Abstract
The crystal structure of an archaeal-type phosphoenolpyruvate carboxylase from Clostridium perfringens has been determined based on X-ray data extending to 3 Ǻ. The asymmetric unit of the structure includes two tetramers (each a dimer-of-dimers) of the enzyme. The precipitant, malonate, employed for the crystallization is itself a weak inhibitor of phosphoenolpyruvate carboxylase and a malonate molecule is seen in the active-site in the crystal structure. The allosteric binding sites for aspartate (an inhibitor) and glucose-6-phosphate (an activator) observed in the Escherichia coli and Zea mays phosphoenolpyruvate carboxylase structures, respectively, are not conserved in the C. perfringens structure. Aspartate inhibits the C. perfringens enzyme competitively with respect to the substrate, Mg++.phosphoenolpyruvate. A mechanism for inhibition is proposed based on the structure and sequence comparisons with other archaeal-type phosphoenolpyruvate carboxylases with differing sensitivity to inhibition by aspartate.
Keywords: X-ray structure, SAD phasing, β-barrel, competitive inhibition
INTRODUCTION
Phosphoenolpyruvate carboxylase (Pepc) catalyzes the formation of oxaloacetate from phosphoenolpyruvate (PEP) and bicarbonate. The reaction proceeds in three steps.[1] An initial transfer of phosphate from PEP to bicarbonate forms carboxyphosphate and the enolate of pyruvate. Removal of a proton from carboxyphosphate facilitates its breakdown to CO2 and phosphate. Finally, the enolate of pyruvate attacks the nucleophilic carbon of CO2, forming the product oxaloacetate (OAA). The reaction requires a divalent cation, typically Mg++. In some plants, the enzyme utilizes Mg++.PEP as substrate, satisfying the requirement for the divalent cation.[2-4] X-ray structures are available for Pepc from E. coli and Z. mays.[5][6] Both enzymes are tetramers with 222 point-group symmetry. The E. coli structures all have the allosteric inhibitor aspartate bound, and thus represent the T-state of the enzyme. The mechanism of inhibition became clear when the R-state Z. mays structure with sulfate occupying the allosteric activator site was reported.[6] Comparison of the T-state E. coli structures with the R-state Z. mays structure showed the active-site residue Arg587 moves 15 Ǻ to form part of the aspartate binding site. Arg587 is essential for activity, as replacement of the Arg by Ser compromises Vmax and the KM values for both PEP and bicarbonate.[7] A second mobile element in the active-site was identified via comparison of the T-state and R-state structures. The allosteric activator site is coupled to His138, a residue known from a mutagenesis study to be dispensable for the initial formation of carboxyphosphate and the enolate of pyruvate, but essential for the subsequent enzymatic steps which result in the formation of OAA.[8] One proposal is that His138 removes a proton from carboxyphosphate and facilitates its breakdown to CO2 and phosphate.[6][8] In the T-state structures, the loop including His138 moves away from the active-site; occupancy of the allosteric activator site repositions the loop with His138 in the active-site. Additional information concerning residues involved in catalysis was provided by the X-ray structure of the T-state E. coli enzyme in complex with the allosteric inhibitor aspartate together with Mn++ and a PEP analog, 3,3-dichloro-2-(phosphonomethyl)prop-2-enoate.[6] While the available E. coli and Z. mays X-ray structures have been informative, many questions remain. A fuller understanding of the R to T-state transition requires X-ray structures of both states from the same Pepc. In addition, none of the X-ray structures reveal how bicarbonate binds, as the residues implicated in bicarbonate binding by a mutagenesis study [5] are present on a loop which is disordered. And although Pepc activity is known to be regulated via phosphorylation in plants, a structural explanation for the regulation is not yet available.[9]
The Pepc in archaea (PepcA) is only distantly related to the eukaryotic and bacterial Pepc sequences [10][11], and an accurate sequence alignment showing the similarities in the PepcA and Pepc folds was only recently published.[12] The archaeal-type PepcA differs from Pepc in its simpler pattern of regulation by metabolites, and in its size (typically 500 residues versus ~900 residues for Pepc). PepcA is widespread in the archaea, rare in bacteria, and absent in eukaryotes.[10][11] Clostridium perfringens is one of a handful of Gram-positive bacteria which express the archaeal-type PepcA rather than Pepc.[10][11] The C. perfringens PepcA has been expressed in E. coli and crystallized [13], and its structure is reported here.
In a bold modeling study, Matsumura et al. [12] created a structural profile based on the E. coli and Z. mays X-ray structures, and then found the best fit of the PepcA sequences to the profile. Their model includes the Pepc eight-stranded β-barrel and is consistent with the secondary structure predictions for the PepcA sequences. The alignment matches 428 of the 537 residues of C. perfringens PepcA with counterparts in the 883 residue E. coli Pepc sequence, and the two aligned sequences share 16% sequence identity. The X-ray structure of C. perfringens PepcA confirms the gross features of the model, and highlights the difficulty in modeling specific interactions and subunit assembly at this low level of sequence conservation.
Matsumura et al. [12] also attempted to explain the differing sensitivity of two PepcA representatives to inhibition by aspartate. Aspartate has been reported as an allosteric inhibitor of PepcA from Sulfolobus acidocaldarius [14] and Sulfolobus solfataricus [11], while PepcA from Methanothermobacter thermautotrophicus and Methanothermus sociabilis is not sensitive to inhibition by aspartate.[19][15] The modeling suggested the sensitivity of some PepcA family members to aspartate inhibition was due to binding of aspartate at an allosteric site, in a mode mimicking aspartate binding to the allosteric site of E. coli Pepc. The C. perfringens PepcA is also sensitive to aspartate inhibition, and the X-ray structure of the enzyme shows that aspartate inhibits activity via a mechanism distinct from the mode of inhibition of the related E. coli and Z. mays enzymes.
