Abstract
Serpins are the dominant group of protease inhibitors in metazoans that control a wide variety of biological processes including major innate immune reactions. One of these inhibitors, SRPN2, controls melanization in mosquitoes – a powerful, arthropod-specific innate immune response. SRPN2 depletion from the hemolymph of adult female mosquitoes significantly reduces longevity and therefore this serpin is a potential target for novel insecticides. We report here the crystal structure of SRPN2 in its native conformation from the African malaria mosquito, Anopheles gambiae to 1.75 Å resolution. SRPN2 adopts a similar fold as observed for other serpins with a core of three β-sheets surrounded by nine α-helices with an exposed reactive center loop (RCL) that extends from the protein body. Similar to other native serpin structures, several residues within the reactive center loop were disordered and could not be modeled. Intriguingly, the N-terminal hinge of the RCL in SRPN2 was found to be inserted into β-sheet A, suggesting a potential activation mechanism analogous to heparin-mediated activation of Antithrombin III.
Keywords: serpin, Anopheles gambiae, mosquito, melanization, malaria
INTRODUCTION
Extracellular proteolytic cascades regulate a wide variety of biological processes in vertebrates including blood clotting and complement activation. Similarly, in invertebrates they regulate major innate immune reactions, including hemolymph coagulation, the Toll pathway, and prophenoloxidase (proPO) activation that leads to melanization. Serpins, members of a large, wide-spread superfamily of serine proteinase inhibitors, play a critical role in these immune reactions by controlling accidental triggering of the cascades and regulating the spread of the signal once the cascades are activated (recently reviewed in 1). Serpins typically consist of 350–400 amino acid residues and adopt a canonical fold of three β-sheets (A–C) in the center of the protein surrounded by up to nine α helices. The reactive center loop (RCL), located at the carboxy terminus, is positioned away from the body of the serpin, highly flexible, and contains the scissile bond (designated P1–P1'), which is cleaved by the target protease during inhibition (recently reviewed in 2).
Most serpins inhibit S1 family serine proteinases while others function as cross-class inhibitors that can also target cysteine proteinases 3. Other serpins do not function as proteinase inhibitors but play a role in e.g. hormone transport 4. Inhibitory serpins function as single-use suicide substrates. Upon cleavage of the scissile bond in the RCL by the target protease, the serpin undergoes a large conformational change. The RCL is inserted into β-sheet A in the core of the serpin, distorting the active center of the protease. This traps the acyl-intermediate of the reaction, resulting in the formation of SDS-stable, covalent complexes between serpin and target protease 5.
We recently described Serpin 2 (SRPN2) from the African malaria mosquito, Anopheles gambiae, to be a key regulator of the melanization response. Depletion of SRPN2 from the hemolymph of adult female mosquitoes causes the formation of melanotic tumors, and killing of allochthonous malaria parasite species 6–8. SRPN2 is a functional inhibitor of serine proteases, and one of its endogenous targets is the clip-serine protease CLIPB9, which was shown to cleave and activate proPO 9. This result indicated that SRPN2 regulates melanization by inhibiting proPO activation. In addition, SRPN2 depletion significantly shortens mosquito longevity, making it an attractive insecticide target for malaria control. Here we report the crystal structure of An. gambiae SRPN2, the first serpin fold from a dipteran insect, in its native conformation to 1.75Å resolution.
