Abstract
MicroRNAs (miRNAs) are key regulators of gene expression that regulate important oncogenes and tumor suppressors. Many miRNAs act as oncogenes or tumor suppressors, and the altered misexpression of miRNAs is a hallmark of many cancer types. Dysregulated miRNAs are a potentially powerful new tool that could be used to enable the characterization of tumor environments and identify novel and important oncogenic pathways. More recently, there has been growing interest in the field of miRNAs as biomarkers of cancer risk, diagnosis and response to therapy. Understanding the associations between miRNA expression and cancer phenotypes, and the potential of miRNA profiling in clinical applications, promises to be highly rewarding in the field of cancer research.
Keywords: microRNA, profiling, cancer, biomarker, diagnosis, prognosis
MicroRNAs in cancer
The last decade has witnessed an explosion of research into small non-coding RNAs. From viruses, to plants, to humans, these ~21 nucleotide RNA regulators of gene expression have been demonstrated to be involved in every biological process examined. The largest family of these noncoding RNAs, microRNAs (miRNAs), constitutes greater than 1% of the human genome, and is expected to regulate up to a third of all genes. MiRNAs are typically excised from a hairpin precursor RNA, originally part of a larger primary transcript, and function by base-pairing with target mRNAs to prevent protein translation either through mRNA repression or degradation (for an extensive review on miRNA biogenesis and function, see ref.[1]).
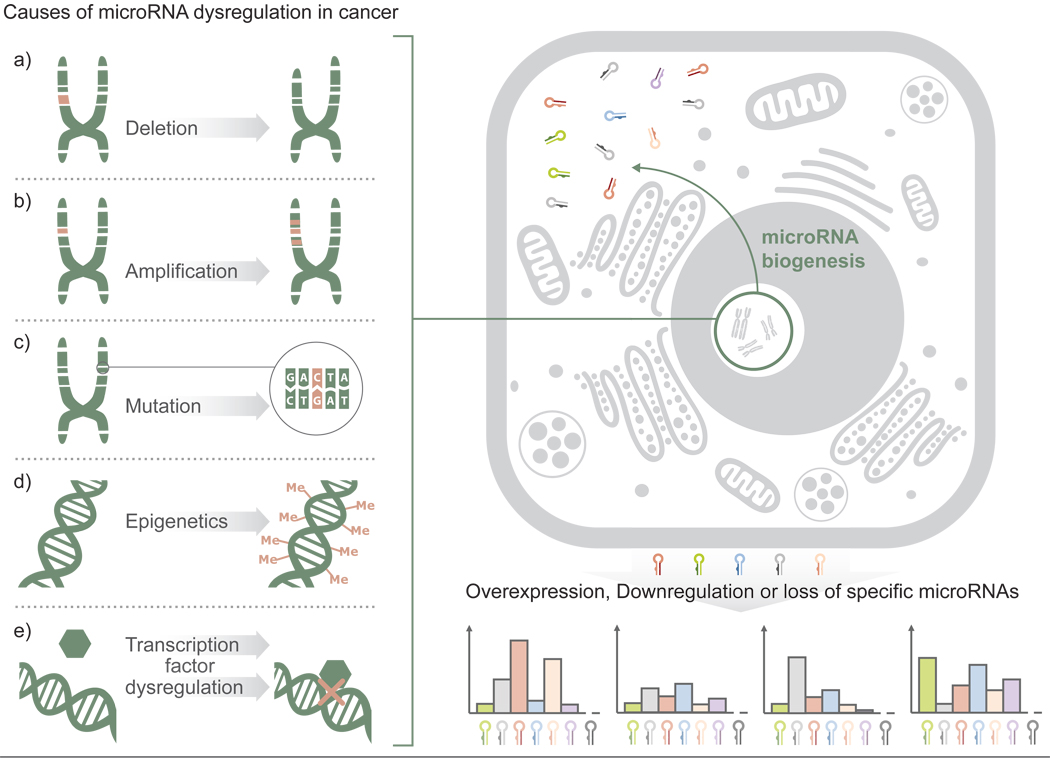
The role of miRNAs in many different types of human cancer has been studied. Cancer is defined by abnormal and uncontrolled cell division, a phenotype that arises from the mis-regulation of any number of genes. miRNAs are major regulators of gene expression, with roles in nearly every area of cell behavior, development and survival; therefore it is not surprising that miRNAs are actively altered in all types of cancers [2] (Fig. 1). Aberrant expression of miRNAs can arise from the deletion or mutation, as well as methylation, of miRNA encoding genes [3–5]. Many miRNAs are located at fragile sites or regions in the genome that are frequently deleted or amplified in cancer [6]. A landmark study from the Croce group found that a frequently deleted region, chromosome 13q14 in chronic lymphocytic leukemia (CLL), harbored two miRNAs, hsa-miR-15a and hsa-miR-16-1 [7]. These miRNAs represented the first tumor suppressor miRNAs to be discovered and spurred the search for other miRNAs with functions in oncogenesis [8]. Additional studies have shown that miRNA expression is commonly altered in cancer cells compared to normal adjacent cells [9, 10], revealing vast potential in the study of miRNAs as a means to further understand the changes that drive cancer phenotypes.