METHODS
Structure solution
As described previously [13], enzymatically-active, full-length PepcA with an N-terminal His-tag was expressed in E. coli and purified via affinity chromatography followed by gel filtration. Crystals of PepcA were grown via hanging-drop vapor diffusion from equal volumes of protein in 20 mM HEPES buffer, pH 7 and 1.25-1.5 M sodium malonate, pH 7. The protein stock solution was 5 mg per ml as determined by the (Bio-Rad) Bradford Coomassie-binding assay. X-ray data were collected from a single PepcA crystal soaked with KAu(CN)2 as described in Dharmarajan et al. [13] (Table I). The X-ray energy was just above the Au LIII edge. Gold atoms were located with Shelxd and an initial map was produced by Shelxe.[16] The anomalously-scattering gold substructure was completed with Phaser.[17] Resolve [18] and Buccaneer [19] were used to auto-build the initial model, and Coot [20] and Xpleo [21] were used to adjust and complete the model. Parts of the model meriting further inspection were identified with Probe [22] and MolProbity.[23] Refinement including tight ncs restraints (except for residues 347-359, with no ncs restraints, and residues 360-387, with loose restraints applied) was performed with Refmac.[24] The reflections used to calculate R-free during refinement were chosen in thin resolution shells using the program Sftools. After initial cycles of refinement with isotropic, restrained, individual-atom temperature factors, TLS groups were added based on results returned from the TLSMD server.[25][26] Residues 349-353 were not observed in the electron density. The main-chain for residues 354-386 following the missing residues is found in either of two slightly different conformations in the eight independent molecules of the asymmetric unit as a consequence of different crystal packing environments. The N-terminal His-tag was not observed in the electron density. The bound gold compound was modeled as either Au(I) or Au(CN)2− depending on the occupancy of the site and the corresponding detail present in the electron density. Density present in the active-site was interpreted as bound malonate. With an assigned occupancy of ~70%, the B-factors of the bound malonate matched the B-factors of the neighboring protein atoms. Overlapping density from a minor (partially occupied) Au(CN)2− was present in the active-site of several molecules. Water molecules were added in positions where compatible hydrogen-bond donors and acceptors supported their placement and electron density was present in both 2mFo-DFc and mFo-DFc maps. Figures were prepared with Ccp4mg [27], which utilizes Pisa [28] for analysis of subunit interfaces.
Table I.
Diffraction Data and Structure Refinement Statistics
| Data Quality | |
|
| |
| BeamLine | SSRL BL11-1 |
| Wavelength | 1.03948 Ǻ |
| Space Group | P212121 |
| Cell edges (Å) | 121.4, 161.6, 280.0 |
| Resolution (Å) | 2.95 (3.03-2.95) |
| Unique Observations | 116,330 |
| Completeness (%) | 99.9 (98.6) |
| I/σ(I) | 17.4 (2.0) |
| Multiplicity | 14.3 (9.6) |
| Rmerge1 | 0.133 (1.076) |
|
| |
| Model Quality | |
|
| |
| Rcryst2 | 17.8% |
| Rfree3 | 22.3% |
| Wilson B factor | 69.2 Ǻ2 |
| RMSD-bonds (Ǻ) | 0.012 |
| RMSD-angles (°) | 1.34 |
| Number of residues | 4217 |
| Numbers of waters | 200 |
| Ramachandran plot PDB entry |
94.9% favored 99.6% allowed 0.4% outliers 3ODM |
Rmerge=Σhkl(|<Ihkl>-Ihkl|)/( Σhkl Ihkl) where <Ihkl> is the average intensity over symmetry related and equivalent reflections and Ihkl is the observed intensity for reflection hkl.
Rcyst=Σhkl(∥Fobs|-|Fcalc∥)/( Σhkl|Fobs|) where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitude for reflection hkl. The sum is carried out over the 115,073 observed reflections which are used in refinement.
Rfree refers to the R factor for the test set (1135 reflections) which was excluded from refinement.
Enzyme assays
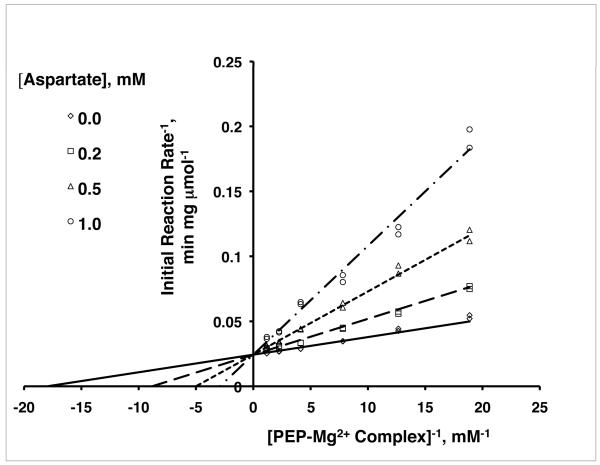
Phosphoenolpyruvate carboxylase activity was assayed aerobically by coupling the reaction to the malate dehydrogenase (MDH) reaction and monitoring the oxidation of NADH spectrophotometrically at 340 nm. Assays were conducted at 37°C, with an assay mixture of the following composition: 50 mM HEPES-NaOH buffer (pH 7.2), 0.2 mM NADH, 30 mM KHCO3, 1.5 mM (or varied concentration of) sodium phosphoenolpyruvate (PEP), 2 mM MgCl2, and 1 U of thermophilic MDH from Thermus flavus (Sigma, St. Louis, MO) per ml. In the amount used, MDH did not limit the reaction rate in any assay. The assay mixture was prepared without PEP. After warming to the reaction temperature, the mixture was supplemented with 1 μg of the purified His-tagged enzyme. The reaction was initiated with the addition of PEP. The concentration of Mg++.PEP in solution was calculated using a dissociation constant of 5.55 mM for the complex.[29] Inhibition of activity was observed at high Mg++ concentrations, complicating the kinetics analysis. Inhibition by Mg++ was not seen after the His-tag was removed from the recombinant enzyme via protease treatment with enterokinase. His-tagged PepcA was used to determine the kinetic constants and generate the data presented in Figure 4. In the assays measuring aspartate inhibition, the Mg++.PEP concentration was varied by holding the total Mg++ concentration constant and varying the concentration of PEP added. The assays were designed this way to avoid inhibition of the His-tagged enzyme by high Mg++ concentrations.
