Abstract
The future of genetic medicine hinges on successful intracellular delivery of nucleic acid-based therapeutics. While significant effort has concentrated on developing nano-carriers to improve the delivery aspects, scant attention has been paid to the synthetic process of poorly controlled nanocomplex formation. Proposed here is a reliable system to better control the complexation process, and thus the physical properties of the nanocomplexes, through microfluidics-assisted confinement (MAC) in picoliter droplets. We show that these homogeneous MAC-synthesized nanocomplexes exhibit narrower size distribution, lower cytotoxicity, and higher transfection efficiency compared to their bulk-synthesized counterparts. MAC represents a physical approach to control the energetic self-assembly of polyelectrolytes, thereby complementing the chemical innovations in nano-carrier design to optimize nucleic acid and peptide delivery.
Keywords: Nanomedicine, Gene delivery, Nanocomplexes, Microfluidics, Nanotoxicity
Nucleic acid-based therapeutics acting at the molecular level require an effective intracellular delivery system. Use of polycations to condense nucleic acids into nanocomplexes facilitates cellular internalization. This has inspired many innovative chemical designs to form nanoparticles with interesting functionalities, ranging from stealth properties through PEGylation to environment-specific unpacking via pH-sensitive or bioreductive degradable bonds in the polymeric carrier. While the carrier design has achieved tremendous progress, the process of assembling the nanocomplexes has received scant attention. Charge neutralization between cationic gene carriers and negatively charged nucleic acid payloads is a highly energetic process. Vortex mixing, a process used by almost all researchers to form these nanocomplexes via electrostatic self-assembly, introduces great variability into the quality of the nanocomplexes because of the metastable preparation and subsequent aggregation.1 This in turn leads to poor biological reproducibility and difficulty in establishing a robust structure-function relationship.2 To improve the physical properties of the nanocomplexes, we propose to control the complexation through microfluidics-assisted confinement (MAC) in picoliter droplets. The hypothesis is that confined diffusion in a small volume (hundreds picoliter) would facilitate the charge neutralization between the oppositely charged polyelectrolytes (diffusion length ~80μm, Dpolymer~10−6 cm2/sec, given Δt=30sec) to reach equilibrium, thereby yielding nanocomplexes that are more uniform and compact. This effective reaction may also exhaust the free polyelectrolytes within the volume, leaving a minimum of unreacted reagents, particularly the polycations that typically generate a polymer corona on surface of the nanocomplexes.
The promise of micro-incubators has been previously demonstrated for nanoparticle synthesis, such as metallic nanoparticles,3 oxide nanoparticles,4 nanocrystals,5 and recently on lipoplexes.6 However, there has been no study on the formulation of polyplexes, the complexation of polycations and DNA, using this approach. In this study, we examine the effects on transfection efficiency and cytotoxicity of a set of distinctive particulate parameters (size, heterogeneity and stability) enabled by MAC for a comparison with nanocomplexes synthesized under conventional bulk mixing. We present the first demonstration that the more homogeneous and compact MAC-synthesized polymeric nanocomplexes exhibit lower cytotoxicity and higher transfection efficiency. In the current formation of polyplexes by bulk mixing, the quality of the product often depends on the experience of the operator. For instance, the manner of mixing and the sequence of adding one polyelectrolyte to the other would make a drastic difference.7, 8 Our findings suggest that MAC is an attractive approach, not available previously, to formulate polyplexes in a robust, reproducible, and scalable manner. This operator-independent process will also benefit the synthesis of other nanoparticulate delivery systems, such as protein nanoparticles formed by complex coacervation.