Figure 1. Causes of microRNA dysregulation in cancer.
Expression changes for any number of miRNA genes in cancer can stem from cancer pathogenesis; however, such changes can also serve as a crucial contributors to malignant transformation. Such alterations can result from various mechanisms including the deletion (a) or amplification of miRNA-coding chromosomal regions (b), mutations within the miRNA or the target site sequence of the respective gene (c), epigenetic silencing of miRNA promoters (d) or the dysregulation of proteins upstream of the RNAi pathway such as cellular signaling and transcription factors. Each of these mechanisms results either in overexpression, downregulation or loss of function of specific miRNAs. Dysregulation of these regulatory units, whether the affected miRNAs act as oncogenes or tumour suppressor genes, can drastically alter cell signaling pathways, metabolism and cell cycle control.
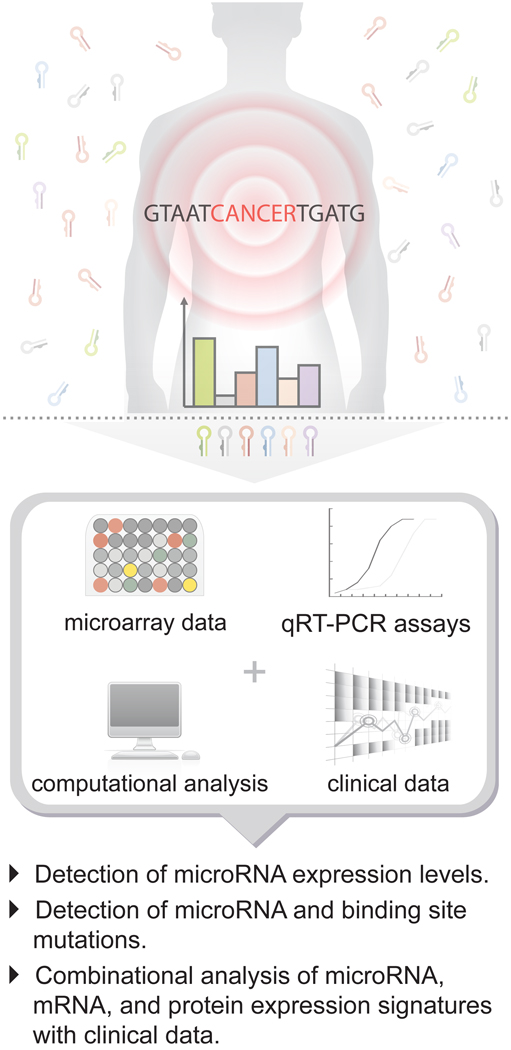
Broad scale expression profiling has proven useful in the initial identification of miRNAs with potential importance for further investigation (Fig. 2). The development of miRNA microarrays [11], high-throughput deep sequencing [12, 13], and bead-based flow cytometric miRNA analysis methods [9] has greatly facilitated this approach. Many of the now recognized mis-regulated miRNAs in cancer were first highlighted with similar techniques. Overall, the combination of miRNA expression profiles and mRNA expression profiles, coupled with knowledge of transcription factor expression patterns, forms a robust partnership that can clarify some of the complex networks occurring in cancer cells [14, 15]. However, in the growing field of genetic diagnostics, the use of miRNA signatures for identification purposes is increasingly being preferred over the traditional use of mRNA profiles. MiRNA profiles have been shown to perform better than mRNA profiles, particularly when isolated from samples that are traditionally difficult to process [16]. This can largely be attributed to their increased stability [17]. Refined techniques for studying miRNAs have also helped to improve their handling and manipulation, and coupled with their robust expression and lack of transcript variants (e.g. mRNA splicing variants and isoforms) gives miRNAs greater reliability. Furthermore, discerning miRNA profiles have the potential to offer greater insight into important gene regulatory networks given that they directly influence mRNA expression. Finally, miRNA misexpression patterns, when directly compared to mRNA misexpression patterns, are better able to identify the origin of tumors of unknown primary [sc1][9], suggesting that tumors more clearly maintain a unique “tissue miRNA expression profile”.
