Abstract
Oral squamous cell carcinoma (OSCC) accounts for more than 90% of the malignant neoplasms that arise in the mucosa of the upper aerodigestive tract. Recent studies of cleft lip/palate have shown the association of genes involved in cancer. WNT pathway genes have been associated with several types of cancer and recently with cleft lip/palate. To investigate if genes associated with cleft lip/palate were also associated with oral cancer, we genotyped 188 individuals with OSCC and 225 control individuals for markers in AXIN2, AXIN1, GSK3β, WNT3A, WNT5A, WNT8A, WNT11, WNT3, and WNT9B. Statistical analysis was performed with PLINK 1.06 software to test for differences in allele frequencies of each polymorphism between cases and controls. We found association of SNPs in GSK3B (p = 0.0008) and WNT11 (p = 0.03) with OSCC. We also found overtransmission of GSK3B haplotypes in OSCC cases. Expression analyses showed up-regulation of WNT3A, GSK3B, and AXIN1 and down-regulation of WNT11 in OSCC in comparison with control tissues (P < 0.001). Additional studies should focus on the identification of potentially functional variants in these genes as contributors to human clefting and oral cancer.
Keywords: WNT pathway genes, cleft lip/palate, oral cancer, squamous cell carcinoma, polymorphism
Introduction
Oral cancer accounts for roughly 2% of all cancers diagnosed annually in the United States. Approximately 35,000 people will be diagnosed with oral cancer each year, and about 7600 will die from the disease (National Institute of Dental and Craniofacial Research, 2010). Oral-facial clefts comprise a large fraction of all human birth defects, affecting approximately 1 in every 500 to 1000 births worldwide, and are notable for their significant lifelong morbidity and complex etiology (Murray, 2002).
Several authors have proposed that cancer and congenital malformations such as cleft lip and palate may occasionally have a common etiology (Nishi et al., 2000; Zhu et al., 2002; Bille et al., 2005; Menezes et al., 2009). The underlying concept is that the same genes may have functions during normal development, as well as in cancer development.
Several genes compose the Wnt signaling pathway, each with a specific role contributing to the complex cascade of events that eventually lead to the normal morphogenesis of the face (Mani et al., 2010). WNT genes have been recently suggested as candidate genes for cleft lip/palate, due to their essential functions during embryonic development (Chiquet et al., 2008). Moreover, mutations in AXIN2, a gene belonging to the Wnt signaling pathway, have also been detected in families segregating cancer and tooth agenesis (Lammi et al., 2004). Single-nucleotide polymorphisms (SNPs) in AXIN2 have been associated with cleft lip/palate in a case-control study (Letra et al., 2009), and in a family study where the incidence of cancer in cleft families was notably higher than in control families (Menezes et al., 2009). To further investigate the hypothesis that genes may have roles in cleft lip/palate and in cancer development, we examined WNT pathway genes that were previously investigated or associated with oral clefts for association with oral squamous cell carcinoma.
Materials & Methods
Study Population
The study was approved by the University of Pittsburgh Institutional Review Board (IRB#010356). In total, 413 individuals of white ancestry from Pittsburgh, PA, were ascertained at the University of Pittsburgh Hillman Cancer Institute. The study population consisted of 188 affected individuals (98 male, 90 female), aged 33-92 (average, 62.5 yrs), diagnosed with oral squamous cell carcinoma, and 225 unrelated control individuals (109 male, 116 female), aged 29-85 (average, 60.3 yrs), without oral squamous cell carcinoma or family history of oral squamous cell carcinoma. Data on smoking and ethanol consumption were not consistently available for all study participants. Participants signed an informed consent and were examined clinically and through their medical records for determination of their individual oral cancer status.
Single-nucleotide Polymorphism Selection and Genotyping
We selected 32 SNPs spanning 9 WNT pathway genes that have been previously suggested as candidate genes for cleft/lip palate based on studies with animal models or association studies in humans (Juriloff et al., 2005, 2006; Lan et al., 2006; Liu et al., 2007; Chiquet et al., 2008; Menezes et al., 2009, 2010) to test for association with oral squamous cell carcinoma in our population. Details of the selected SNPs and genes investigated are presented in Table 1.
Table 1.