Picoliter droplets were generated by a cross-flow microfluidic droplet generator shown in Figure 1a, through a competition between the continuous phase (carrier fluid, oil) and the disperse phase (the aqueous reagents).9 The DNA payloads and polycations were confined in individual water-in-oil droplets and subsequently self-assembled through electrostatic interaction, denoted as MAC-nanocomplexes (Figure 1b). Confining the self-assembly to within discrete droplets effectively eliminated dispersion and reduced non-specific adsorption to the channel surface, allowing precise control of the amount of reagents entrapped inside the droplets.9 Additionally, a center channel infused with buffer was included to avoid aggregation, as the immediate electrostatic interaction might cause aggregation to clog the channel.10 The design of a serpentine channel (Figure 1c) ensured the mixing between the reagents, even under laminar flow.9 A high-density carrier fluid (FC-40, fluorocarbon oil, 3M) and a neutral surfactant (RainDance, proprietary) were selected for the ease of final product collection and the elimination of non-specific interaction between the polycations and DNA, respectively. The nanocomplexes collected were then directly used for subsequent characterization or cellular investigation without any purification or separation.
Figure 1. Microfluidics-Assisted Self-Assembly in Picoliter Droplets.
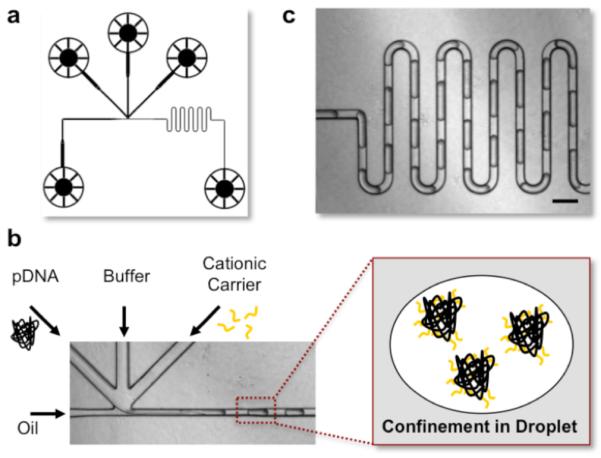
(a) Design of the cross-flow type of droplet generator. (b) Plasmid DNA, buffer, cationic gene carrier and oil were introduced into each channel with syringe pumps. Oil/surfactant was used to generate monodisperse water-in-oil droplets. The DNA and polycation solutions were confined into individual droplets and subsequently self-assembled through electro-static interaction, forming DNA nanocomplexes. (c) The droplets were then introduced into a serpentine channel to ensure complete mixing. Scale bar: 200μm.
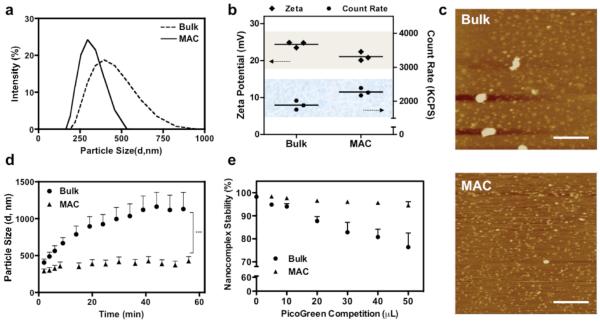
Plasmid DNA encoding GFP, as a reporter gene, was complexed with a commercially available polymeric transfection reagent of Turbofect (poly(2-hydroxypropyleneimine), pHP) or jetPEI (linear polyethylenimine, PEI, 20kD). Consistent with our hypothesis and previous findings with lipoplexes,6 MAC enabled a generation of small (ZAve, MAC= 289.8nm Versus ZAve, Bulk= 406.6nm) and more monodispersed ( PDIMAC= 0.125 Versus PDIBulk= 0.161) nanocomplexes (Figure 2a), regardless of the solvent conditions and polymer to DNA ratios (N/P ratio) (Supplementary Figure 1). Under defined polymer and DNA concentrations, the higher count rates of particle size measurement indicated that MAC produced higher concentration of smaller complexes compared to the bulk-prepared counterparts (Figure 2b). The lower surface charge on MAC-nanocomplexes suggested an exterior without a polymer corona, in contrast to excess polymer loosely attaching to a nanocomplex in bulk preparation (Figure 2b).11 In addition to particle size analysis, nanocomplexes were investigated using atomic force microscopy (AFM) in Figure 2c. Although discrepancy in particle sizes between characterizations of dynamic light scattering and AFM is common,12 quantification of surface area showed that MAC preparation (463±242 nm2) produced smaller and relatively homogeneous nanocomplexes than bulk counterpart (3106±6106 nm2). Bulk-synthesized polymeric nanocomplexes are prone to aggregation or flocculation due to the imbalanced surface charge resulting from the heterogeneity in size or composition.13 Typical solutions to produce colloidal stability are to operate the reaction at low ionic strength (electrostatic stabilization),1, 14 or to apply stabilizing additives (steric stabilization).15 The MAC-produced nanocomplexes were highly resistant to aggregation (Figure 2d), presumably benefitting from their more uniform surface charge to satisfy the grounding principle of electrostatic stabilization. MAC proposed herein is a particularly valuable strategy that does not require any additional treatment or stabilization to avoid the complication from aggregation, which may confound the structure-function correlation.