Figure 2. Detection and analysis of cancer-specific microRNA expression patterns.
Microarrays and quantitative RT-PCR techniques have been instrumental in the identification of unique miRNA expression signatures as well as mutations that affect the function of miRNAs that correlate with cancer. Combination approaches involving miRNA, mRNA, protein and clinical diagnosis have enabled more thorough characterization of many cancer regulatory networks.
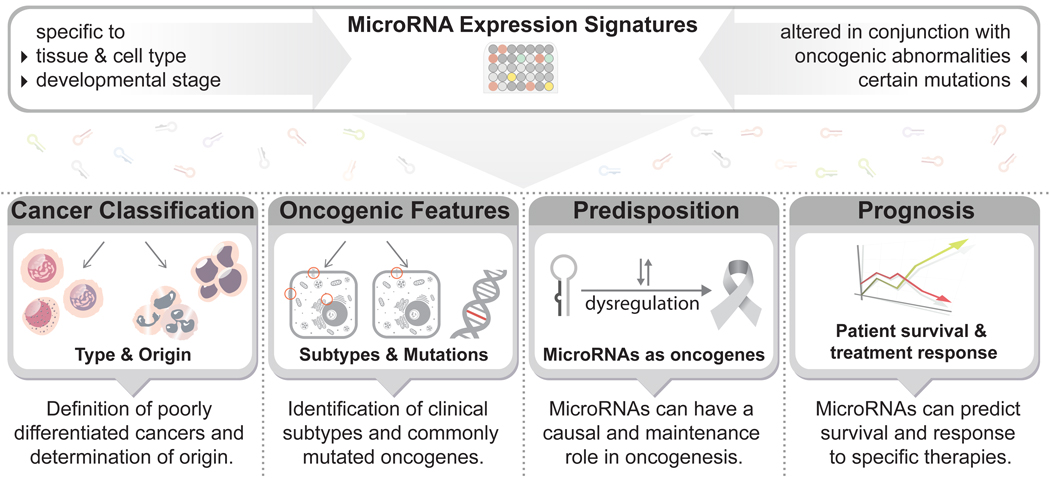
MiRNA profiles can distinguish between similar tumor environments and can provide insights into the type of cancer, associated mutations and even response to therapy (Fig. 3). In this review, we explore the associations that have been found between miRNA expression and cancer phenotypes and discuss the potential use of miRNA profiling in clinical applications.
Figure 3. MicroRNA-based diagnosis and prognosis in cancer.
The expression signature of many miRNAs is specific to cell type and function, and also correlates with common oncogenic features. This specificity is useful as a diagnostic tool in the discrimination of tumor origins, subtypes, and oncogenic mutations. Individual miRNAs can also act as markers of cancer susceptibility; specifically, mutations in miRNAs and their binding sites are commonly associated with increased cancer risk. Furthermore, miRNA-associated mutations and aberrant expression can predict cancer prognosis and response to therapy. These features make miRNAs an appealing tool for clinical use.
Cancer classification by microRNA profiling
Several large scale tumor miRNA profiling studies have shown that miRNA signatures are significantly different between cancerous and matched non-cancerous tissue [9, 10, 18, 19]. More importantly, miRNAs are often tissue-specific [20, 21] and developmental- or differentiation-stage specific [22], and hence miRNA profiles can also help to define poorly differentiated cancers and determine their origin. Indeed, one study developed a classifier of 48 miRNAs from a sample of 336 primary and metastatic tumors, and was able to use this classifier to accurately predict tissue origin in 86% of an independent blind test set, including 77% of the metastatic tumors [21]. Similarly, 12 out of 17 histologically undefined tumors were correctly diagnosed through the use of a miRNA-based classifier generated from the miRNA profiles of 68 tumors [9]. By contrast, mRNA based classifiers were only able to identify the tissue of origin in 1 out of these 17 samples [9, 23]. This clearly demonstrates that miRNAs have greater diagnostic value in identifying poorly differentiated tumors.