FIGURE 4.
Double-reciprocal plots of PepcA activity versus [Mg++.PEP] showing the characteristic pattern of competitive inhibition. The lines fit the competitive inhibition equation V = Vmax[Mg++.PEP]/(KM(1+[aspartate]/Ki)+[Mg++.PEP]).
Quaternary structure in solution
Gel filtration was performed using 100 mM sodium phosphate buffer (pH 7.0) containing 100 mM NaCl. The protein concentration of the loaded sample was 0.9 mg per ml. Separation employed an analytical TSK-GEL G3000SWXL column (TosoHaas, Montgomeryville, PA).[30] Analytical ultracentrifugation experiments were performed in a Beckman/Coulter XL-I ultracentrifuge using an An60Ti rotor at the Center for Analytical Ultracentrifugation of Macromolecular Assemblies, at the University of Texas Health Science Center at San Antonio, TX. Sedimentation velocity experiments were performed on protein solutions of PepcA ranging in concentration between 50 μg per ml and 3 mg per ml. All experiments were performed at 20 °C, 30 krpm in 10 mM sodium phosphate buffer, pH 7.0, containing 25 mM NaCl. The velocity data was analyzed with UltraScan software, version 9.9, release 976.[31] Two-dimensional spectrum analysis was performed to remove time- and radially-invariant noise, and to provide shape- and size-distributions of the components in the system.[32] Diffusion-corrected sedimentation coefficient distributions were generated via enhanced van Holde-Weischet analysis.[33]
RESULTS AND DISCUSSION
PepcA structure determination and fold
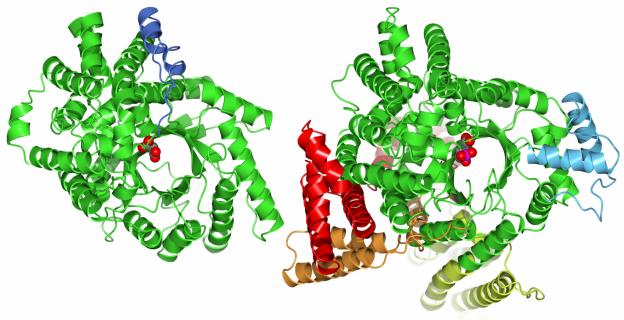
Crystals of PepcA diffracting to 3 Ǻ were obtained using 1.25-1.5 M sodium malonate, pH 7, as precipitant. Data from a single crystal of PepcA soaked in KAu(CN)2 yielded SAD phases of sufficient quality to solve the X-ray structure (Table I). 532 of the 537 residues could be modeled. Although no substrates were included in the crystallization cocktail, the precipitant itself is known to be a weak (mM) inhibitor of the E. coli Pepc.[34] Residual electron density in the active-site of PepcA was modeled as bound malonate. Comparison of the X-ray structure with the models of Matsumura et al. [12] shows the predicted eight-stranded β-barrel is present. The sequence alignment of Matsumura et al. is however out of register for β-strands β2 (by one residue) and β4 (by two residues). The PepcA fold is relatively simple compared to that of Pepc, with β-α-β secondary structure elements forming the β-barrel, followed by an all α-helical domain (Figure 1). The C-terminus, rather than being flexible or exposed, folds back towards the active-site and forms a salt-bridge with Arg246 (Figure 2A). All of the potential hydrogen-bonds of the Arg side-chain are satisfied; Arg246 also hydrogen-bonds with the side-chains of Ser201 and Gln270, orienting the two hydrogen-bond donating side-chains in the active-site. The main-chain NH and the hydroxyl group on the side-chain of Ser201 interact with one of the carboxylate moieties of the bound malonate anion in the crystal structure (Figure 2A). The side-chain of Gln270 is near the same carboxylate of the bound malonate, however the distance (3.5 Ǻ) is not optimal for hydrogen-bonding. The malonate’s other carboxylate interacts with the side-chains of His11 and Arg82. In contrast to the simpler PepcA fold, the Pepc fold is decorated with additional α-helices before the start of the barrel and after β-strands β1, β2, and β3 (see Figure 1 and the structure-based sequence alignment shown in Supplementary Figure 1). The C-terminus of the Z. mays Pepc, in the active R-state, forms a salt-bridge with Arg647 (the equivalent of Arg246 in C. perfringens), which in turn helps position Gln673 in the active-site (the equivalent of Gln270 in C. perfringens). In the 3.0 Ǻ Z. mays structure, not all of the potential hydrogen-bonds to Arg647 are satisfied, leading to the proposal that the side-chain may reposition in the active-site to participate directly in catalysis.[6][12] Mutation of the corresponding Arg587 in the E. coli Pepc lowers Vmax dramatically and raises the KM values for both PEP and HCO3−.[7] The C. perfringens structure argues the Arg does not move during catalysis, and its mutation is deleterious due to the loss of positioning of residues Ser201 and Gln270 in the active-site. The sets of interactions involving the C-terminus in the Z. mays Pepc and C. perfringens PepcA structures are very similar. The interactions of the C-terminus in the inactive T-state E. coli Pepc structure are quite different, as described below. Z. mays Pepc is activated by glucose-6-phosphate, and the allosteric binding site for glucose-6-phosphate includes three Arg residues located on two α-helices following β1.[6] The simpler β-barrel of PepcA lacks these α-helices and hence the allosteric activator binding site is not present, as correctly predicted by the modeling study of Matsumura et al.[12] Glucose-6-phosphate has not been reported as an activator of any PepcA, nor did it activate C. perfringens PepcA when added to the standard assay mixture at 2 mM. A comparison of the E. coli and Z. mays Pepc structures shows that activator binding at the glucose-6-phosphate site leads to a rearrangement of the loop connecting the β-strand β1 to the following α-helix.[6] In the R-state Z. mays structure, a sulfate occupies the glucose-6-phosphate regulatory site, and the loop residue His177 is oriented with its side-chain in the active-site. In the T-state E. coli structure, the corresponding His138 faces away from the active-site. In the C. perfringens PepcA structure His11 faces the active-site and its side-chain interacts with the bound malonate anion.