Figure 2. Representative Characterizations of Nanocomplexes Produced with Turbofect and Plasmid DNA.
(a) Intensity-based size distribution obtained under the reaction condition of 2μL Turbofect reagent per μg pDNA (ZAve, Bulk= 406.6nm, ZAve, MAC= 289.8nm; PDIBulk= 0.161, PDIMAC= 0.125). (b) Surface charge measured as zeta potential and count rate obtained from particle analysis, (c) Visualization of nanocomplexes under AFM (Scale Bar: 500nm), (d) Aggregation kinetics, and (e) Nanocomplex stability resolved by PicoGreen competition assay. Mean ± s.e.m. (n = 3).
Along with colloidal stability, an increase in nanocomplex stability was observed from MAC preparation. PicoGreen, a positively charged dye that fluoresces upon intercalating with DNA, was selected to assay the nanocomplex stability. Clearly seen from Figure 2e, the MAC nanocomplexes remained highly stable under the PicoGreen competition, especially when compared to the bulk counterparts. To explain the experimental results, we established a coarse-grained model16 to describe the confined diffusion of oppositely charged polymers in micro-incubator. Preliminary results, considering diffusion only, showed that the reactor volume alters the time-to-FMS (fully mixed state) significantly (Supplementary Figure 2). Taken together, these findings underpin our hypothesis that confining random motions in a finite domain enhances the charge neutralization between polycations and nucleic acid payloads, thereby reaching a fully mixed state within a greatly reduced amount of time. As a result, MAC is able to generate small, homogeneous and tightly bound nanocomplexes.
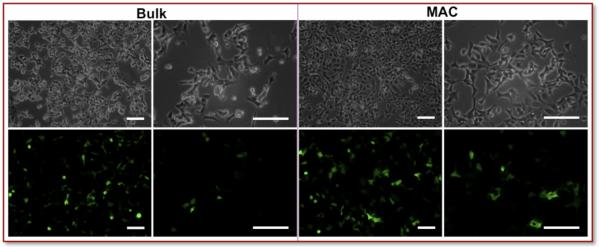
Upon incubation with human embryonic kidney cells (HEK293), MAC nanocomplexes showed higher transfection efficiency and lower cytotoxicity compared to bulk-prepared controls. At 24h post-transfection, the morphology of HEK293 cells transfected by bulk-prepared nanocomplexes suggested the onset of cytotoxicity (Figure 3). In contrast, the cells transfected with MAC nanocomplexes looked healthy (Figure 3).
Figure 3. Microscopic Observation.
After 24h post-transfection, the HEK293 cells transfected by Turbofect nanocomplexes were examined by microscopy and assayed using flow cytometry. The morphology of cells transfected by bulk-prepared nanocomplexes suggests onset of cytotoxicity. In contrast, the cells transfected with MAC-prepared nanocomplexes appeared relatively healthy. Scale bar = 100 μm.