More recently, the marriage of experimental data with computational studies has enabled the creation of a novel classification technique called SFSSClass (Simultaneous Feature (miRNA) and Sample (tissue) Selection) [24]. The authors developed this classification technique by building on a previous study where they developed a cancer-miRNA network by mining the literature of experimentally verified cancer-miRNA relationships, and identifing ‘modules’ of commonly dysregulated miRNAs in different cancers [25]. This cancer-miRNA network enabled the identification of ~100 tissue specific miRNAs. SFSSClass was subsequently generated by the integration of a biclustering technique for simultaneous feature and sample selection (SFSS) with the cancer-miRNA network and an algorithm-based classifier. The authors then applied this integrated approach to 17 poorly differentiated tumor samples and reported that the combined analysis offers improved prediction accuracy in comparison to studies using only miRNA expression data [9, 26]. This is an example of the first attempts to develop a system that amalgamates the latest miRNA-cancer research findings and integrates them directly into a diagnostic tool. Given that cancers of undefined origin account for ~4% of all malignancies and are associated with poor prognosis [27, 28], the continued development of miRNA classifiers has foreseeable benefits in aiding clinical diagnosis and subsequent treatment.
MicroRNAs define oncogenic features
Given that miRNA profiles distinguish between normal and cancerous tissue and identify tissues of origin, it becomes important to ask if miRNA profiles can also distinguish between the subtypes of a particular cancer, or even specific oncogenic abnormalities. Gene expression profiling has already demonstrated its effectiveness at subtyping various cancers and offers insights into the biological pathways at work beyond traditional histopathological analysis of single markers. For example, breast cancer is typically classified into one of 4 subtypes; basal-like, luminal A, luminal B and HER2 positive, and gene expression signatures suggest that each subtype arises from separate cell types which are thought to be molecularly distinct with variable morphological features [29, 30]. miRNA profiles are equally discriminatory and can be far more informative as changes in their expression can provide insights into the myriad of gene permutations observed in various cancer subtypes. Indeed, many miRNAs have been shown to associate with various cancer subtypes, or to correlate with the presence or absence of specific oncogenic mutations (Table 1). Links have also been made between mis-regulated miRNAs and the target genes that are affected, thus unraveling some of the unique gene networks involved [31].
Table 1.
Cancer subtypes that can be distinguished by microRNA profiles.
| Cancer type | MicroRNAsa | Ref. |
|---|---|---|
| Breast | ||
| ER status | miR-26a/b, miR-30 family, miR-29b, miR-155, miR-342, miR-206, miR-191 |
[38–40, 42] |
| PR status | let-7c, miR-29b, miR-26a, miR-30 family, miR-520g |
[41, 42] |
| Her2/neu status | miR-520d, miR-181c, miR-302c, miR-376b, miR-30e |
[38, 41] |
| Lung | ||
| squamous vs non-squamous cell | miR-205 | [33] |
| small cell vs non-small cell | miR-17-5p, miR-22, miR-24, miR-31 |
[32] |
| Gastric | ||
| diffuse vs intestinal | miR-29b/c, miR-30 family, miR-135a/b |
[35] |
| Endometrial | ||
| endometrioid vs uterine papillary | miR-19a/b, miR-30e-5p, miR-101, miR-452, miR-382, miR-15a, miR-29c |
[37] |
| Renal | ||
| clear cell vs papillary | miR-424, miR-203, miR-31, miR-126 |
[34, 36] |
| oncocytoma vs chromophobe | miR-200c, miR-139-5p | [36] |
| Myeloma | ||
| with t(14;16) | miR-1, miR-133a | [60] |
| with t(4;14) | miR-203, miR-155, miR-375 | [60] |
| with t(11;14) | miR-125a, miR-650, miR-184 | [60] |
| Acute myeloid leukemia | ||
| with t(15;17) | miR-382, miR-134, miR-376a, miR-127, miR-299-5p, miR-323 |
[52] |
| with t(8;21) orf inv(16) | let-7b/c, miR-127 | [52] |
| with NPM1b mutations | miR-10a/b, let-7, miR-29, miR-204, miR-128a, miR-196a/b |
[51, 52] |
| with FLT3 ITD | miR-155 | [51, 52, 54] |
| Chronic lymphocytic leukemia | ||
| ZAP-70 levels and IgVH status | miR-15a, miR-195, miR-221, miR-155, miR-23b |
[50] |
| Melanoma | ||
| with BRAF V600E | miR-193a, miR-338, miR-565 | [56] |