MATERIALS AND METHODS
Protein production and purification
The pET28a_His-SRPN2 plasmid 9 was used to transform Escherichia coli BL-21 DE3 pRare; two colonies were picked and inoculated in 15 ml of LB medium containing 50 μg/ml kanamycin and 25 mg/ml chloramphenicol. A starter culture was transferred to 1.5 l of LB medium containing antibiotics and grown to an OD600 ~0.8 at 37°C. Induction was carried out with 0.1mM IPTG at 15°C overnight with shaking. Cells were harvested via centrifugation at 8,500 rpm for 10 minutes. Cell pellets were resuspended in 50 mL of buffer A (50mM NaCl, 20mM Tris-HCl, pH of 8.0) supplemented with protease inhibitor cocktail (Roche) and lysed by sonication. The insoluble material was pelleted by centrifugation at 19,000 rpm for 30 minutes and the clarified lysate was loaded onto a 5mL HisTrap HP column. All purification steps were conducted using an AKTA Xpress purification system (GE Healthcare) at 4°C. Non-specifically bound proteins were washed using 10% buffer B (500mM Imidazole, 50mM NaCl, 50mM Tris-HCl, pH 8.0). Elution was carried out with a linear gradient from 10% to 100% buffer B and all elution peaks of interest were collected and analyzed by SDS-PAGE. Fractions containing SRPN2 were pooled and loaded onto a HiTrap Q HP anion exchange column equilibrated with buffer A. Elution was carried out with a linear gradient from 0% to 100% Buffer C (1M NaCl, 20mM Tris, pH 8.0) and the purity was analyzed by SDS-PAGE. SRPN2 fractions, which eluted at approximately 50% buffer C, were pooled and concentrated to 6.2 mg/mL for crystallization screening.
Crystallization and data collection
Crystallization screening was conducted in Compact Jr. (Emerald biosystems) sitting drop vapor diffusion plates at 20°C using 0.5μl of protein and 0.5μl of crystallization solution equilibrated against 100μl of the latter. Prismatic, hexagonal crystals were obtained in two days from the Index Screen (Hampton Research) condition #24 (2.8 M Sodium Acetate, pH 7.0). Samples were transferred to a solution containing 70% crystallization solution and 30% glycerol for approximately 30 seconds before freezing in liquid nitrogen for data collection. X-ray diffraction data were collected at 100K at the Advanced Photon Source (APS) IMCA-CAT beamline 17ID using an ADSC Quantum 210r CCD detector. Intensities were integrated and scaled using the HKL2000 package 10.
Structure Solution and Refinement
Structure solution was carried out by molecular replacement with BALBES 11 in the space group P63 searching for one molecule in the asymmetric unit, which produced a homology model for the rotation and translation searches from PDB: 2GD4 (Antithrombin-S195A factor Xa heparin structure). The R/Rfree converged at 44%/49% following initial refinement. At this stage, main chain atoms of the model fit the electron density maps reasonably well. However, the side chains were difficult to discern. The side chains were then removed, the model refined and the resulting model was improved using the automated model building package in PHENIX 12. The refined model obtained from the autobuilding step converged at R=22%/Rfree=27%. This model was then used for molecular replacement search with PHASER 13 against higher resolution diffraction data that was subsequently collected. The final model was obtained by iterative rounds of refinement and manual model building with PHENIX and COOT 14, respectively. Anisotropic displacement parameters were modeled using TLS 15 refinement, in which the protein chain was partitioned into 3 groups determined from the TLSMD server 15. Disordered side chain atoms were truncated to the point where electron density could be observed. Figures were prepared using the CCP4MG package 16. Relevant crystallographic data are provided in Table 1. Structure comparison was conducted using secondary structure matching with the program Superpose 17 via the CCP4 interface 18. Coordinates and structure factors have been deposited to the Protein Databank with the accession code 3PZF.
Table 1.
Crystallographic data for SRPN2 refined to 1.75Å resolution.
| SRPN2 | |
|---|---|
| Data Collection | |
| Unit-cell parameters (Å, o) | a = 96.54, c = 78.29 |
| Space group | P63 |
| Resolution (Å)1 | 30.0–1.75 (1.81–1.75) |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Observed reflections | 448,554 |
| Unique reflections | 40,410 |
| <I/σ(I)>1 | 35.9 (2.3) |
| Completeness (%)1 | 96.5 (78.7) |
| Redundancy1 | 11.1 (8.6) |
| Rsym (%)1, 2 | 5.9 (41.5) |
| Refinement | |
| Resolution (Å) | 26.25–1.75 |
| Reflections (working/test) | 38,251 / 2,042 |
| Rfactor / Rfree (%)3 | 17.3 / 20.7 |
| No. of atoms (protein/water) | 2,953 / 165 |
| Model Quality | |
| R.m.s deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (o) | 1.233 |
| Average B factor (Å2) | |
| All Atoms | 35.7 |
| Protein | 35.6 |
| Water | 37.2 |
| Coordinate error, maximum likelihood (Å) | 0.19 |
| Ramachandran Plot | |
| Most favored (%) | 97.0 |
| Additionally allowed (%) | 3.0 |
Values in parenthesis are for the highest resolution shell.