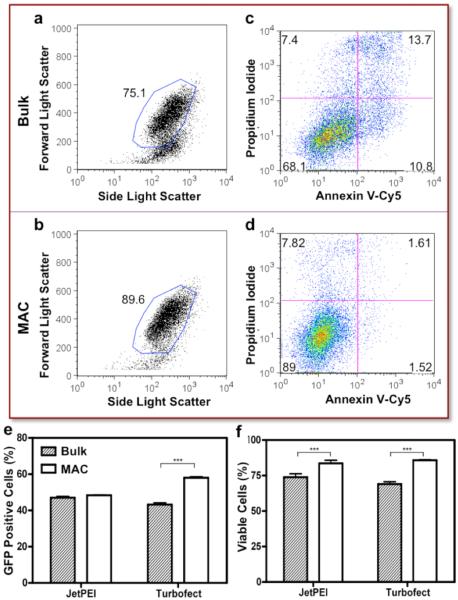
Consistent with the microscopic observation, the FSC/SSC plot suggested the MAC-prepared nanocomplexes induced minimum cell death (as gated in Figure 4a, 4b, ~96% cells were gated in the negative controls, data not shown). Cell viability was further evaluated by apoptosis, a major route of polyplex-mediated cytotoxicity,17 through PI and Annexin V-Cy5 staining. Clearly shown in Figure 4c, the bulk-prepared nanocomplexes induced significant cell death (PI+) and apoptosis (PI−, Annexin V+). Strikingly, only minimal alteration of cell integrity was observed in the MAC-prepared counterparts (Figure 4d). The transfection efficiency (Figure 4e) of MAC nanocomplexes was not compromised by the high cell viability (Figure 4f).
Figure 4. Quantification of Transfection Efficiency and Toxicity.
Consistent with the microscopic observation, (a, b) the FSC/SSC plot suggested that the MAC-prepared nanocomplexes induce minimum cell death (~96% cells were gated with untreated cells, data not shown). (c, d) Bivariate plots showing the fluorescence of PI and Annexin V-Cy5 staining were used to quantitatively evaluate cytotoxicity. Clearly, the bulk prepared nanocomplexes induced significant cell death (PI+) and apoptosis (PI−, Annexin V+). On the other hand, only minimum alteration of cell integrity was observed in the MAC counterparts. Quantification of (e) GFP expression level and (f) cell viability. Mean ± s.e.m. (n = 3). ***p<0.001 (Unpaired t-test, CI 95%, two-tailed p-value).
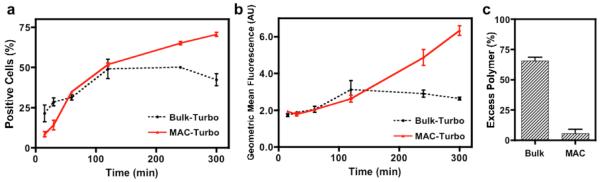
Although the optimum dimension and geometry of a nanocomplex for cellular uptake are still a topic of debate, it is generally believed a size-dependent process.18 A higher percentage of uptake, shown as percentage of fluorescently positive cells in Figure 4a, was observed from MAC at a typical transfection period of 4hr (~65% versus ~50% of bulk). Such a finding resonates with previous reported linear relationship between cellular uptake and transfection efficiency.12 The interesting crossover of the two uptake curves is likely due to the size variation, elaborated in the following: Large nanocomplexes (500nm to 1μm) deposit on the culture well quicker, generate higher local concentration and thus induce higher uptake at the early time point;14, 19 However, smaller nanocomplexes, typically below 500 nm, are favored for consistent and efficient uptake.20 Notably, the bulk-synthesized nanocomplexes almost doubled to micronsize within an hour (Figure 2d), while their uptake declined accordingly. In Figure 5b, greater MAC uptake was witnessed in geometric mean fluorescence, a preferable indication of total uptake, because the internalized genetic contents (pDNA in this study) may vary based on the size of nanocomplexes. In line with our previous assumption that MAC enables an exhaustive reaction, minimum of free polyelectrolytes was detected in the MAC preparation (Figure 5c), whereas the detected value from bulk preparation was close to those reported previously.21 While free-floating polycations commonly seen in conventional bulk preparation are often believed to contribute to the higher cytotoxicity (Figure 3 and 4f),11 we cannot rule out that the different physical properties observed between the bulk and MAC preparations may also have played a role in affecting the cytotoxicity.