FIGURE 1.
The C. perfringens PepcA fold (left) and the E. coli Pepc fold viewed from above the active-site at the C-terminal end of the β-barrel. Insertions in the folds are indicated via the color scheme (compare to Figure 3). The insertions in the E. coli fold are at the N-terminus (red), after β-strands β1 (orange), β2 (yellow) and β3 (light blue). The helix-turn-helix insertion in the C. perfringens fold follows residue 360 (blue). Malonate is bound in the active-site of PepcA; a PEP analog is bound in the active-site of Pepc.
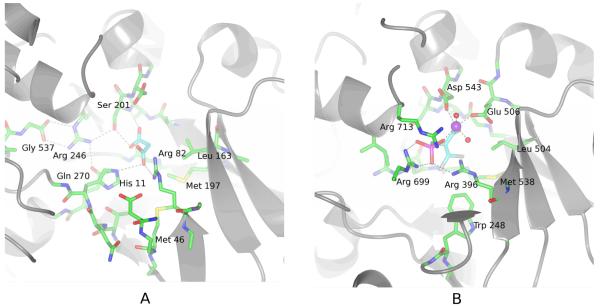
FIGURE 2.
A Malonate binds to the active-site of PepcA via interactions with His11, Arg82, Ser201 and Gln270. Several strands of the β-barrel are visible in the lower right of the figure. The C-terminal Gly537 positions Arg246, which in turn positions Ser201 and Gln270 (visible under His11) in the active-site.
B An inhibitory PEP analog binds to the active-site of the E. coli Pepc via interactions with Arg396, Arg699, Arg713 and a Mn++ ion (shown in purple). In this view into the active-site, the carboxylate of the inhibitor is directly below the Mn++ ion. His138 and Arg587 (corresponding to C. perfringens His11 and Arg246) are missing from the active-site in this T-state structure.
Inhibitors bound at the active-sites of Pepc and PepcA
Malonate was observed bound to the active-site of PepcA, with the binding site comprised of the conserved active-site residues His11, Arg82, Ser201, and Gln270 (Figure 2A). The corresponding Arg396 in the E. coli enzyme is involved in binding an inhibitory PEP analog (Figure 2B), as revealed by the X-ray structure of the E. coli enzyme in complex with Mn++, the inhibitory PEP analog, and the allosteric inhibitor, aspartate (Protein Data Bank entry 1jqn [6]). With the exception of Arg396, the bound inhibitors share no other interactions with the active-sites. The phosphonate moiety of the inhibitor bound to the E. coli Pepc interacts with two Arg side-chains, Arg 396 and Arg699, while one of the carboxylate groups of the bound malonate interacts with a single Arg residue, Arg82 (corresponding to Arg396 of the E. coli Pepc) and His11 (His138 in the E. coli Pepc). Arg699 is located just before the disordered bicarbonate-binding loop in the X-ray structures from C. perfringens, E. coli and Z. mays, and Arg699 appears to be mobile. In comparing the position of the Arg in the active-sites of the X-ray structures, the side-chain appears to move by ~3 Ǻ to interact with the bound PEP analog’s phosphonate group in the E. coli structure. In the C. perfringens structure, the side-chain of the corresponding Arg344 is ~9 Ǻ from the bound malonate. Arg713 is near the phosphonate as well in the E. coli structure, although the distance (3.4 Ǻ) and the orientation of the side-chain are not optimal for hydrogen-bonding. The E. coli structure includes a bound Mn++ ion while the C. perfringens structure does not. The coordination sphere of the Mn++ ion is octahedral. Two completely conserved residues, Asp543 and Glu506, each contribute one carboxylate oxygen atom to the octahedral coordination sphere of the Mn++ ion; the inhibitory PEP analog contributes two oxygen atoms to the coordination sphere, one each from the phosphonate and carboxylate groups; and two water molecules occupy the remaining two positions. The carboxylate moiety of the PEP analog has no interactions with the enzyme apart from the single interaction with the Mn++. Due to the absence of Arg587 and His138 from the active-site in the T-state E. coli structure, as well as the larger size of the inhibitory PEP analog compared to PEP, the interactions observed in the X-ray structure probably do not represent the binding mode of PEP to the active, R-state enzyme. Studies of a series of inhibitory PEP analogs have shown the presence of a bridging O atom is important for potent inhibition, and this feature is lacking in the phosphonate PEP analog co-crystallized with the E. coli enzyme.[35][36] The C. perfringens PepcA complex with malonate may be a better starting point for understanding the roles of individual residues in catalysis than either the T-state E. coli complex with a PEP analog or the R-state Z. mays structure. In the E. coli T-state structure Arg587 is trapped at the aspartate-binding site and His138 is missing from the active-site, rendering the enzyme inactive, while in the Z. mays R-state structure no substrates or substrate analogs are present in the active-site.