Summary of Candidate Genes and SNPs Studied
| Gene | CHR | SNP | Base Pair Position | Base Change | SNP Type | MAF |
|---|---|---|---|---|---|---|
| WNT3A | 1 | rs708111 | 228191365 | A/G | 5′ upstream | 0.47 (G) |
| WNT3A | 1 | rs3094912 | 228209815 | T/A | Intron | 0.47 (A) |
| WNT3A | 1 | rs752107 | 228247351 | T/C | UTR 3 | 0.31 (T) |
| WNT3A | 1 | rs1745420 | 228251732 | C/G | 3′ upstream | 0.13 (C) |
| WNT5A | 3 | rs566926 | 55520778 | A/C | Intron | 0.2 (A) |
| GSK3B | 3 | rs13314595 | 119631098 | C/T | Intron | 0.24 (T) |
| GSK3B | 3 | rs4072520 | 119635393 | A/C | Intron | 0.24 (A) |
| GSK3B | 3 | rs7620750 | 119643181 | A/G | Intron | 0.24 (A) |
| GSK3B | 3 | rs6769435 | 119684407 | A/C | Intron | 0.25 (C) |
| GSK3B | 3 | rs6771023 | 119693611 | C/T | Intron | 0.25 (C) |
| GSK3B | 3 | rs9879992 | 119712721 | A/G | Intron | 0.25 (G) |
| WNT8A | 5 | rs2040862 | 137419989 | C/T | Intron | 0.16 (T) |
| WNT11 | 11 | rs1533767 | 75905800 | A/G | Exon | 0.22 (A)* |
| AXIN1 | 16 | rs214249 | 348687 | A/C | Intron | 0.4 (C) |
| AXIN1 | 16 | rs7359414 | 362638 | G/T | Intron | 0.48 (T) |
| WNT3 (NSF) | 17 | rs142167 | 44795234 | A/G | Intron | 0.2 (G) |
| WNT3 | 17 | rs199498 | 44865603 | C/T | Intron | 0.17 (C) |
| WNT3 | 17 | rs111769 | 44871987 | C/T | Intron | 0.35 (T) |
| WNT3 | 17 | rs9890413 | 44901449 | A/G | Intergenic | 0.32 (G)* |
| WNT9B | 17 | rs2165846 | 44941366 | A/G | Intron | 0.44 (G) |
| WNT9B/GOSR2 | 17 | rs197915 | 44990522 | A/G | Intergenic | 0.4 (G)* |
| AXIN2 | 17 | rs7591 | 63525082 | A/T | UTR 3 | 0.42 (A) |
| AXIN2 | 17 | rs7224837 | 63528123 | A/G | Intron | 0.13 (G) |
| AXIN2 | 17 | rs4128941 | 63531331 | A/G | Intron | 0.06 (A) |
| AXIN2 | 17 | rs11867417 | 63537898 | C/T | Intron | 0.3 (T) |
| AXIN2 | 17 | rs4791171 | 63541497 | A/G | Intron | 0.3 (A) |
| AXIN2 | 17 | rs3923087 | 63549261 | C/T | Intron | 0.21 (T) |
| AXIN2 | 17 | rs3923086 | 63549488 | A/C | Intron | 0.39 (A) |
| AXIN2 | 17 | rs2240307 | 63554307 | A/G | Exon | 0.02 (G) |
| AXIN2 | 17 | rs2240308 | 63554591 | A/G | Exon | 0.47 (G) |
| AXIN2 | 17 | rs740026 | 63561681 | A/G | Intergenic | 0.46 (A) |
| AXIN2 | 17 | rs1017020 | 63563906 | C/A | Intergenic | 0.11 (C) |
MAF, minor allele frequency according to HapMap CEPH.
According to Applied Biosystems.
Genomic DNA was obtained from blood samples as previously described (Andrade Filho et al., 2010). Genotyping of selected polymorphisms was carried out with Taqman assays and reagents in an ABI 7900 automatic instrument (Applied Biosystems, Foster City, CA, USA). We used PLINK 1.06 software (Purcell et al., 2007) to test for differences in allele frequencies of each polymorphism between cases with oral squamous cell carcinoma and controls. Haplotype analysis was also performed with PLINK. We considered significant the P-values adjusted with Bonferroni correction as implemented in PLINK.