Figure 5. Uptake Kinetics and Excess Polymer.
(a) Percentage of positive cells was monitored for 5 hr, beyond a typical length of transfection time. Larger nanocomplexes from bulk preparation initiated with a higher uptake at early time point. As nanocomplexes aggregated, their uptake reached a plateau. In contrast, small and stable MAC prepared nanocomplexes were favorable for a consistent uptake. (b) Because the amount of genetic materials in different size of nanocomplexes varied, geometric mean fluorescence may be a preferable indication of total uptake. (c) Significantly lower excess polymer remained in the MAC preparation, whereas ~70% of polymer at a ratio of 2μL Turbofect transfection reagent per μg of DNA was unreacted in the bulk preparation. Mean ± s.e.m. (n = 3).
Vast information has been gathered on the size- and shape- dependency of particle transport in biological compartments, especially in the context of uptake efficiency, circulation time and nanotoxicity.18, 20, 22, 23 However, a precise structure-function correlation requires a reliable technique to produce particles of well-defined properties. We have shown that MAC can produce small and homogeneous nanocomplexes. Made possible by the increased consistency, reduced variances in cellular processing will provide more predictable pharmacokinetic and pharmacodynamic properties for the rational design of the next generation of gene carriers.
Nucleic acid decondensation, or unpacking of the nanocomplex, in relation to endosomal escape is considered a major delivery hurdle in nonviral gene transfer, since premature dissociation or overly stable binding would be detrimental to their cellular uptake and therapeutic efficacy.24, 25 Adjustment of unpacking heretofore has mainly relied on chemical modification of the carrier.26 Herein, MAC represents a complementary strategy that modulates nanocomplex stability through a physical approach. Together with existing development on intracellular trafficking,12, 27, 28 MAC may accelerate the progress on the mechanistic insights into the “stability-function” relationship. In addition to serving as a new tool to optimize the potency of polyplexes, MAC also represents a unique tool to study DNA condensation in a previously unavailable experimental format,29 that is, homogeneous water-in-oil emulsions mimicking biological microreactors.30
Nanomedicine will continue to demand more sophisticated nanocomplexes for progress. To complement the innovative chemical and biological approaches to create multifunctional nanoparticles, this study indicates that MAC can serve as an interdependent strategy to modulate and optimize the physical characteristics of DNA or RNA nanocomplexes. Finally, the unexpectedly low cytotoxicity observed in this study may also facilitate development of safe and effective carriers for nucleic acid or peptide therapeutics.
Supplementary Material
ACKNOWLEDGMENT
This work is supported by NIH (HL89764), NSF EEC-0425626, and an award from the American Heart Association (CLG). We thank the Flow Cytometry Shared Resource at the Duke Cancer Institute (Dr. Mike Cook and Mr. Thusitha Dissanayake) for their support. We also thank RainDance Technologies for providing the EA Surfactant and Droplet Destablizer.