Rsym = Σ hklΣi |Ii(hkl) - <I(hkl)>| / ΣhklΣi Ii(hkl), where Ii(hkl) is the intensity measured for the ith reflection and <I(hkl)> is the average intensity of all reflections with indices hkl.
Rfactor = Σhkl ||Fobs (hkl) | - |Fcalc (hkl) || / Σhkl |Fobs (hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
RESULTS AND DISCUSSION
Crystallization screening of SRPN2 yielded two crystal forms at 20°C. The first is a primitive hexagonal form (P63) yielding hexagonal plates obtained in 2.8 M Sodium Acetate, pH 7.0 from a sitting drop vapor diffusion experiment. The same crystallization conditions in a microbatch experiment under paraffin oil produced isomorphous crystals with a modified habit in which the crystals were elongated along the crystallographic c-axis. This particular crystal form consistently yielded diffraction to at least 2.0 Å resolution or better. A second crystal form of SRPN2 was obtained in 2M Li2SO4, 100mM Hepes pH 7.6 that belonged to the space group I4132 with a = 282.9 Å. These crystals diffracted to significantly lower resolution to a maximum of 3.6 Å.
The structure of the hexagonal form (P63) of SRPN2 was solved by molecular replacement using diffraction data to 1.75Å resolution and contained a single molecule in the asymmetric unit. Data collection and refinement statistics are provided in Table 1. The structure of the I4132 form was solved by molecular replacement searching for 4 molecules in the asymmetric unit. Two sets of dimers were located using Molrep in the space group I4132. However, no solution was found in I432. Following refinement, the average B-factors were approximately 2-fold higher for one of the subunits and the resulting electron density maps were ambiguous for certain regions making it difficult to build an accurate model. Therefore, the structure was not refined further.
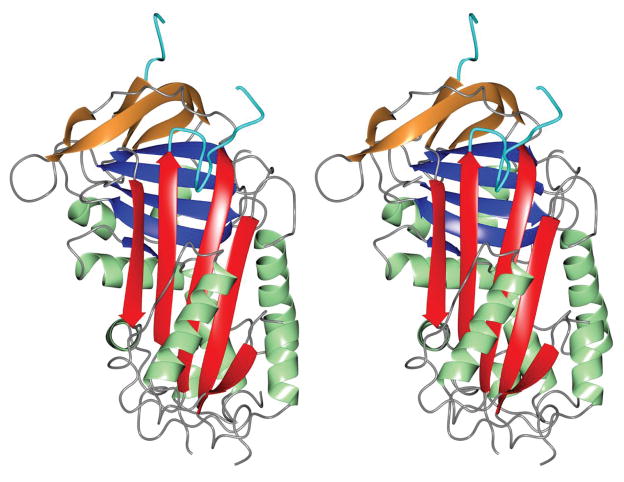
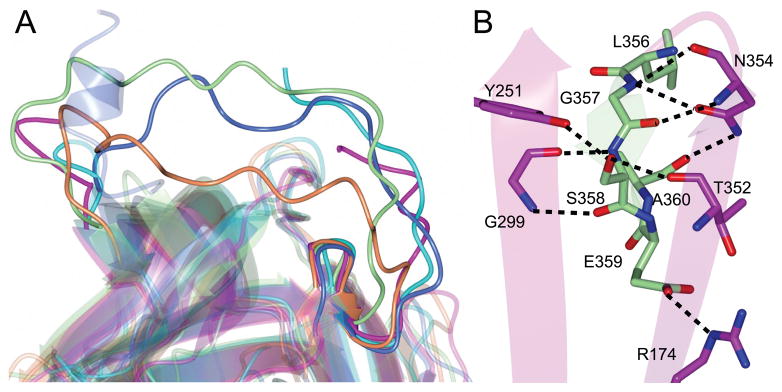
SRPN2 adopts a similar fold as observed for other serpins (Fig. 1), however significant differences are present which is indicated by the RMS deviations calculated between Cα atoms for homologs (Table S1). Similar to several other native serpin structures 19, the RCL residues between I366 to D375 in SRPN2 were disordered and could not be modeled. The most notable difference between SRPN2 and most native serpin folds is the conformation of the N-terminal hinge region of the RCL, which is partially inserted between strands 2 and 3 of β-sheet A (Fig. 2A). This conformation has so far only been seen in a small number of serpins, non-heparin bound antithrombin III (ATIII) 20, heparin cofactor II 21, murine anti-chymotrypsin (mACT) 22, and Spn48 from Tenebrio molitor 23. The N-terminal hinge of the RCL that is inserted into and interacts with the β-sheet is mainly localized to the residues from Leu 356 to Ala 360. This region could be accurately modeled to the electron density maps (Fig. S1). The insertion is stabilized by several