Not all distinguishing microRNAs are represented in this table
nucleophosmin1
Distinguishing clinical subtypes
Many groups have characterized miRNA signatures that associate with cancer subtypes [32–37], and may even infer mechanisms specific to individual subtypes. MiRNA profiling studies have identified a number of miRNAs that are differentially expressed between basal and luminal breast cancer subtypes [38, 39], and can specifically classify estrogen receptor (ER), progesterone receptor (PR) and HER2/neu receptor status [38, 40–42]. Some of the miRNAs that associate with the luminal or basal subtype reflect their epithelial and myoepithelial origins, respectively. For instance, miR-200 associates with the luminal subtype; miR-200 family members regulate the epithelial phenotype by inhibiting zinc finger E-box binding homeobox 1 (ZEB1) and ZEB2, genes that promote epithelial to mesenchymal transition (EMT) [43, 44]. Low miR-200 expression associates with tumor progression [45], likely as a result of failure to regulate EMT, a process that can drive the transformation of tumor cells into metastatic cells [43]. Lack of miR-200 in basal-type breast cancer might provide one explanation for the increased metastatic potential observed in this subtype. Additionally, the miR-200 family also plays a role in negatively regulating epidermal growth factor (EGF)-driven breast cancer cell invasion; specifically, individual members can differentially arrest cells at G1 or G2/M stages of the cell cycle [46]. EGF receptor (EGFR) is also frequently over-expressed in basal-type breast cancer, and thus compounding the effect of low miR-200.
Interestingly, miR-145 is preferentially expressed in normal myoepithelial cells, but there is a dramatic reduction observed in basal-like triple negative tumors (ER−/PR−/HER2−); this expression change might be a consequence of disease progression in this subtype [39]. MiR-145 is a well-characterized tumor suppressor [47]. It has a pro-apoptotic effect when ectopically expressed in breast cancer cell lines [48], and can also suppress cell invasion and metastasis by silencing the metastasis gene, mucin 1 [49]. Decreased miR-145 expression is apparent in hyperplastic ducts and appears to signal compromised myoepithelial architecture. This finding suggests that loss of miR-145 could be a marker of early onset basal-type breast cancer [39].
The ability of a single miRNA to indicate a breast cancer subtype, and furthermore to predict tumor biology, in contrast to the requirement of hundreds of mRNAs to do the same, again indicates the greater predictive power of miRNAs compared to mRNAs.
Distinguishing mutational status
miRNAs have been shown to be effective in predicting the mutational status of several commonly mutated genes involved in oncogenesis. The first such association came from the Croce group, who discovered that 13 miRNAs distinguished zeta-chain associated protein kinase 70kDa (ZAP-70) protein expression and IgVH mutation status in CLL and were highly accurate as a predictive marker in an independent test set [50]. Likewise in acute myeloid leukemia, there is an association between miR-155 expression and FLT3-ITD (internal tandem dupliacations); however, miR-155 expression appears to be independent of FLT3 signaling [51–54]. Proteins involved in the mitogen-activated protein kinase (MAPK) signaling pathway are among the most commonly altered in a variety of cancers, particularly BRAF and members of the RAS family [55]. MiR-193a, a known tumor-suppressor is down-regulated in both melanoma [56] and thyroid cancers [57] which harbor the BRAF V600E oncogenic amino acid subsitution, and is a strong discriminator for this in melanomas [56]. Some predicted targets of miR-193a are linked to the MAPK pathway [56], though there has been no direct target validation studies to support this. These examples suggest a connection in the regulation of the miRNA either directly by the proteins themselves or their downstream effectors, and further studies will be needed to clarify these correlations. In contrast to the activating mutations detailed above, similar studies involving loss of function mutations, such as profiling of miRNAs in high grade serous ovarian carcinomas with breast cancer 1, early onset (BRCA1) and/or BRCA2 mutations [58], indicated no significant expression changes.