Quantitative Real-time PCR
Frozen tumor samples (n = 10) and normal mucosal tissues (n = 4) collected after surgeries for tumor biopsies and amygdalectomies, respectively, were obtained from the University of Pittsburgh Hillman Cancer Institute. Total RNA was isolated with the RNeasy Mini Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s protocol. The mucosa was carefully dissected from subjacent muscle or lymphoid tissue before RNA extraction, to completely isolate the normal squamous epithelium. The RNA pellet was dried under a vacuum and re-suspended in 30-50 μL of DEPC-treated water. We confirmed the integrity of RNA samples by analyzing 1 μg of total RNA on 1.2% (w/v) denaturing formaldehyde-agarose gels. Prior to DNA synthesis, RNA was treated with deoxyribonuclease I, Amplification Grade (Invitrogen, Carlsbad, CA, USA) for 15 min at room temperature to avoid DNA contamination. DNase I was inactivated by incubation with 25 mM EDTA at 65°C for 10 min. Complementary DNA was synthesized by the use of 3 μg of RNA through a reverse-transcription reaction as previously described (Garlet et al., 2004). Real-time polymerase chain-reaction (qPCR) quantitative messenger RNA (mRNA) analyses were performed in an ABI 7900 automatic instrument (Applied Biosystems, Foster City, CA, USA) with SYBR-green chemistry (Applied Biosystems) and specific primers (Appendix Table). The determination of the relative levels of gene expression was performed by the cycle threshold (CT) method, in reference to the internal control gene beta-actin, as described elsewhere (Garlet et al., 2004). Reaction conditions were 95°C for 10 min, 40 cycles at 94°C for 1 min, annealing at 56°C for 1 min, and at 72°C for 2 min. Results are depicted as the mean average mRNA expression from triplicate measurements normalized by the internal control gene beta-actin. Statistical analyses included analysis of variance followed by Bonferroni correction in GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA, USA). A P ≤ 0.05 was considered statistically significant.
Immunohistochemical Localization of GSK-3B
We used immunohistochemistry to investigate the presence of GSK-3B in human oral squamous cell carcinoma samples. Briefly, paraffin-embedded oral squamous cell carcinoma tissues obtained from the archives of the Department of Diagnostic Sciences at the University of Pittsburgh School of Dental Medicine were sectioned into 5-μm slices, deparaffinized, and rehydrated by standard techniques. Immunohistochemical localization of GSK-3B was performed by the immunoperoxidase (avidin-biotin-peroxidase) method. Antigen retrieval was performed with citrate buffer, pH 6.0, at 96°C for 20 min. Sections were incubated in a rabbit polyclonal GSK-3B antibody (SC-9166; Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 1:100 dilution at 4°C overnight, followed by incubation in biotinylated secondary antibody for 1 hr at room temperature. Negative controls were obtained by substitution of the primary antibody with PBS.
Results
The most significant genetic association results are summarized in Table 2. Genotype and allele distributions were within Hardy-Weinberg equilibrium (data not shown). SNPs in 2 genes, GSK3B (rs9879992) and WNT11 (rs1533767), showed association with oral squamous cell carcinoma (p = 0.0007 and p = 0.03, respectively). Alternatively, a SNP in AXIN2 (rs3923087) also showed association with oral squamous cell carcinoma (p = 0.001), although toward a protective effect [OR = 0.57 (95%CI: 0.41-0.8)]. Borderline association was also observed for SNPs located in WNT3A and AXIN1 genes (Table 2).
Table 2.