hydrogen bonds, with the side chains of Arg 174, Tyr 251, Asn 354, Ser 358 and Glu 359 forming the majority of the main contacts (Fig. 2B). Similarly, the electron density maps of the Cubic I form obtained following refinement clearly showed that the residues Leu 356 to Ala 360 are inserted into β-sheet A (Fig. S2). As was observed for the initial crystal form, no crystal contacts were present near these residues which indicated that this observed conformation is not an artifact of crystal packing.
Figure 1. Stereoview of the overall structure of SRPN2.
The A-sheet is colored red, B-sheet blue, C-sheet in tan, α-helices are shown in green and the RCL is colored cyan. PDB: 3PZF
Figure 2. The hinge region of SRPN2 is partially inserted into β-sheet A.
(A) Superposition of serpin structures that contain loop insertions at the N-terminal hinge region of the RCL. SRPN2 (magenta, PDB: 3PZF), non-heparin bound ATIII (coral, PDB: 1T1F), heparin cofactor II (blue, PDB: 1JMJ), mACT (green, PDB: 1YXA), and SPN48 (cyan, PDB: 3OZQ). (B) Hydrogen bond interactions between the RCL loop insertion and strands 2 and 3 of β-sheet A in SRPN2.
Partial insertion of the hinge region can reduce serpin activity as it moves the RCL closer to the main body of the protein, making the scissile bond less accessible to the protease target as compared to a fully exposed loop. In some cases, this can be overcome by binding of heparin to the serpin molecule. In ATIII, and most likely also HCII, heparin binding on or near to homologous regions of helix D favors expulsion of the hinge and increased inhibitory activity of the serpin 21,24,25. Similarly, heparin binding to Spn48 increases its activity 23. However, the lysine and arginine residues responsible for binding of heparin in ATIII and HCII are not conserved in Spn48, and heparin binding might be conferred by basic residues on helix I 23 (Fig. S3). In SRPN2, the basic residues in helices D and I are not conserved (Fig. S3) and calculations of surface charges on the protein do not indicate clearly an alternative heparin binding site. In accordance with these observations, heparin pentasaccharide does not increase the inhibitory activity of SRPN2 on its cognate protease, AgCLIPB9 (An and Michel, unpublished). Future work is needed to elucidate whether the partial hinge insertion in the native SRPN2 molecule is indicative of a novel regulatory mechanism of serpin function.
Supplementary Material
Acknowledgments
We thank Dr. Na Zhang for SRPN2 purification. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. The project described and the use of the KU COBRE Protein Structure Laboratory was supported by NIH Grant Number P20 RR-17708 from the National Center for Research Resources. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Center of Biomedical Research Excellence in Protein Structure and Function or NIH. This is contribution 11-179-J from the Kansas Agricultural Experiment Station.
