Introduction
Major advancements have been made in the past decade towards understanding the regulatory role of small non-coding RNAs (ncRNAs) in diverse development processes1. An integral part of this regulation is the conserved PIWI/Argonaute protein family. Phylogenetic analysis indicates that this protein family can be divided into two sub-families: the Piwi sub-family, named after Drosophila melanogaster protein PIWI (P-element induced wimpy testis), and the Ago sub-family, based on its founding member in Arabidopsis thaliana (Ago). Ago proteins are involved in RNA-mediated gene silencing complexes and utilize short guide RNAs to target specific nucleic acids1-4, whereas Piwi proteins bind to a novel class of ncRNAs called PIWI-interacting RNAs (piRNAs) that play diverse functions in germline development and gametogenesis5-11.
Piwi/Ago proteins have a molecular weight of about 90 kDa and are defined by three protein motifs: the PAZ, MID and PIWI domains. The N-terminal PAZ domain is accountable for binding the 3′-end of the guide strand RNA; the middle MID domain is responsible for binding the 5′-phosphate RNA; and the following C-terminal PIWI domain has an RNase H endonuclease fold that is responsible for the RNA cleavage. The best-characterized small RNA partners of Ago proteins are microRNAs (miRNAs). Ago proteins utilize miRNAs to silence genes post-transcriptionally or use small-interfering RNAs (siRNAs) in both transcription and post-transcription silencing mechanisms12-17. However, Piwi proteins are restricted to germline cells and stem cells, and have been found to interact with a novel class of piRNAs (∼28-33 nucleotides) longer than miRNAs and siRNAs (∼20 nucleotides), suggesting novel functions distinct from those of Ago proteins18-22.
piRNAs are 28 to 33 nucleotides in length and have been characterized by the cloning of small RNAs from anti-Piwi immunoprecipitates prepared from mammalian testes7-9. As the proportion of repeat elements able to generate piRNAs is actually smaller within the piRNA regions than the frequency of repeat sequences in the mouse genome, they are not believed to be derived from repeat sequences. It is believed that piRNAs are processed from single-stranded primary transcripts that are transcribed from defined genomic regions and have a preference for a uridine at their 5′ terminal6-10,23. Mammalian piRNAs are a highly complex mix of sequences, with tens of thousands of distinct piRNAs generated from the 50 to 100 defined primary transcripts. This may suggest that mammalian piRNAs, unlike miRNAs, are not processed in a precise manner. However, approximately 20% of all piRNA sequences were cloned three or more times, and many piRNA sequences from the same strand are partially overlapping, suggesting a quasi-random mechanism. The mechanism of biogenesis of D. melanogaster rasiRNAs is beginning to be elucidated, and may offer parallels for a specific mode of processing for piRNAs as well. piRNA biogenesis is thought to be Dicer-independent and they appear to be 2′-O'methylated at their 3′ terminal24-26. Between mammals, mature piRNAs are not conserved; however, the genomic regions, from which they derive, in particular the promoter sequences, are conserved. Mammalian piRNAs are strongly expressed in the male germline, their total number per cell obtained from testis tissue reaching up to two million – about 10-fold higher than the miRNA content of these cells.
Despite their importance in developmental biology, a mechanistic understanding of the biogenesis of piRNAs is lacking, and the specific functional role PIWI proteins play in piRNA generation remains elusive. To understand the molecular basis of piRNA recognition by PIWI proteins, we have determined the three-dimensional solution structures of the human PIWI-like 1 (hPIWIL1) PAZ domain in its free form, as well as bound to a ssRNA (5′-pUGACA) oligonucleotide using nuclear magnetic resonance (NMR) spectroscopy methods.
Materials and Methods
Sample preparation
The PAZ domain of human PIWI-like 1 (residue 268-395) was cloned into a pET19b vector and expressed in Escherichia coli BL21(DE3) cells as an N-terminal His10-tagged protein. Uniformly 15N- and 15N/13C-labelled proteins were prepared by growing bacteria in M9 minimal medium with 15NH4Cl and/or 13C6-glucose as the sole nitrogen and carbon sources. Deuterated protein was generated by cell growth in 95% 2H2O. The PAZ domain was purified by nickel-NTA affinity chromatography. After cleavage of the His10-tag with TEV protease, the protein was further purified by ion-exchange and size-exclusion chromatography. Protein NMR samples (approximate 0.5mM) were prepared in 100mM phosphate buffer of pH 6.5, 150 mM NaCl and 5 mM dithiothreitol (DTT)-d10 in H2O/2H2O (9/1) or 2H2O. The complex protein sample was titrated with ssRNA (5′-pUGACA) oligonucleotide to 1:1.5 molar ratio.