Results of Association Tests with WNT Genes in OSCC and Control Individuals
| Gene | SNP | A1 | MAF_A | MAF_U | P | OR (95%CI) |
|---|---|---|---|---|---|---|
| WNT3A | rs708111 | A | 0.53 | 0.47 | 0.08 | 1.28 (0.97-1.69) |
| WNT3A | rs3094912 | T | 0.46 | 0.53 | 0.04 | 0.75 (0.57-0.99) |
| WNT3A | rs752107 | T | 0.31 | 0.39 | 0.02 | 0.7 (0.52-0.94) |
| WNT3A | rs1745420 | C | 0.11 | 0.13 | 0.29 | 0.8 (0.52-1.23) |
| WNT5A | rs566926 | T | 0.27 | 0.29 | 0.42 | 0.88 (0.65-1.20) |
| GSK3B | rs13314595 | T | 0.22 | 0.19 | 0.19 | 1.25 (0.89-1.76) |
| GSK3B | rs4072520 | A | 0.22 | 0.20 | 0.42 | 1.15 (0.82-1.62) |
| GSK3B | rs7620750 | A | 0.22 | 0.18 | 0.24 | 1.23 (0.87-1.75) |
| GSK3B | rs6769435 | C | 0.21 | 0.19 | 0.44 | 1.15 (0.81-1.62) |
| GSK3B | rs6771023 | C | 0.23 | 0.19 | 0.14 | 1.29 (0.92-1.82) |
| GSK3B | rs9879992 | G | 0.27 | 0.17 | 0.0007 | 1.78 (1.27-2.50) |
| WNT8A | rs2040862 | T | 0.19 | 0.20 | 0.59 | 0.91 (0.64-1.29) |
| WNT11 | rs1533767 | A | 0.33 | 0.26 | 0.03 | 1.4 (1.03-1.90) |
| AXIN1 | rs214249 | G | 0.38 | 0.37 | 0.69 | 1.06 (0.80-1.41) |
| AXIN1 | rs7359414 | T | 0.45 | 0.51 | 0.08 | 0.78 (0.59-1.03) |
| WNT3 (NSF) | rs142167 | G | 0.24 | 0.24 | 0.90 | 0.98 (0.71-1.35) |
| WNT3 | rs199498 | C | 0.24 | 0.21 | 0.35 | 1.17 (0.84-1.62) |
| WNT3 | rs111769 | T | 0.42 | 0.39 | 0.53 | 1.09 (0.83-1.45) |
| WNT3 | rs9890413 | G | 0.30 | 0.31 | 0.71 | 0.94 (0.70-1.27) |
| WNT9B | rs2165846 | G | 0.43 | 0.38 | 0.17 | 1.22 (0.92-1.62) |
| WNT9B | rs197915 | G | 0.41 | 0.44 | 0.46 | 0.9 (0.68-1.19) |
| AXIN2 | rs7591 | A | 0.42 | 0.42 | 0.9 | 0.98 (0.74-1.30) |
| AXIN2 | rs7224837 | G | 0.09 | 0.11 | 0.3 | 0.78 (0.48-1.26) |
| AXIN2 | rs4128941 | A | 0.03 | 0.05 | 0.13 | 0.57 (0.27-1.19) |
| AXIN2 | rs11867417 | T | 0.37 | 0.34 | 0.52 | 1.10 (0.82-1.48) |
| AXIN2 | rs4791171 | T | 0.33 | 0.30 | 0.29 | 1.17 (0.87-1.58) |
| AXIN2 | rs3923087 | T | 0.1885 | 0.29 | 0.001 | 0.57 (0.41-0.80) |
| AXIN2 | rs3923086 | A | 0.4601 | 0.47 | 0.82 | 0.97 (0.73-1.28) |
| AXIN2 | rs2240307 | A | 0.484 | 0.47 | 0.76 | 1.04 (0.79-1.37) |
| AXIN2 | rs2240308 | A | 0.4798 | 0.5 | 0.58 | 0.92 (0.69-1.22) |
| AXIN2 | rs740026 | A | 0.4472 | 0.4 | 0.18 | 1.21 (0.91-1.61) |
| AXIN2 | rs1017020 | C | 0.1317 | 0.1 | 0.21 | 1.31 (0.85-2.01) |
A1: Minor allele (based on whole sample).
MAF_A : minor allele frequency in cases.
MAF_U : minor allele frequency in controls.
The results of the haplotype analysis further support the associations found for the individual SNPs (Table 3). We observed overtransmission of GSK3B haplotypes containing the associated SNP rs9879992: rs6771023-rs9879992 (p = 0.0008) and rs6769435-rs6771023-9879992 (p = 3.00e-005).
Table 3.
