cilium-dependent receptor signal transduction. Shh signaling requires enrichment of Smo in the ciliary membrane, and signal transduction results in increased Gli1 and Patched1 (Ptc1) mRNA levels (14, 30). In SEPT2-depleted cells, Smo accumulation in cilia was reduced, although total cellular levels were unaffected (Fig. 4, D and F, and fig. S5C). After induction with Shh or Smo agonist (SAG), Gli1 and Ptc1 mRNA levels were reduced significantly in SEPT2-depleted cells compared with controls (Fig. 4E). Thus, the SEPT2 diffusion barrier is required for cilium-dependent Shh signal transduction.
Here, we have identified a septin-containing diffusion barrier at the base of the ciliary membrane that is required to retain receptor-signaling pathways in the primary cilium. This diffusion barrier restricts the diffusion of ciliary membrane proteins between the ciliary and periciliary membrane but permits the diffusion of ciliary transport proteins (IFT88). This implies that newly synthesized ciliary membrane proteins are most likely inserted by IFT and Bardet-Biedl Syndrome proteins into the ciliary membrane above the diffusion barrier (7). Thus, septins appear to have evolutionarily conserved roles from fungi to animals in the functional compartmentalization of membrane domains.
Supplementary Material
Acknowledgments
We thank M. Kinoshita, S. Munro, G. J. Pazour, K. Mykytyn, D. R. Beier, C. Janke, J. Christianson, R. Kopito, and T. Stearns for reagents, and J. Atkins (Applied Precision, Inc.) for technical support. Supported by NIH grant GM35527 (W.J.N.), Department of Defense Breast Cancer Research Program Predoctoral Fellowship BC083077 (Q.H.), and Drexel CURE award from the State of Pennsylvania Tobacco Settlement Funds (E.T.S.). Supported in part by grants (M.V.N.) from the American Heart Association, the March of Dimes, NIH (GM089933), and the Sloan, Klingenstein and Searle Foundations. L.M. is a Research Specialist, and M.P.S. an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/science.1191054/DC1
Materials and Methods
Figs. S1 to S5
Tables S1 to S3
References
Movies S1 to S23
References and Notes
- 1.Eggenschwiler JT, Anderson KV. Annu Rev Cell Dev Biol. 2007;23:345. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall WF, Nonaka S. Curr Biol. 2006;16:R604. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Lancaster MA, Gleeson JG. Curr Opin Genet Dev. 2009;19:220. doi: 10.1016/j.gde.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emmer BT, Maric D, Engman DM. J Cell Sci. 2010;123:529. doi: 10.1242/jcs.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sloboda RD, Rosenbaum JL. J Cell Biol. 2007;179:575. doi: 10.1083/jcb.200709039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunnicutt GR, Kosfiszer MG, Snell WJ. J Cell Biol. 1990;111:1605. doi: 10.1083/jcb.111.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachury MV, Seeley ES, Jin H. Annu Rev Cell Dev Biol. 2010;26 doi: 10.1146/annurev.cellbio.042308.113337. 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musgrave A, et al. Planta. 1986;167:544. doi: 10.1007/BF00391231. [DOI] [PubMed] [Google Scholar]
- 9.Vieira OV, et al. Proc Natl Acad Sci U S A. 2006;103:18556. doi: 10.1073/pnas.0608291103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caudron F, Barral Y. Dev Cell. 2009;16:493. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Hu Q, Nelson WJ, Spiliotis ET. J Biol Chem. 2008;283:29563. doi: 10.1074/jbc.M804962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Mol Biol Cell. 2008;19:1540. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Follit JA, Li L, Vucica Y, Pazour GJ. J Cell Biol. 2010;188:21. doi: 10.1083/jcb.200910096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohatgi R, Milenkovic L, Scott MP. Science. 2007;317:372. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 15.Huang K, et al. J Cell Biol. 2007;179:501. doi: 10.1083/jcb.200704069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran PV, et al. Nat Genet. 2008;40:403. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pazour GJ, et al. J Cell Biol. 2000;151:709. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calvert PD, Schiesser WE, Pugh EN., Jr J Gen Physiol. 2010;135:173. doi: 10.1085/jgp.200910322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barral Y, Mermall V, Mooseker MS, Snyder M. Mol Cell. 2000;5:841. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- 20.Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Science. 2000;290:341. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 21.Ihara M, et al. Dev Cell. 2005;8:343. doi: 10.1016/j.devcel.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Kissel H, et al. Dev Cell. 2005;8:353. doi: 10.1016/j.devcel.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 23.Spiliotis ET, Hunt SJ, Hu Q, Kinoshita M, Nelson WJ. J Cell Biol. 2008;180:295. doi: 10.1083/jcb.200710039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinoshita M, Field CM, Coughlin ML, Straight AF, Mitchison TJ. Dev Cell. 2002;3:791. doi: 10.1016/s1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- 25.Graser S, et al. J Cell Biol. 2007;179:321. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa H, Kubo A, Tsukita S, Tsukita S. Nat Cell Biol. 2005;7:517. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 27.Gillingham AK, Munro S. EMBO Rep. 2000;1:524. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, et al. Curr Biol. 1999;9:1458. doi: 10.1016/s0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka-Takiguchi Y, Kinoshita M, Takiguchi K. Curr Biol. 2009;19:140. doi: 10.1016/j.cub.2008.12.030. [DOI] [PubMed] [Google Scholar]
- 30.Corbit KC, et al. Nature. 2005;437:1018. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