NMR spectroscopy
The human PIWI-like 1 PAZ domains containing residues 268-395 was used for structure determination. All NMR spectra were acquired at 25°C on BRUKER 500, 600, and 800 MHz spectrometers equipped with z-gradient triple-resonance cryo-probe. The backbone 1H, 13C and 15N resonances were assigned using standard three-dimensional triple-resonance HNCA, HN(CO)CA, HN(CA)CB and HN(COCA)CB experiments27. The side-chain atoms were assigned from three-dimensional HCCH-TOCSY, HCCH-COSY and (H)C(CO)NH-TOCSY data28. The NOE derived distance restrains were obtained from 15N- or 13C-edited three-dimensional NOESY spectra. The ssRNA atoms were assigned from two-dimensional HTOCSY, HNOESY, HROESY and 13C/15N-filtered HTOCSY. The intermolecular NOEs used in defining the structure of the complex were detected in 13C-edited (F1), 13C/15N-filtered (F3) three-dimensional NOESY spectra29. Spectra were processed with nmrPipe and analyzed using NMRVIEW30-31.
Structure Calculations
Structures of the PAZ domain were calculated with a distance-geometry simulated annealing protocol with CNS. Initial protein structure calculations were performed with manually assigned NOE-derived distance constraints. Hydrogen bond distance, φ and ψ dihedral-angle restraints were from the TALOS prediction, and they were added at later stage of structure calculations for residues with characteristic NOE patterns32. The converged structures were used for the iterative automated NOE assignment by ARIA refinement. Structure quality was assessed with ARIA analysis. Total 43 intermolecular NOE-derived distance restraints were added in the structure determination of the PAZ domain-ssRNA complex. A family of 200 structures was generated and 20 structures with the lowest energies were selected for final analysis33-36.
Results and Discussion
The three-dimensional solution structures of the human PIWI-like 1 PAZ domain in the free form, and in the presence of an ssRNA (5′-pUGACA) oligonucleotide, were solved using NMR methods to a final RMSD at 0.33 Å and 0.25 Å for secondary backbone atoms, respectively (Table 1, Fig. 1a-1b). The structure of the hPIWIL1 PAZ domain (residues 266-399) is composed of a left-handed, six-stranded β-barrel core consisting of amino acid sequences that is conserved within the PAZ domain family12. The structure of hPIWIL1 PAZ domain is similar to the structures of other PAZ domains12, but attachments surrounded the β-barrel core show variations (Supplementary Fig. 1-2). For example, hPIWIL1 PAZ domain has one α-helix and β-hairpin in the protein appendage unlike two α-helices for PfAGO PAZ domain (Supplementary Fig. 2); hPIWIL1 PAZ has two α-helices that are the anti-parallel to each other, whereas gDicer PAZ domain has no N-terminal α-helix, and the dAGO1, dAGO2 and hAGO1 PAZ domains have two α-helices that are aligned nearly perpendicular in comparison to those in hPIWIL1 PAZ domain.
Table 1. Summary of Restraints and Statistics of the final 20 structures of human PIWI-like PAZ domain.
| Free Protein | RNA Complex | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance constraints | ||
| Total NOE | 2554 | 2497 |
| Intra-residue | 1039 | 1044 |
| Inter-residue | 1515 | 1453 |
| Sequential (|i - j| = 1) | 485 | 482 |
| Medium-range (|i - j| ≤ 5) | 379 | 354 |
| Long-range (|i - j| > 5) | 651 | 617 |
| Hydrogen bonds | 48 | 48 |
| Total dihedral angle restraints | ||
| Φ angle | 62 | 62 |
| Ψ angle | 62 | 62 |
| Ramachandran Map Analysis (%) | ||
| Most favored regions | 66 | 68.7 |
| Additional allowed regions | 32.6 | 27.5 |
| Generously allowed regions | 1.3 | 2.4 |
| Disallowed regions | 0.1 | 1.5 |
| Structure statistics | ||
| Violations (mean +/- s.d.) | ||
| Distance constraints (A) | 0.092 +/- 0.012 | 0.087 +/- 0.0095 |
| Dihedral angle constraints (°) | 1.39 +/- 0.072 | 0.95 +/- 0.101 |
| Max. dihedral angle violation (°) | 1.49 | 1.2 |
| Max. distance constraint violation (A) | 0.13 | 0.11 |
| Deviations from idealized geometry | ||
| Bond lengths (A) | 0.0075 +/- 0.00021 | 0.0063 +/- 0.000104 |
| Bond angles (°) | 0.84 +/- 0.013 | 0.76 +/- 0.018 |
| Impropers (°) | 0.71 +/- 0.025 | 0.67 +/- 0.024 |
| Average pairwise r.m.s. Deviation ** (A) | ||
| Heavy | 0.61 +/- 0.065 | 0.66 +/- 0.095 |
| Backbone | 0.24 +/- 0.057 | 0.37 +/- 0.105 |
The residue number ranges used for RMSD calculations are 278-364 and 380-389. Pairwise r.m.s. deviation was calculated among 20 refined structures.