Most Significant Sliding Window Haplotype Association Results Observed for Markers in GSK3β and WNT3A Genes Located in Chromosomes 3 and 1, Respectively, and OSCC in a Pittsburgh, PA, USA, Population
| GSK3β | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs13314595 |
rs4072520 |
rs7620750 |
rs6769435 |
rs6771023 |
rs9879992 |
|||||
| p = 0.2 (TA) | ||||||||||
| p = 0.2 (AA) | ||||||||||
| p = 0.2 (AC) | ||||||||||
| p = 0.2 (CC) | ||||||||||
| p = 0.0008 (TG) | ||||||||||
| p = 0.17 (TAA) | ||||||||||
| p = 3.00E-05 (ATG) | ||||||||||
| p = 0.00005 (CCGATG) | ||||||||||
| WNT3A | ||||||||||
| rs708111 |
rs3094912 |
rs752107 |
rs1745420 |
|||||||
| p = 0.06 (AA) | ||||||||||
| p = 0.1 (AC) | ||||||||||
| p = 0.03 (CG) | ||||||||||
| p = 0.08 (AAC) | ||||||||||
| p = 0.05 (ACG) | ||||||||||
| p = 0.06 (AACG) | ||||||||||
Results of Expression Analyses
To support the potential association of WNT3A, WNT11, GSK3B, AXIN1, and AXIN2 genes with the pathogenesis of oral squamous cell carcinoma, we investigated their expression in diseased vs. healthy tissues (Fig.). WNT3A, GSK3B, and AXIN1 mRNA expression was significantly higher in cases with oral squamous cell carcinoma compared with control tissues (P < 0.001), whereas expression of AXIN2 and WNT11 mRNA was similar in both groups, with WNT11 showing even lower expression in tumor tissues (Fig.).
Figure.
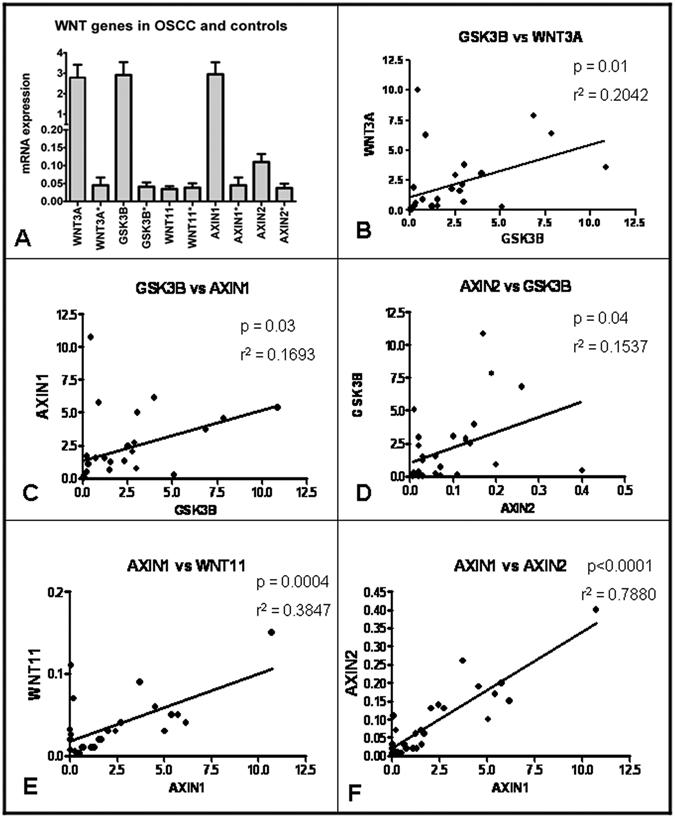
Summary of gene expression results. mRNA expression in tumor samples and controls (A). Most significant positive correlations observed: GSK3B vs. WNT3A (B); GSK3B vs. AXIN1 (C); AXIN2 vs. GSK3B (D); AXIN1 vs. WNT11 (E); and AXIN1 vs. AXIN2 (F).