Subtyping studies have also revealed associations of miRNAs with specific cytogenic features, particularly in hematological malignancies where complex karyotypes involving various chromosomal deletions and translocations characterize the majority of subtypes. In CLL, miRNAs discriminate the 11q deletion, 17p deletion, trisomy 12, 13q deletion, and normal karyotype subgroups [59]. Retinoblastoma (RB) deletion and translocations in multiple myeloma (MM) linked to miRNA expression, such as t(4;14), t(14;16), and t(11;14) have also been recently reported [60]. A number of these links can be attributed to the location of the miRNAs at or near these aberrant translocations. For example, the miRNAs that are under-expressed in correlation with retinoblastoma (RB) deletion in MM are located on chromosome 13, and likewise, monosomy 13 is also a feature of RB deletion, thus providing an explanation for the loss of these miRNAs [60]. Similarly, miR-125b-1, which is over-expressed in acute lymphoblastic leukemia (ALL) patients with t(11;14), is located in the breakpoint vicinity of this translocation, and has been shown to be relocated as a result [61]. However, in the majority of observed miRNA-translocation relationships, the respective locations do not match [52, 60]. Therefore, it is tempting to speculate that chromosomal rearrangements affect miRNA expression as a result of the translocation of miRNA regulatory elements. Indeed, in Burkitt lymphoma (BL), c-myc is frequently relocated from chromosome 8 to chromosome 14, where it falls under the control of the immunoglobulin enhancer locus that subsequently alters c-Myc expression [62]. Not surprisingly, c-Myc regulates many of the miRNAs that are associated with BL [63]; however, few experiments have documented similar findings for other cytogenic abnormalities.
MicroRNAs as cancer predisposing genes
Most studies have identified miRNAs that are down-regulated in cancers, in comparison to normal tissue [9, 18, 64–66]. Such trends can be a result of disruptions to components of the miRNA processing machinery [67], rather than individual miRNAs themselves, but nevertheless hints at innate tumor suppressor functions for miRNAs as a whole. This was effectively demonstrated in the Dicer knockout mouse: the resultant global knockdown of miRNAs in the mouse and in mouse embryonic fibroblasts (MEFs) enhanced tumorigenesis and transformation, respectively [68]. These findings also suggest that the expression of at least some miRNAs has a causal role in cancer. Indeed, one study has shown a correlation between the chromosomal location of miRNAs and cancer susceptibility loci that influence tumor development in mouse models [69].
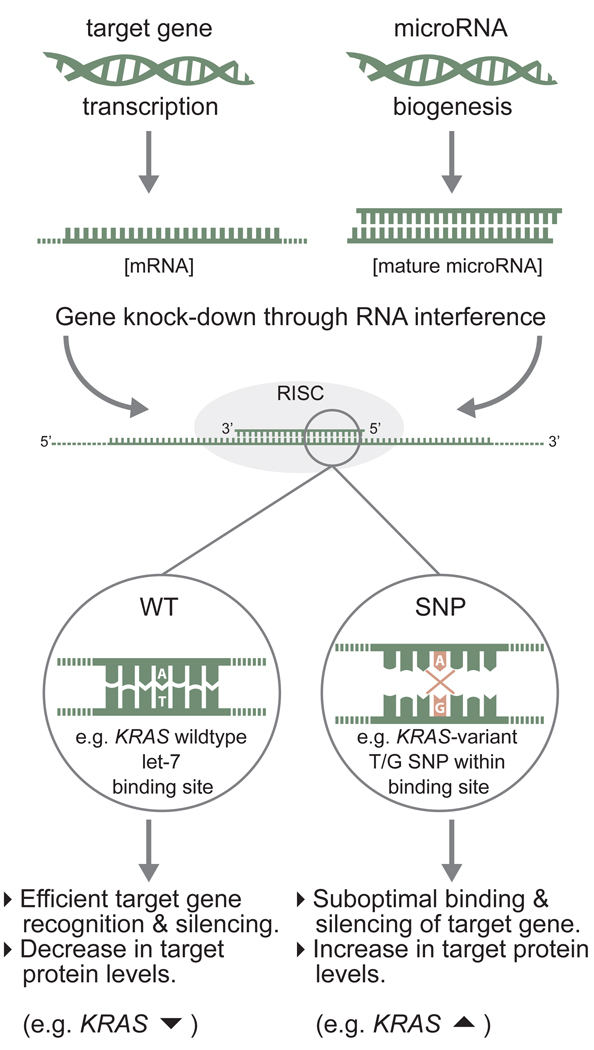
The early discovery of a germline mutation in the hsa-miR-16-1/hsa-miR-15a primary transcript (that associates with CLL) provides additional support for the role of miRNAs as cancer predisposing genes [50, 70]. Morevoer, a germline mutation in hsa-miR-125a has been reported to associate with breast cancer tumorigenesis [71]. Subsequently, a host of variants in the precursor or primary transcript of miRNAs that alter secondary structure, and consequently mature miRNA expression, have been shown to be associated with various cancer types, including familial breast and ovarian cancer lacking BRCA1/2 mutations [72–75], papillary thyroid carcinoma [76], gastric cancer [77], lung cancer [78, 79], prostate cancer [80], head and neck squamous cell carcinoma [81], and bladder cancer [82]. Similarly, mutations in the miRNA binding sites of target genes linked to cancer have been extensively reported (for reviews see [83, 84]). Many of these significantly influence cancer susceptibility [85], and one such hereditary mutation, in a putative let-7 binding site in the 3’UTR of the KRAS oncogene, significantly increases lung and ovarian cancer risk [86, 87]. This increased risk can, in part, be attributed to the modulation of mRNA regulation as a result of altered miRNA–target binding sites affinities (Fig. 4). Identified hereditary miRNA-associated variants greatly outnumber similarly inherited mRNA/protein coding variants that are associated with increased cancer risk.[sc2]
Figure 4. Single nucleotide polymorphisms (SNPs) within microRNA binding sites can lead to inefficient gene silencing.