Figure 1.
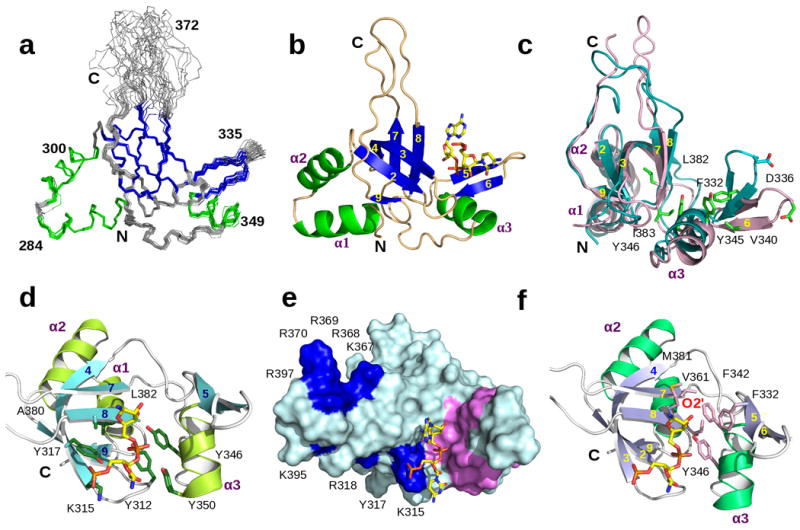
Three-dimensional structures of the human PIWI-like PAZ domain. (a) The ensemble of 20 lowest-energy NMR structures of the PAZ domain (residues 275-393). The β-strands are indicated in blue and α-helices in green. The N-terminal (268-274) and C-terminal (394-398) unstructured residues are omitted for clarity. (b) Ribbon representation of a NMR structure of PAZ in ssRNA complex. The orientation is similar to (a). The β-strands are indicated in blue and α-helices are shown in green. (c) Ribbon depiction of superimposed PAZ-RNA structures (free protein vs. complex). The free protein is in blue and the complex in brown. (d) The human PIWI-like 1 PAZ domain recognizes the 2nt 3′–OH end of the single-stranded RNA and buries the two 3′-terminal nucleotides in the hydrophobic cleft. Side-chains of amino acids are colored in green and labeled. (e) Surface representation of the human PIWI-like PAZ domain bound to a 5′ phosphorylated ssRNA (5′-pUGACA; PAZ:ligand molar ratio 1:1.5). The blue surface shows positively charged residues on the surface β7-8 loop and the C-terminal of colors indicating a possible alignment of RNA on the protein surface. The surface highlighted in red is for residues that show further perturbation upon adding methylated ssRNA compared to unmodified ssRNA. (f) Ribbon representation of the view of the PAZ domain binding-pocket. Side-chains of amino acids in red and labeled are involved in further structures changes upon adding methylated ssRNA compared to unmodified ssRNA.
Other differences lie in loop regions. For instance, hPIWIL1 and gDicer PAZ domains have a long extended loop between β7 and β8 (Supplementary Fig. 2), which contains positively charged residues that are not conserved, while AGO PAZ domains have short loops. The cleft-like structure formed between the β–barrel core and a conserved α-β appendage is the nucleic acid-binding surface of the protein (Fig. 1b). The 3′-end of the RNA is inserted into the central pocket formed between the β-barrel and the α-β appendage and sandwiched by β8 strand and α3 helix. The 3′-end of the RNA in bound is a partial hydrophobic cage formed by a positively charged Lys315, hydrophobic Ala380, Leu382 and aromatic Tyr312, Tyr317, Tyr346 and Tyr350 (Fig. 1d). The hydrophobic protein-nucleic-acid interactions were observed by NMR. Intermolecular tertiary interactions are formed from Lys315, Ala380 and Leu382 to the backbone of C4 and A5. The non-bridge oxygens of the phosphodiester backbone linking A3 and C4 point toward to side-chain Hζ of Lys315 and Hη of Tyr317, whereas phosphodiester oxygens linking C4 and A5 interface with Hη atoms of Tyr346, Tyr350 and Tyr312.