An interesting feature of the malonate binding site in the new PepcA structure is the involvement of the backbone NH group of Ser201. The backbone NH of Asp202 is also available as a hydrogen-bond donor, as both residues 201 and 202 are located at the start of an α-helix. The known role of Asp202 is its side-chain’s involvement in divalent cation binding. A proposal suggested by comparison of all the X-ray structures is that the substrate PEP might bind such that the phosphate is positioned to interact with both the divalent cation and the two NH groups at the N-terminus of the α-helix beginning at residue 201. The N-terminus of an α-helix provides a favorable dipole-moment contribution to binding anions, and tetrahedral anions such as phosphate and sulfate are known to bind at such sites. This mode of binding would place the carboxylate of PEP near Arg344, rather than the phosphate (reversing the relative positions of the carboxylate and phosphonate observed in the inhibited E. coli structure with a PEP analog bound). In comparison to the binding mode of the PEP analog in the E. coli structure, the proposed ‘reversed’ binding mode places the phosphate moiety of PEP nearer to the likely position of the (disordered) bicarbonate-binding loop. Following binding of bicarbonate and formation of carboxyphosphate, the backbone NH groups of residues 201 and 202 would help anchor the phosphate group in place, and thereby position the protonated carboxy group near His11, the active-site base thought to accept a proton from the carboxyphosphate intermediate.[6][8] In the C. perfringens X-ray structure, malonate binds with one carboxylate interacting with the NH of Ser201 and the other with the side-chain of His11. The C. perfringens structure shows the distance spanning the anion binding-site at the N-terminus of the α-helix starting at Ser201 and the side-chain of His11 is appropriate for the proposed binding mode of carboxyphosphate. Breakdown of carboxyphosphate would release CO2 in the hydrophobic pocket bordered by residues Met46, Leu163 and Met197, immediately adjacent to His11 in the R-state C. perfringens structure (Figure 2A). These hydrophobic residues form a pocket equivalent to that formed by Trp248, Leu504 and Met538 of the E. coli PEPC (Figure 2B), which was proposed to have the same role.[6] Further support for the hypothesis that the substrate PEP binds productively in an orientation with the carboxylate and phosphate reversed with respect to the binding mode of the PEP analog comes from analysis of the structure-activity relationship of PEP analogs substituted with hydrophobic groups at C3.[36] The Z-isomers of PEP analogs with substituents at C3 are more potent than the E-isomers, and binding of these analogs in the proposed ‘reversed’ mode places the hydrophobic substituents in the pocket defined by residues Trp248, Leu504 and Met538. Finally, binding of PEP in the ‘reversed’ mode places the si side of the plane defined by the substituents about the double bond directly over the hydrophobic pocket. Work with radiolabeled substrates has shown that carboxylation of PEP occurs via addition of CO2 to the si face of PEP.[37]
Bicarbonate-binding site is disordered
The missing residues in the X-ray structure (349-353) are part of a loop which is involved in HCO3− binding [5]; the corresponding loop is also missing from the E. coli and Z. mays Pepc structures (Supplementary Figure 1). The sequence alignment based on the Pepc structural profile [12] incorrectly aligned the sequences corresponding to the bicarbonate-binding loop residues 349-353 of PepcA. The two Arg residues in the loop implicated in bicarbonate binding in the E. coli enzyme, Arg703 and Arg704, are not conserved in PepcA from C. perfringens, M. thermautotrophicus, or S. acidocaldarius (Supplementary Figure 1). Lys546 of the E. coli Pepc has also been proposed to be part of the bicarbonate binding-site based on site-directed mutagenesis.[38] Replacement of the Lys by Arg had little effect on the KM for bicarbonate, while replacement by Thr raised the KM ten-fold. The corresponding residue in the PepcA sequences aligned in Supplementary Figure 1 is Met205. The lack of conservation of the residues implicated in bicarbonate binding by the E. coli mutagenesis studies (Lys 546, Arg703 and Arg704) suggests these residues may not interact directly with bicarbonate, but rather perform a supporting structural role. Another possibility, consistent with the lack of sequence conservation and the difference in the KMs for bicarbonate binding by PepcA and Pepc, is that the interactions responsible for binding bicarbonate are different in Pepc and PepcA.[5][10][39]
Quaternary structure – in the crystal
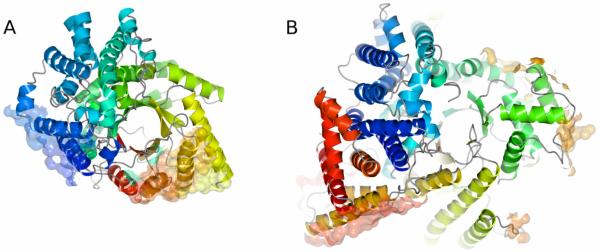
The X-ray structure of PepcA was determined for a crystal form in which two independent tetramers comprise the asymmetric unit. The overall dimensions of the tetramer are 122 × 105 × 79 Å. The tetramer shows approximate 222 symmetry and can be described as a dimer-of-dimers. Within a tetramer, two interfaces on each monomer are involved in intersubunit contacts (Figure 3A, Figure 5). The more extensive interface buries 1160 Å2 of the protein surface upon tetramer formation, while the second, less extensive interface buries 650 Å2. Neither interface is structurally equivalent to the intersubunit interfaces seen in the E. coli and Z. mays tetramers. The extensive dimerization interface includes residues from α-helices α1 (21-22), α2 (55-57), α3 (90-93, 95-96, 99, 102-103), and α4 (144-145, 148-149), as well as residues 53 and 343. The smaller interface apposes a helix-turn-helix motif from two monomers, forming a bundle of four α-helices. The residues involved are in α22 (489, 492-493, 495-496, 499-500, 503) and α24 (517-518, 521, 524-525, 528) at the C-terminal end of the sequence. The sequence conservation in the PepcA family for the residues in these two α-helices is very low (Supplementary Figure 1). For comparison, the more extensive subunit interfaces in Pepc from E. coli and Z. mays involve several helices following β1 and β2 which have no counterparts in PepcA (Figure 1, Figure 3B). The second interface involved in tetramerization of the E. coli and Z. mays proteins is also built from parts of the structure that are elaborations of the β-barrel fold missing in PepcA. The modeling study of Matsumura et al. [12] predicted the surfaces involved in tetramerization would be similarly located in Pepc and PepcA, and this is clearly not the case. Thus, although both Pepc and PepcA crystallize as tetramers, the interfaces involved in tetramer formation are distinct. Any regulation of enzyme activity that is linked to changes at the subunit interfaces is not likely to be broadly conserved across the Pepc and PepcA families.
FIGURE 3.
A The surfaces of PepcA involved in tetramer formation are shown in semi-transparent, all-atom representation. The more extensive interface is at the lower right, and the smaller interface involving the C-terminal α-helices is at the left. The ribbon representing the protein is color-ramped from red at the N-terminus to dark blue at the C-terminus.
B The surfaces of Pepc involved in tetramer formation are shown in the same representation for the E. coli enzyme. The most extensive interface is at the lower left.
FIGURE 5.