MicroRNAs regulate gene expression by binding to regions containing sufficient sequence complementary in their target mRNAs, often with internal loops (not depicted). A single mismatch in base-pairing, particularly at the 5’ seed region of the miRNA, can be sufficient to reduce the binding affinity between miRNA and its target mRNA and can lead to inefficient downregulation, and subsequently, an increase in protein expression.
Perhaps the most convincing evidence comes from a recent study which showed that the overexpression of a single miRNA was sufficient to give rise to pre-B-cell lymphoma in mouse models [88]. Conditional expression of miR-21 in vivo gave rise to extreme lymphadenopathy and various other clinical signs of haematological malignancies. Furthermore, the tumors were highly dependent on miR-21 and showed rapid regression with the loss of miR-21 expression, a process referred to as ‘oncomiR addiction’. This study effectively demonstrates the role of miRNAs not only in the initiation of oncogenesis, but also in the maintenance of the oncogenic phenotype.
MicroRNAs and prognosis
Given that miRNAs are individually able to discriminate tumor origins, subtypes, oncogenic mutations, and cancer predisposition, it is logical to hypothesize that miRNAs might be able to predict cancer prognosis. A number of groups in recent years have addressed this issue simultaneously with general miRNA profiling, and have reported success in utilizing miRNAs as prognostic markers to predict outcome. For example, in gastric cancer a robust 7-miRNA signature can predict overall survival and relapse-free survival [89]. Similarly, low miR-191 and high miR-193a levels were associated with a significantly shorter survival time as measured by Kaplan-Meier curves in melanomas [56].
Although the prediction of survival might be important in a more general sense, the prediction of response to specific therapies is of far greater clinical value. For example, low miR-26 expression is an independent predictor of poor survival in patients with hepatocellular carcinoma (HCC); more importantly, however, patients with low miR-26 responded well to interferon alpha treatment with subsequent improved survival [90]. Thus, miR-26 expression might be a useful marker in selecting for patients that could benefit most from interferon alpha therapy. A number of miRNAs have also been correlated with a negative effect on patients’ response to specific treatments. In various cancers, increased miR-21 expression is an indictor of poor outcome [91–93], and is also sufficient to predict poor response to adjuvant chemotherapy in adenocarcinomas [93]. High levels of miR-125b in breast cancer predict poor response to Taxol-based treatments in vitro [94], and a similar finding has been reported for miR-21 in pancreatic cancer patients treated with Gemcitabine [95]. These associations highlight an alternative focus for tackling drug resistance. For example, miR-21 expression in gemcitabine-resistant cells can be attenuated by treatment with a curcumin analogue, giving rise to an increase in phosphatase and tensin homolog (PTEN) protein levels [92]. Furthermore, the authors note an up-regulation of miR-200c following curcumin treatment; high miR-200c levels have also been associated with better prognosis in pancreatic cancer, possibly due to up-regulation of E-cadherin, which is often silenced in metastatic disease [96]. Thus, combination treatments that factor in the adverse effects of misregulated miRNAs might have marked efficacy in cancer therapy.