We observed some interesting correlations with regard to the expression levels of the investigated genes. We noted that GSK3B expression levels showed positive correlation with WNT3A (p = 0.01, r2 = 0.2042), AXIN1 (p = 0.03, r2 = 0.1693), and AXIN2 (p = 0.04, r2 = 0.1537) levels. In addition, AXIN1 levels showed positive correlation with WNT3A (P < 0.0001,r2 = 0.8011), WNT11 (p = 0.0004, r2 = 0.3847), and AXIN2 (p < 0.0001, r2 = 0.7880) levels. Further, AXIN2 expression showed positive correlation with WNT3A (p < 0.0001, r2 = 0.8671) and WNT11 (p < 0.0001, r2 = 0.7035), and WNT3A expression was positively correlated with WNT11 (p < 0.0001, r2 = 0.4680).
We also investigated the GSK3-B protein expression in oral squamous cell carcinoma tissues, due to the significant results found in the SNP association analysis. We detected the marked presence of GSK3-B in the cytoplasm of tumor cells (Appendix Fig.).
Discussion
In this study, we investigated the association of Wnt signaling pathway genes that had previously been suggested as candidates for oral-facial clefting for their association with oral squamous cell carcinoma. We found association of GSK3B and WNT11 genes with oral squamous cell carcinoma. This study provides evidence that variations in WNT genes shown to be involved in oral-facial clefting may also increase an individual’s susceptibility of developing oral squamous cell carcinoma.
The importance of WNT signaling for proper craniofacial development in the mouse embryo has been reported by several authors (Juriloff et al., 2005, 2006; Clevers, 2006; Lan et al., 2006). Activation of Wnt signaling has also been demonstrated as the underlying etiology of some oral cancers (Liu and Millar, 2010). Increasing evidence also exists to support the hypothesis that some genes display essential functions during embryogenesis and at some point may be involved in cancer development (Christensen et al., 2004; Bille et al., 2005; Menezes et al., 2009). We have previously reported that individuals with oral-facial clefts present a family history of cancer much more frequently than do individuals without clefts. Further, we verified that variations in AXIN2, a gene that belongs to the WNT pathway and which, when mutated, increases susceptibility to colon cancer, were also associated with a cleft phenotype in those families (Menezes et al., 2009). Additional studies in mice and humans support the involvement of WNT genes in the etiology of oral-facial clefts. In an inbred A/WySn mouse strain characterized for the presence of cleft palate, Wnt3 and Wnt9B genes were found to be located in the clf1 cleft locus, suggesting their possible involvement in the cleft phenotype (Juriloff et al., 2005). Recently, variations in WNT genes were associated with non-syndromic oral-facial clefts in different populations, further supporting the notion that WNT genes may be involved in the etiology of oral clefts in humans as well (Chiquet et al., 2008; Menezes et al., 2010).
GSK3B is part of the Wnt signaling pathway and plays a major role in epithelial cell homeostasis (Kim et al., 2007), and because GSK3B interacts with many different pathways, its specific developmental roles remain unclear. Our results showed a significant association of an intronic SNP in GSK3B (rs9879992) with oral squamous cell carcinoma. Analysis of GSK3B combined marker haplotypes also showed association of haplotypes containing the associated SNP. In addition, we observed GSK3B to be highly expressed in the oral squamous cell carcinoma biopsies. It has been proposed that the manipulation of the activated GSK3B may provide hope for treating oral cancer (Mishra, 2010). Interestingly, a role for GSK3B in craniofacial defects has also been demonstrated, since homozygous null mice display cleft palate, incomplete fusion of the ribs at the midline and bifid sternum, and delayed sternal ossification (Liu et al., 2007).
WNT11 also appears to be involved with oral squamous cell carcinoma risk. We found association of a synonymous coding mutation in WNT11 (rs1533767, Pro136Pro) so far not predicted to have any deleterious effect on protein function. Using the software FAS-ESS (http://genes.mit.edu/fas-ess/), we verified that this mutation is involved in splicing regulation and the ancestral allele G attributes splicing silencing properties. Moreover, the results of WNT11 mRNA expression analysis showed significant down-regulation in the tumor tissues when compared with healthy tissues as measured by quantitative real-time PCR, suggesting a role for WNT11 as a tumor suppressor gene during the development of oral squamous cell carcinoma. These findings are further supported by a recent study where loss of WNT11 expression was shown to promote the malignant phenotype via both canonical and non-canonical Wnt signaling pathways (Toyama et al., 2010).