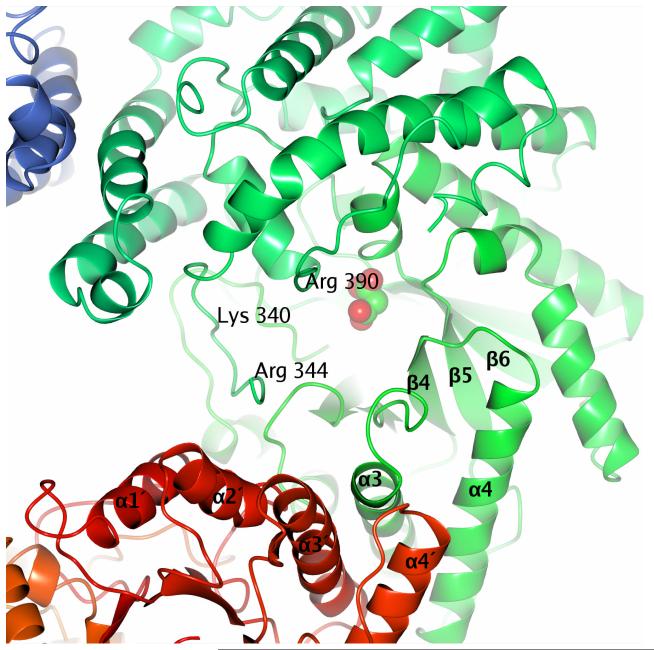
Residues which may be involved in aspartate binding are Lys340, Arg344 and Arg390. Three subunits of the PepcA tetramer are visible in this view. The larger of the two interfaces is at the bottom of the figure, and the bundle of four α-helices defining the smaller interface is at the upper left.
The two independent tetramers in the asymmetric unit themselves associate to form an octameric higher-order assembly. At the tetramer – tetramer interface, reciprocal interactions between adjacent dimers shift the positions of residues 354-386 following the missing loop 349-353 in two subunits. Since these residues immediately follow the missing loop responsible for HCO3− binding, the higher order oligomerization could potentially influence enzyme activity. Interestingly, an octameric assembly of plant Pepc has recently been reported.[40] The helix-turn-helix motif spanning residues 360-380 is missing in Pepc sequences (Figure 3, Supplementary Figure 1). Hence, in plants, potential regulation of HCO3− binding via assembly of tetramers into octamers seems unlikely.
Quaternary structure – in solution
The predominant form of C. perfringens PepcA in solution at concentrations below 3 mg per ml is a dimer, as shown by both gel filtration chromatography [13] and analytical ultracentrifugation (data not shown). Tetramers and higher molecular-weight species were also detected by analytical ultracentrifugation. Sedimentation velocity experiment at the highest PepcA protein concentration of 3 mg per ml showed a very heterogeneous composition with sedimentation coefficient values ranging between 1-40 S. Two-dimensional spectrum analysis consistently revealed the presence of globular monomer, dimer and tetrameric species (about 18, 30 and 20%, respectively, of the total concentration) and the remainder were higher molecular-weight species covering a wide molecular mass range reaching up to 1.8 million Daltons; since the amounts of the individual larger species were rather low, their oligomeric states and relative amounts could not be determined precisely. Addition of the inhibitor aspartate to the gel filtration running buffer (at a concentration ten-fold higher than the Ki) did not change the elution volume. Aspartate binding thus affects PepcA and Pepc differently, as aspartate shifts the equilibrium between dimers and tetramers toward the tetrameric form of E. coli Pepc.[41]
Inhibition by aspartate
Bacterial and eukaryotic Pepc are inhibited by aspartate with half-maximal inhibition occurring at 60 μM aspartate for the E. coli enzyme.[42] The known X-ray structures of the E. coli enzyme all include aspartate and thus represent the T-state of the enzyme. An R-state structure of the Z. mays enzyme revealed an Arg residue moves 15 Å to form either part of the aspartate-binding site or the active-site. In the R-state structure, Arg647 interacts with the C-terminal glycine’s carboxylate group and positions Ser602 and Gln673 in the active-site. In the T-state structures, the corresponding Arg587 is a partner in three interactions with aspartate, while Arg832 is involved in a second bidentate salt bridge to the aspartate, Asn881 provides two hydrogen bonds, and Lys773 provides one interaction. Movement of Arg587 to the aspartate-binding site is accompanied by shifts in the positions of residues 588 - 590 which extend the α-helix α21 between β-strands β7 and β8 by one residue at its N-terminal end. The C. perfringens enzyme lacks the equivalent of Arg832, and neither Asn881 nor Lys773 is conserved. The side-chain of Arg246 is anchored in the active site, as in the R-state Z. mays structure, and it interacts with the C-terminal carboxylate of Gly537 and the side-chains of Ser201 and Gln270. The dramatic rearrangements seen in the E. coli versus Z. mays comparison seem unlikely to occur in PepcA. Phe249 packs well in a hydrophobic environment and would be energetically costly to move. The corresponding residues in the E. coli and Z. mays structures are Ala590 and Gly650, respectively.
Thus, the C. perfringens structure more closely resembles the R-state of Pepc, with both elements which are mobile in the Pepc structures positioned at the active-site. Neither of the allosteric regulatory sites seen in the bacterial and eukaryotic Pepcs is present in the PepcA structure.
Enzyme Kinetics
Activity assays of the C. perfringens enzyme were performed to determine the mode of inhibition by aspartate. C. perfringens PepcA catalyzed formation of OAA in the presence of Mg++, Mn++, or Co++ but not in the absence of a divalent metal ion. Initial studies to determine the optimal concentration of divalent metal ion employed His-tagged PepcA purified in one step via affinity chromatography. Specific activity measured at the optimal concentration for each added metal ion fell in the range of 50-70 μmol OAA formed / minute / mg enzyme. Although the specific activity of His-tagged PepcA purified via affinity chromatography and gel filtration was slightly lower, the enzyme purified over two column chromatography steps was used for further kinetic studies. Initial velocity data were measured in the direction of OAA formation as a function of Mg++ and PEP concentrations. Formation of the Mg++.PEP complex in solution must be considered, as a substantial fraction of PEP is present as the metal complex at the concentrations used. The kinetics results suggest Mg++.PEP is the substrate, rather than PEP, with KM 60μM and Vmax 40 μmol−1 min−1 mg. Mg++.PEP has also been reported to be the substrate for Pepc from several plants.[2-4] Aspartate inhibition was competitive with Mg++.PEP, with a Ki of 0.2 mM (Figure 4).