Footnotes
Supporting Information Available: (1) Supplementary Figures and (2) Experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Sharma VK, Thomas M, Klibanov AM. Biotechnol. Bioeng. 2005;90(5):614. doi: 10.1002/bit.20444. [DOI] [PubMed] [Google Scholar]
- 2.Xu Y, Hui SW, Frederik P, Szoka FC. Biophys. J. 1999;77(1):341. doi: 10.1016/S0006-3495(99)76894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner J, Kohler JM. Nano Lett. 2005;5(4):685. doi: 10.1021/nl050097t. [DOI] [PubMed] [Google Scholar]
- 4.Abou Hassan A, Sandre O, Cabuil V, Tabeling P. Chem. Commun. 2008;(15):1783. doi: 10.1039/b719550h. [DOI] [PubMed] [Google Scholar]
- 5.Chan EM, Mathies RA, Alivisatos AP. Nano Lett. 2003;3(2):199. [Google Scholar]
- 6.Hsieh AT-H, Hori N, Massoudi R, Pan PJ-H, Sasaki H, Lin YA, Lee AP. Lab Chip. 2009;9(18):2638. doi: 10.1039/b823191e. [DOI] [PubMed] [Google Scholar]
- 7.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. P. Natl. Acad. Sci. USA. 1995;92(16):7297. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boussif O, Zanta MA, Behr JP. Gene Ther. 1996;3(12):1074. [PubMed] [Google Scholar]
- 9.Song H, Chen DL, Ismagilov RF. Angew. Chem. Int. Edit. 2006;45(44):7336. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho Y-P, Chen HH, Leong KW, Wang T-H. Nanotechnology. 2009;20(9):95103. doi: 10.1088/0957-4484/20/9/095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. J. Gene Med. 2004;6(10):1102. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 12.Ulasov AV, Khramtsov YV, Trusov GA, Rosenkranz AA, Sverdlov ED, Sobolev AS. Mol. Ther. 2010 doi: 10.1038/mt.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter RJ. Introduction to Modern Colloid Science. Oxford University Press; New York: 1994. [Google Scholar]
- 14.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. Gene Ther. 1998;5(10):1425. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasachari S, Liu Y, Zhang G, Prevette L, Reineke TM. J. Am. Chem. Soc. 2006;128(25):8176. doi: 10.1021/ja0585580. [DOI] [PubMed] [Google Scholar]
- 16.Rabani E, Reichman DR, Geissler PL, Brus LE. Nature. 2003;426(6964):271. doi: 10.1038/nature02087. [DOI] [PubMed] [Google Scholar]
- 17.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. Mol. Ther. 2005;11(6):990. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Jiang W, Kim BYS, Rutka JT, Chan WCW. Nat. Nanotechnol. 2008;3(3):145. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 19.Okuda T, Kidoaki S, Ohsaki M, Koyama Y, Yoshikawa K, Niidome T, Aoyagi H. Org. Biomol. Chem. 2003;1(8):1270. doi: 10.1039/b212086k. [DOI] [PubMed] [Google Scholar]
- 20.Prabha S, Zhou WZ, Panyam J, Labhasetwar V. Int. J. Pharm. 2002;244(1-2):105. doi: 10.1016/s0378-5173(02)00315-0. [DOI] [PubMed] [Google Scholar]
- 21.Clamme JP, Azoulay J, Mély Y. Biophys. J. 2003;84(3):1960. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Adv. Mater. 2009;21:419. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adler A, Leong KW. Nano Today. 2010;5:553–569. doi: 10.1016/j.nantod.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitehead KA, Langer R, Anderson DG. Nature Reviews Drug Discovery. 2009;8(2):129. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grigsby CL, Leong KW. Journal of the Royal Society, Interface / the Royal Society. 2010;7(Suppl 1):S67. doi: 10.1098/rsif.2009.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putnam D. Nat. Mater. 2006;5(6):439. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 27.Chen HH, Ho Y-P, Jiang X, Mao H-Q, Wang T-H, Leong KW. Molecular Therapy. 2008;16(2):324. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen HH, Ho Y-P, Jiang X, Mao H-Q, Wang T-H, Leong KW. Nano Today. 2009;4(2):125. doi: 10.1016/j.nantod.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bloomfield VA. Curr. Opin. Struct. Biol. 1996;6(3):334. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 30.Swami A, Espinosa G, Guillot S, Raspaud E, Boué F, Langevin D. Langmuir. 2008;24(20):11828. doi: 10.1021/la802233e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.