A comparison of the structures of the RNA-bound and free-form hPIWIL1 PAZ domains (Fig. 1c) show that the main structures are similar, indicating no major changes of the overall protein conformation upon RNA binding. The backbone atoms of secondary residues can be superimposed with an RMSD of 1.04 Å. However, the β–hairpin module of the bound protein is pushed away from the β–barrel core by 4.5 Å at its tip (Cα of Asp336). The local structure of the β–hairpin is preserved and only β5 and β6 strands open up allowing the binding 3′-end of the RNA in the core. In the unbound structure, NOEs were observed in the NMR spectra between Phe332 and Tyr345; Phe332 and Val340; Tyr345 and Val340 (Fig. 1c). But when the protein is bounded to the RNA, there is no long-range NOE from Phe332 to this group of residues. Phe332 also has no observed intermolecular interactions to the RNA. This is contrast to the opposed conserved residues Phe72 in AGO2 PAZ and Phe369 in AGO1 PAZ complex structures, which showed strong interactions to the RNA12-13.
Although there is no overall conformational change in α3-helix upon RNA binding, the inserting 3′-end A5 perturbs local tertiary interactions between the β7-β8 sheet and α3-appendage, resulting in loss of long-range NOEs between Tyr345and Leu382; Tyr346 and Leu382; Tyr346 and Ile383. The structure of hPIWIL1 PAZ domain shows that the 3′-OH end of small RNA is satisfactorily accommodated in the pocket. The pull-away of the β–hairpin and the disappearance of long-range tertiary interactions from conserved residues on α-β appendage could be explained by A5 base edge of the RNA, which is larger in size than those in dAGO1, dAGO2 and hAGO1 complexes. The A5 base edge in hPIWIL1 PAZ complex is facing outward towards solvent and has no contact to the protein, but its large aromatic ring forces the binding pocket to open a wider space, which results in further distance from α-β appendage to β7-β8 interface and causes the loss of interactions.
The structure of hPIWIL1 PAZ bound to RNA reveals that two 3′-terminal nucleotides insert into the pocket sandwiched by β-barrel and α-β appendage, which is similar to those of dAGO1, dAGO2 and hAGO1 PAZ domains, but lacks of interactions between other nucleotides to the protein. However, how the RNA strand is aligned on the protein surface is not fully understood (Supplementary Fig. 2). The crystal structure of hAGO1 PAZ domain bound to siRNA-like duplex shows that the PAZ domain secures the RNA binding through hydrogen-bond and electrostatic interactions between five phosphodiester groups of the strand bound at the 3′-terminal, and the positively charged Arg275, Lys333, Arg278, Lys264 and C-terminal residues. The RNA strand aligns horizontally along the surface of β2, β3, β7 and β8 strands. These residues are conserved among PAZ domains of dAGO1 and dAGO2, so RNAs should have the similar β2-3-7-8 alignment on both PAZ domains to dAGO1 and dAGO2 PAZ. However, hPIWIL1 and gDicer PAZ domains only have one conserved Arg on the top of β7 sheet and do not have positive residues on β2, β3 and β8 strands that are conserved among dAGO1, dAGO2 and hAGO1 PAZ domains. This implies that bound RNAs do not align precisely the same way among PAZ domains even though the general composition of side-chains in the binding pocket is maintained and the interaction occurs across the same PAZ binding cleft.
hPIWIL1 PAZ domain has basic residues Lys and Arg on its extended β7-8 loop and C-terminal region (Fig. 1e). The distance between these amino acid residues matches the distance between phosphodiester groups of the RNA. The positively charged residues on both the extended loop and C-terminus likely form the hydrogen bonds and electrostatic interactions to the phosphodiester groups of RNA, thus likely playing an important role in aligning the RNA strand vertically along the β7-8 loop and C-terminal surface. The extended loop between β7-β8 also shows significant sequence variances from other PAZ domains. Its positive charged Arg and Lys residues are not conserved in other PAZ domains. This extra loop changes the electrostatic potential and molecular surface surrounding the 3′-OH terminal RNA-binding pocket relative to the AGO PAZ domains. It could substantially affect the RNA recognition.