AXIN2 is a negative regulator of the WNT-beta-catenin pathway. We observed the association of AXIN2 with a lower risk of oral squamous cell carcinoma, thus suggesting that the associated variant may have a protective effect in these cases. We also detected lower AXIN2 mRNA expression in the tumor tissues when compared with healthy control tissues. Our results corroborated those from previous studies showing that epigenetic silencing of AXIN2 is relevant for hypermethylation and histone deacetylation, leading to a decreased mRNA/protein expression of AXIN2 gene in lung cancer (Tseng et al., 2008). We may speculate that a similar mechanism may occur during oral squamous cell carcinoma development.
In addition to the individual genetic and expression results for GSK3B, WNT11, and AXIN2 that implicate a possible role for these genes in the etiology of oral squamous cell carcinoma, the correlation of their expression levels revealed interesting findings that warrant further investigations. Except for the exonic WNT11 mutation, both GSK3B and AXIN2 SNPs are located in introns and are less likely to be considered disease-causing variants. Instead, they are likely to be in linkage disequilibrium with the true causal variant, and may contribute indirectly to the disease risk. A substantial proportion of disease-associated SNPs is not located within or near known genes, suggesting that non-coding DNA variation may impart functional effects (Notaridou et al., 2011). In regard to gene expression in the tumor tissues, several positive correlations were observed between the associated genes and additional genes in the WNT pathway. WNT3A, GSK3B, and AXIN1 were all significantly up-regulated in oral squamous cell carcinoma cases in comparison with controls, whereas AXIN2 was slightly up-regulated and WNT11 was down-regulated. Further, the positive correlations observed for GSK3B with WNT3A, AXIN1 and AXIN2, where GSK3B and WNT3A are considered “tumor-promoters”, and AXIN1 and AXIN2 are considered “tumor-suppressors”, highlight the importance of studying genetic pathways instead of one single gene as causative for complex diseases such as cancer and craniofacial anomalies.
Even though numerous common genetic variants have now been shown to be associated with the risk of several cancer types, this study provides evidence that variation in WNT pathway genes, in particular GSK3B, might contribute to an individual’s susceptibility to oral squamous cell carcinoma. We must note that in addition to individual predisposition to cancer, we should also consider environmental factors that may modulate each individual’s risk to develop the disease, such as smoking and alcohol intake, among other factors. Unfortunately, data on these environmental variables were not conclusive to be included in our analyses, which was a limitation of this study. Nevertheless, our findings associating WNT genes such as GSK3B and WNT11 and oral squamous cell carcinoma reinforce the suggested implications for this gene family in cancer. In addition, considering the recently reported associations of these genes in oral-facial clefts, the results of this study corroborate previous suggestions that the same genes may have different roles during different biological processes such as embryonic development and cancer. Additional studies should focus on the identification of potentially functional variants in these genes, while approaching cell-signaling networks as contributors to human clefting and oral cancer.
Supplementary Material
Acknowledgments
The authors are indebted to the participants of the study. This work was supported by NIH grants R00DE018413 (to RM) and R00DE018954 (to AL). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research or the National Institutes of Health.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Andrade Filho PA, Ito D, Deleo AB, Ferris RL. (2010). CD8+ T cell recognition of polymorphic wild-type sequence p53 (65-73) peptides in squamous cell carcinoma of the head and neck. Cancer Immunol Immunother 59:1561-1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bille C, Winther JF, Bautz A, Murray JC, Olsen J, Christensen K. (2005). Cancer risk in persons with oral cleft: a population-based study of 8,093 cases. Am J Epidemiol 161:1047-1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet BT, Blanton SH, Burt A, Ma D, Stal S, Mulliken JB, et al. (2008). Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum Mol Genet 17:2212-2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Juel K, Herskind AM, Murray JC. (2004). Long term follow up study of survival associated with cleft lip and palate at birth. BMJ 328:1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. (2006). Wnt/beta-catenin signaling in development and disease. Cell 127:469-480 [DOI] [PubMed] [Google Scholar]