Two studies have reported that aspartate is an allosteric inhibitor of archaeal PepcAs. One was based on an activity assay at a single concentration of aspartate.[11] The other, more extensive study of the Sulfolobus acidocaldarius PepcA showed that PEP does not compete with aspartate, indicating the binding sites for PEP and aspartate are distinct.[14] In that study, however, Mg++ and aspartate were present at 2 mM while the PEP concentration never exceeded 10 mM. Assuming Mg++.PEP is the substrate for the S. acidocaldarius enzyme, with KM 0.2 mM, and that aspartate binds competitively with Ki 0.2 mM, then increasing the PEP concentration to 10 mM will only bring the initial velocity of the reaction to 36% of Vmax. The hallmark of competitive inhibition, complete recovery of activity in the presence of saturating substrate, would not be observed under the experimental conditions of Sako et al.[14] The data of Sako et al.[14] appear to be consistent with a kinetic model which need not invoke allostery, and thus none of the kinetic studies of aspartate inhibition rule out simple competitive binding at the PepcA active-site.
It can be difficult to identify a regulatory binding site based on a crystal structure of an enzyme with no bound ligands. The Z. mays Pepc structure is a cogent example. Although the enzyme is inhibited strongly by aspartate, the aspartate binding site is not present in the R-state structure. Nonetheless, it is tempting to speculate based on sequence comparisons that Lys340 may form part of the aspartate binding site in PepcA (Figure 5). Residue 340 is an Arg in the aspartate-sensitive PepcA from S. acidocaldarius, and a Gly in the aspartate-resistant PepcA from M. thermoautotrophicus (unfortunately the sequence of PepcA from M. sociabilis is not known). The nearby residues Arg344 and Arg390 could also be involved in aspartate binding. Arg699 in the E. coli enzyme (corresponding to Arg344 of C. perfringens) is directly involved in binding the inhibitory PEP analog, and Arg713 in the E. coli enzyme (corresponding to Arg390 of C. perfringens) is 3.4 Ǻ from the PEP analog. Thus in PepcA inhibition by aspartate could be due to direct competition for residues involved in PEP binding.
Summary
The first crystal structure of an archaeal-type PepcA shows that the underlying basis for inhibition by aspartate does not involve a dramatic rearrangement of the active-site, as observed in the E. coli and Z. mays Pepc structures. Neither of the allosteric mechanisms used to activate or inhibit Pepc activity are supported by the PepcA structure. Further work will be required to pin down the basis for the differing sensitivity of PepcA family members to inhibition by aspartate.
Supplementary Material
Acknowledgements
Analytical ultracentrifugation was performed by Borries Demeler at the University of Texas Health Science Center at San Antonio. LD received a graduate fellowship from the Virginia Tech Genetics, Bioinformatics and Computational Biology Ph.D. program. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. The projects described were partially supported by Grant Number 5 P41 RR001209 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
REFERENCES
- 1.Chollet R, Vidal J, O’Leary MH. Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:273–298. doi: 10.1146/annurev.arplant.47.1.273. [DOI] [PubMed] [Google Scholar]
- 2.Wedding RT, Rustin P, Meyer CR, Black MK. Kinetic studies of the form of substrate bound by phosphoenolpyruvate carboxylase. Plant Physiol. 1988;88:976–979. doi: 10.1104/pp.88.4.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer CR, Rustin P, Black MK, Wedding RT. The influence of pH on substrate form specificity of phosphoenolpyruvate carboxylase purified from Crassula argentea. Arch Biochem Biophys. 1990;278:365–372. doi: 10.1016/0003-9861(90)90272-z. [DOI] [PubMed] [Google Scholar]
- 4.Tovar-Méndez A, Rodriguez-Sotres R, Lopez-Valentin DM, Munoz-Clares RA. Re-examination of the roles of PEP and Mg2+ in the reaction catalysed by the phosphorylated and non-phosphorylated forms of phosphoenolpyruvate carboxylase from leaves of Zea mays. Effects of the activators glucose 6-phosphate and glycine. Biochem J. 1998;332:633–642. doi: 10.1042/bj3320633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kai Y, Matsumura H, Inoue T, Terada K, Yoshinaga T, Kihara A, Tsumura K, Izui K. Three-dimensional structure of phosphoenolpyruvate carboxylase: A proposed mechanism for allosteric inhibition. Proc Natl Acad Sci USA. 1999;96:823–828. doi: 10.1073/pnas.96.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumura H, Xie Y, Shirakata S, Inoue T, Yoshinaga T, Ueno Y, Izui K, Kai Y. Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylase. Structure. 2002;10:1721–1730. doi: 10.1016/s0969-2126(02)00913-9. [DOI] [PubMed] [Google Scholar]
- 7.Yano M, Terada K, Umiji K, Izui K. Catalytic role of an arginine residue in the highly conserved and unique sequence of phosphoenolpyruvate carboxylase. J Biochem (Tokyo) 1995;117:1196–1200. doi: 10.1093/oxfordjournals.jbchem.a124844. [DOI] [PubMed] [Google Scholar]
- 8.Terada K, Izui K. Site-directed mutagenesis of the conserved histidine residue of phosphoenolpyruvate carboxylase. His138 is essential for the second partial reaction. Eur J Biochem. 1991;202:797–803. doi: 10.1111/j.1432-1033.1991.tb16435.x. [DOI] [PubMed] [Google Scholar]
- 9.Izui K, Matsumura H, Furumoto T, Kai Y. Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu Rev Plant Biol. 2004;55:69–84. doi: 10.1146/annurev.arplant.55.031903.141619. [DOI] [PubMed] [Google Scholar]