piRNA is known to be 2′-O'methylated at the 3′ end. However, whether and how this methyl group contributes to the PAZ domain binding has remained elusive. To address this question, we performed 2D 1H-15N-HSQC NMR binding of hPIWIL1 PAZ domain with ssRNA (5′-pUGACA-3′) and 2′-O'methylated ssRNA (5′-pUGACA-me-3′) (see Supplementary Figure 3). We noticed that the protein residues that exhibited major differences in chemical shift changes upon binding to the methylated ssRNA versus to the unmodified ssRNA are mainly located in β8 and α3 (Fig. 1e-1f). This observation confirms that 2′-O'-methylation at 3′ end indeed affects ssRNA (5′-pUGACA) binding to the protein. As shown in the ribbon structure of hPIWIL1 PAZ domain (Fig. 1f), the 3′-methylation group is located in the position sandwiched between β8 and α3, and the HO2′ is surrounded (within 6 Å distance) by hydrophobic residues Ala380, Met381 and Leu382 on β8, Phe342 and Tyr346 on α3, Val361 on β7. Since these residues are conserved among all PIWI PAZ domains, the hydrophobic pocket created by these residues is likley conserved as well for the methyl group binding.
In summary, the human PIWI-like 1 PAZ domain is a typical OB fold and specifically recognizes the 2nt 3′–OH end of the single-stranded RNA and buries the two 3′-terminal nucleotides in the hydrophobic cleft. Its binding pocket is similar to those of Ago PAZ domains and the binding residues are conserved, however, the pocket accommodates an extra cavity surrounded by four hydrophobic residues that is available for the 2′-O methyl group binding. hPIWIL-PAZ also differs from other PAZ domains in its lack of key positively charged residues along the second and third β-strands that, in Ago PAZ, forms hydrogen bonds with RNA. Instead, hPIWIL1 PAZ domain has a long extended β7 and β8 loops consisting of positively charged residues which implies that hPIWIL1 PAZ may orientate RNA on the protein surface differently from Ago PAZ domains even though it shares the 2nt 3′–OH RNA binding motif.
Supplementary Material
Acknowledgments
We wish to acknowledge the use of the NMR facility at the New York Structural Biology Center. We thank Zach Charlop-Powers for comments. This work was supported by the grants from the National Institutes of Health (M.-M.Z.). Resonance assignments and 3D NOESY peak lists for corresponding NMR structures have been deposited at the BioMagResBank as 17272 and 17273, the structural coordinates have been deposited in the Protein Data Bank (2L5C, 2L5D).
References
- 1.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipowicz W. The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Hutvagner G, Simard Mj. Argonaute proteins: Key players in RNA silencing. Nature Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 4.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nature Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 5.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel of evolutionary conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12(23):3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox DN, Chao A, Lin H. Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline setm cells. Development. 2000;127(3):503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 7.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Inovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 8.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 9.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20(13):1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imani H. Idetification and characterization of two novel classes of small RNAs in the mouse germline: restrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20(13):1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the Paz domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 13.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2004;426(6965):465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 14.Ma JB, Ye K, Patel DJ. Structural Basis for over-hang-specific small interfering RNA recognition by the Paz domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 Paz domain. Nature Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 16.Song JJ, Smith SK, Hannon GJ, Jishua-Tor L. Crystal Structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 17.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 Paz domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 18.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Seto AG, Kingston RE, Lau NC. The coming age for Piwi proteins. Mol Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 20.Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- 21.O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin H, Thomson T. The biogenesis and function PIWI proteins and piRNAs: progress and prospect. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 24.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, Van Den Elst H, Filippov DV, Blaser H, Moens CB, et al. A role for PIWI and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 25.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. Adistinct small RNA pathway silences selfish genetic elements in the germline. Sience. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 26.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 27.Grzesiek S, Bax A. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 28.Bax A, Clore GM, Driscoll PC, Gronenborn AM, Ikura M, Kay LE. J Magn Reson. 1990;87:620–627. [Google Scholar]
- 29.Lee W, Revington MJ, Arrowsmith C, Kay LE. A pulse filed gradient isotope-filtered 3D 13C-HMQC-NOESY experiment for extracting intermolecular NOE contacts in moldecular complexes. FEBS Letters. 1994;350:87–90. doi: 10.1016/0014-5793(94)00740-3. [DOI] [PubMed] [Google Scholar]
- 30.Johnson BA, Blevins RA. Journal of Biomolecular NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F, Grzesjek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 33.Brunger AT. X-PLOR Version 3.1: A system for X-Ray crystallography and NMR, version 3.1 ed. Yale University Press; New Haven, CT: 1993. [Google Scholar]
- 34.Rieping W, et al. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–92. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, et al. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