- Garlet GP, Martins W, Jr, Fonseca BA, Ferreira BR, Silva JS. (2004). Matrix metalloproteinases, their physiological inhibitors and osteoclast factors are differentially regulated by the cytokine profile in human periodontal disease. J Clin Periodontol 31:671-679 [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, Dewell SL, Brown CJ, Mager DL, Gagnier L, et al. (2005). Investigations of the genomic region that contains the clf1 mutation, a causal gene in multifactorial cleft lip and palate in mice. Birth Defects Res A Clin Mol Teratol 73:103-113 [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, McMahon AP, Carroll TJ, Lidral AC. (2006). Wnt9b is the mutated gene involved in multifactorial nonsyndromic cleft lip with or without cleft palate in A/WySn mice, as confirmed by a genetic complementation test. Birth Defects Res A Clin Mol Teratol 76:574-579 [DOI] [PubMed] [Google Scholar]
- Kim M, Datta A, Brakeman P, Yu W, Mostov KE. (2007). Polarity proteins PAR6 and aPKC regulate cell death through GSK-3beta in 3D epithelial morphogenesis. J Cell Sci 120(Pt 14):2309-2317 [DOI] [PubMed] [Google Scholar]
- Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, et al. (2004). Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet 74:1043-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, et al. (2006). Expression of Wnt9b and activation of canonical Wntsignaling during midfacial morphogenesis in mice. Dev Dyn 235:1448-1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A, Menezes R, Granjeiro JM, Vieira AR. (2009). AXIN2 and CDH1 polymorphisms, tooth agenesis, and oral clefts. Birth Defects Res A Clin Mol Teratol 85:169-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Millar SE. (2010). Wnt/β-catenin signaling in oral tissue development and disease. J Dent Res 89:318-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KJ, Arron JR, Stankunas K, Crabtree GR, Longaker MT. (2007). Chemical rescue of cleft palate and midline defects in conditional GSK-3beta mice. Nature 446:79-82 [DOI] [PubMed] [Google Scholar]
- Mani P, Jarrell A, Myers J, Atit R. (2010). Visualizing canonical Wnt signaling during mouse craniofacial development. Dev Dyn 239:354-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes R, Marazita ML, Goldstein McHenry T, Cooper ME, Bardi K, Brandon C, et al. (2009). Axis inhibition protein 2, orofacial clefts and a family history of cancer. J Am Dent Assoc 140:80-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes R, Letra A, Kim AH, Küchler EC, Day A, Tannure PN, et al. (2010). Studies with Wnt genes and nonsyndromic cleft lip and palate. Birth Defects Res A Clin Mol Teratol 88:995-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R. (2010). Glycogen synthase kinase 3 beta: can it be a target for oral cancer. Mol Cancer 9:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JC. (2002). Gene/environment causes of cleft lip and/or palate. Clin Genet 61:248-256 [DOI] [PubMed] [Google Scholar]
- National Institute of Dental and Craniofacial Research (2010). Oral cancer. URL available at: http://www.nidcr.nih.gov/OralHealth/Topics/OralCancer/OralCancer.htm (accessed January 27, 2011).
- Nishi M, Miyake H, Takeda T, Hatae Y. (2000). Congenital malformations and childhood cancer. Med Pediatr Oncol 34:250-254 [DOI] [PubMed] [Google Scholar]
- Notaridou M, Quaye L, Dafou D, Jones C, Song H, Hogdall E, et al. (2011). Common alleles in candidate susceptibility genes associated with risk and development of epithelial ovarian cancer. Int J Cancer [Epub ahead of print, July 15, 2010] (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. (2007). PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 81:559-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama T, Lee HC, Koga H, Wands JR, Kim M. (2010). Noncanonical Wnt11 inhibits hepatocellular carcinoma cell proliferation and migration. Mol Cancer Res 8:254-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng RC, Lin RK, Wen CK, Tseng C, Hsu HS, Hsu WH, et al. (2008). Epigenetic silencing of AXIN2/betaTrCP and deregulation of p53-mediated control lead to wild-type β-catenin nuclear accumulation in lung tumorigenesis. Oncogene 27:4488-4496 [DOI] [PubMed] [Google Scholar]
- Zhu JL, Basso O, Hasle H, Winther JF, Olsen JH, Olsen J. (2002). Do parents of children with congenital malformations have a higher cancer risk? A nationwide study in Denmark. Br J Cancer 87:524-528 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