References
- 1.Silverman GA, Whisstock JC, Bottomley SP, Huntington JA, Kaiserman D, Luke CJ, Pak SC, Reichhart JM, Bird PI. Serpins flex their muscle: I. Putting the clamps on proteolysis in diverse biological systems. J Biol Chem. 2010;285:24299–24305. doi: 10.1074/jbc.R110.112771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whisstock JC, Silverman GA, Bird PI, Bottomley SP, Kaiserman D, Luke CJ, Pak SC, Reichhart JM, Huntington JA. Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerization, and transport functions. J Biol Chem. 2010;285:24307–24312. doi: 10.1074/jbc.R110.141408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komiyama T, Ray CA, Pickup DJ, Howard AD, Thornberry NA, Peterson EP, Salvesen G. Inhibition of interleukin-1 beta converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J Biol Chem. 1994;269:19331–19337. [PubMed] [Google Scholar]
- 4.Zhou A, Wei Z, Read RJ, Carrell RW. Structural mechanism for the carriage and release of thyroxine in the blood. Proc Natl Acad Sci U S A. 2006;103:13321–13326. doi: 10.1073/pnas.0604080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gettins PG. Mechanisms of serpin inhibition. In: Silverman GA, Lomas DA, editors. Molecular and cellular aspects of the serpinopathies and disorders in serpin activity. Singapore: World Scientific Publishing Co; 2007. pp. 67–100. [Google Scholar]
- 6.Michel K, Budd A, Pinto S, Gibson TJ, Kafatos FC. Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6:891–897. doi: 10.1038/sj.embor.7400478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michel K, Suwanchaichinda C, Morlais I, Lambrechts L, Cohuet A, Awono-Ambene PH, Simard F, Fontenille D, Kanost MR, Kafatos FC. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum. Proc Natl Acad Sci U S A. 2006;103:16858–16863. doi: 10.1073/pnas.0608033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou Z, Shin SW, Alvarez KS, Kokoza V, Raikhel AS. Distinct Melanization Pathways in the Mosquito Aedes aegypti. Immunity. 2010;32:41–53. doi: 10.1016/j.immuni.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 9.An C, Budd A, Kanost MR, Michel K. Characterization of a regulatory unit that controls melanization and affects longevity of mosquitoes. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-010-0543-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology: Macromolecular Crystallography, part A. Vol. 276. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 11.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 15.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 16.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 17.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.Stout TJ, Graham H, Buckley DI, Matthews DJ. Structures of active and latent PAI-1: a possible stabilizing role for chloride ions. Biochemistry. 2000;39:8460–8469. doi: 10.1021/bi000290w. [DOI] [PubMed] [Google Scholar]
- 20.McCoy AJ, Pei XY, Skinner R, Abrahams JP, Carrell RW. Structure of beta-antithrombin and the effect of glycosylation on antithrombin's heparin affinity and activity. J Mol Biol. 2003;326:823–833. doi: 10.1016/s0022-2836(02)01382-7. [DOI] [PubMed] [Google Scholar]
- 21.Baglin TP, Carrell RW, Church FC, Esmon CT, Huntington JA. Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism. Proc Natl Acad Sci U S A. 2002;99:11079–11084. doi: 10.1073/pnas.162232399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath AJ, Irving JA, Rossjohn J, Law RH, Bottomley SP, Quinsey NS, Pike RN, Coughlin PB, Whisstock JC. The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins. J Biol Chem. 2005;280:43168–43178. doi: 10.1074/jbc.M505598200. [DOI] [PubMed] [Google Scholar]
- 23.Park SH, Jiang R, Piao S, Zhang B, Kim EH, Kwon HM, Jin XL, Lee BL, Ha NC. Structural and functional characterization of a highly specific serpin in the insect innate immunity. J Biol Chem. 2010 doi: 10.1074/jbc.M110.144006. (epub 3 Nov 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huntington JA, Olson ST, Fan B, Gettins PG. Mechanism of heparin activation of antithrombin. Evidence for reactive center loop preinsertion with expulsion upon heparin binding. Biochemistry. 1996;35:8495–8503. doi: 10.1021/bi9604643. [DOI] [PubMed] [Google Scholar]
- 25.Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. The anticoagulant activation of antithrombin by heparin. Proc Natl Acad Sci U S A. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.