- 10.Patel HM, Kraszewski J, Mukhopadhyay B. The phosphoenolpyruvate carboxylase from Methanothermobacter thermautotrophicus has a novel structure. J Bacteriol. 2004;186:5129–5137. doi: 10.1128/JB.186.15.5129-5137.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ettema TJ, Makarova KS, Jellema GL, Gierman HJ, Koonin EV, Huynen MA, de Vos WM, van der Oost J. Identification and functional verification of archaeal-type phosphoenolpyruvate carboxylase, a missing link in archaeal central carbohydrate metabolism. J Bacteriol. 2004;186:7754–7762. doi: 10.1128/JB.186.22.7754-7762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumura H, Izui K, Mizuguchi K. A novel mechanism of allosteric regulation of archaeal phosphoenolpyruvate carboxylase: a combined approach to structure-based alignment and model assessment. Protein Eng. 2006;19:409–419. doi: 10.1093/protein/gzl025. [DOI] [PubMed] [Google Scholar]
- 13.Dharmarajan L, Kraszewski JL, Mukhopadhyay B, Dunten PW. Expression, purification and crystallization of an archaeal-type phosphoenolpyruvate carboxylase. Acta Cryst. 2009;F65:1193–1196. doi: 10.1107/S1744309109042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sako Y, Takai K, Nishizaka T, Ishida Y. Biochemical relationship of phosphoenolpyruvate carboxylases (PEPCs) from thermophilic archaea. FEMS Microbiol Lett. 1997;153:159–165. [Google Scholar]
- 15.Sako Y, Takai K, Uchida A, Ishida Y. Purification and characterization of phosphoenolpyruvate carboxylase from the hyperthermophilic archaeon Methanothermus sociabilis. FEBS Lett. 1996;392:148–152. doi: 10.1016/0014-5793(96)00805-8. [DOI] [PubMed] [Google Scholar]
- 16.Sheldrick GM. A short history of SHELX. Acta Cryst. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 17.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Cryst. 2003;D59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Cryst. 2006;D62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 20.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Bedem H, Lotan I, Latombe J-C, Deacon AM. Real-space protein-model completion: an inverse-kinematics approach. Acta Cryst. 2005;D61:2–13. doi: 10.1107/S0907444904025697. [DOI] [PubMed] [Google Scholar]
- 22.Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogens. J Mol Biol. 1999;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- 23.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skubák P, Murshudov GN, Pannu NS. Direct incorporation of experimental phase information in model refinement. Acta Cryst. 2004;D60:2196–2201. doi: 10.1107/S0907444904019079. [DOI] [PubMed] [Google Scholar]
- 25.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Cryst. 2006;39:109–111. [Google Scholar]
- 26.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Cryst. 2006;D62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 27.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Cryst. 2004;D60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Dawson RMC, Elliot DC, Elliot WH, Jones KM. Data for Biochemical Research. 3rd edition Oxford University Press; Oxford: 1986. [Google Scholar]
- 30.Kato Y, Komiya K, Sasaki H, Hashimoto T. Separation range and separation efficiency in high-speed gel-filtration on TSK-gel SW columns. Journal of Chromatography. 1980;190:297–303. 1980. [Google Scholar]
- 31.Demeler B. UltraScan: A comprehensive data analysis software package for analytical ultracentrifugation experiments. In: Scott D, Harding S, Rowe A, editors. Modern Analytical Ultracentrifugation: Techniques and Methods. Royal Society of Chemistry; Cambridge, U.K.: 2005. pp. 210–229. [Google Scholar]
- 32.Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39:423–35. doi: 10.1007/s00249-009-0413-5. [DOI] [PubMed] [Google Scholar]
- 33.Demeler B, van Holde KE. Sedimentation velocity analysis of highly heterogeneous systems. Anal Biochem. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 34.Corwin LM, Fanning GR. Studies of parameters affecting the allosteric nature of phosphoenolpyruvate carboxylase of Escherichia coli. J Biol Chem. 1968;243:3517–3525. [PubMed] [Google Scholar]
- 35.O’Leary MH. Mechanism of action of phosphoenolpyruvate carboxykinase. Physiol Vég. 1983;21:883–888. [Google Scholar]
- 36.González DH, Andreo CS. The use of substrate analogues to study the active-site structure and mechanism of PEP carboxylase. TIBS. 1989;14:24–27. [Google Scholar]
- 37.Rose IA, O’Connell EL, Noce P, Utter MF, Wood HG, Willard JM, Cooper TG, Benziman M. Stereochemistry of the enzymatic carboxylation of phosphoenolpyruvate. J Biol Chem. 1969;244:6130–6133. [PubMed] [Google Scholar]
- 38.Gao Y, Woo KC. Site-directed mutagenesis of Lys600 in phosphoenolpyruvate carboxylase of Flaveria trinervia: its roles in catalytic and regulatory functions. FEBS Letters. 1995;375:95–98. doi: 10.1016/0014-5793(95)01189-l. [DOI] [PubMed] [Google Scholar]
- 39.Dong L-Y, Masuda T, Kawamura T, Hata S, Izui K. Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: Comparison with the C4-form enzyme. Plant Cell Physiol. 1998;39:865–873. doi: 10.1093/oxfordjournals.pcp.a029446. [DOI] [PubMed] [Google Scholar]
- 40.O’Leary B, Rao SK, Kim J, Plaxton WC. Bacterial-type phosphoenolpyruvate carboxylase (PEPC) functions as a catalytic and regulatory subunit of the novel class-2 PEPC complex of vascular plants. J Biol Chem. 2009;284:24797–805. doi: 10.1074/jbc.M109.022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coomes MW, Mitchell BK, Beezley A, Smith TE. Properties of an Escherichia coli mutant deficient in phosphoenolpyruvate carboxylase catalytic activity. J Bacteriol. 1985;164:646–652. doi: 10.1128/jb.164.2.646-652.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yano M, Izui K. The replacement of Lys620 by serine desensitizes Escherichia coli phosphoenolpyruvate carboxylase to the effects of the feedback inhibitors l-aspartate and l-malate. Eur J Biochem. 1997;247:74–81. doi: 10.1111/j.1432-1033.1997.t01-1-00074.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.