The challenges and future of microRNAs in cancer
For the purposes of risk predictors, diagnostics and prognostics, miRNAs have already been shown to be, by themselves, robust and important biomarkers, a reality that has not been achieved with mRNAs except in rare circumstances. Success at narrowing down the biology of a specific tumor to enable more accurate diagnoses is of immense benefit to both doctors and patients. As a result, there is understandably a strong drive in the scientific community to push forward the use of miRNA biomarkers into clinics. But like all big innovations, there are some kinks that need to be addressed. Ultimately, the clinical use of miRNA signatures to determine tumor cause, origin or subtype will influence patient treatment, and thus their accuracy is of paramount importance. Reported lack of consistency between similar studies therefore gives rise to some concern. Notably, Blenkiron et al. [38] found little agreement among miRNAs that they identified as being associated with clinicopathological factors and miRNAs identified in this context in a previous study [42]. Such differences typically arise from sample selection or preparation, experimental design and/or data analysis [97]. Indeed, the use of a different control for the normalization of data can explain some of the observed variability across studies [98, 99]. Another possibility that must be considered is the dynamic and immediate regulation in miRNA levels in the stress response [100] and in hypoxia [101]; thus time of collection and processing could impact miRNA levels. Nevertheless, current studies give us cause for optimism, and if more comprehensive validation is the primary concern, then it is unlikely to remain a barrier to the development of miRNAs in diagnostics.
In terms of the use of miRNAs to predict specific oncogenic mutations, the benefits are somewhat more ambivalent. Essentially, this type of diagnostic tool might be redundant. Although it could provide secondary validation, miRNAs are an indirect method of assessing specific gene mutations that could, and likely would regardless, be independently genotyped. Nevertheless, there might be an argument for the development of this type of miRNA diagnostic test as the first port-of-call, to simultaneously gauge the status of a variety of cancer-related genes, subsequently enabling more thorough and directed testing. More importantly, miRNA abnormalities caused by these acquired genetic mutations could provide insight into the biological function(s) of these mutations that lead to oncogenesis. Investigating these connections to identify additional mis-regulated gene networks in cancer might ultimately be more rewarding than their direct use as biomarkers.
Unlike miRNA signatures that determine tumor type or characteristics, the use of miRNA biomarkers that predict cancer risk fills a greater clinical need, as there are few known predictors of cancer risk. Some groups have reported that common miRNA mutations associated with cancer, such as hsa-miR-16-1 in CLL [50, 70], is not an informative marker when evaluated in different ethnic cohorts or in other cancers [102, 103]. It is therefore possible that the aptitude of certain miRNA markers could be confined to the patient groups from which their study was derived. Similar limitations exist for many of the diagnostic tests currently available in the market, which is why appropriate education and genetic counseling is essential to prevent mis-diagnosis [104]. Although immense benefit can still be derived from their use, understanding the appropriate applications of these miRNA cancer risk markers is a priority in their development.
The discovery that miRNAs have prognostic value was largely a by-product of efforts to determine their diagnostic potential, but now promises to become one of the most significant clinical applications of miRNAs. That patients with seemingly identical cancer phenotypes can have contrasting responses to the same treatment is widely observed in clinics. For those patients who either do not respond, or respond negatively, to treatment, the time lost could be irrecoverably damaging. miRNAs or miRNA alterations that can predict responses to specific treatments offer a unique and highly promising opportunity for clinicians to be able to determine from the onset what the most effective treatment for an individual patient is likely to be. This presents significant benefits for improving clinical care and is as valuable an application of miRNAs as cancer risk and tumor biology diagnostics.
Concluding remarks
The knowledge surrounding miRNAs and their roles in carcinogenesis has vastly improved and expanded over recent years, and it has become clear that understanding the gene regulatory networks governed by miRNAs is vital to understanding the complex processes contributing to malignancy. While significant advances have been made for the future role of miRNAs in diagnostics, there have been far fewer reported successes in the development of miRNAs for use in therapy. This is to be expected given the more complex requirements, specifically, the challenges facing delivery, efficacy and safety of any novel therapeutic. While successful employment of miRNA therapeutics remains a promising goal, with the limited positive in vivo studies to date, this will unlikely precede the widespread clinical use of miRNAs first in diagnostics. Still, miRNAs are making significant strides for the benefit of cancer patients; they already help us to understand much of what we see in the clinic today and help to shape our approaches to tackling cancer for tomorrow. Thus, miRNAs remain at the forefront of disease research, and it is with great anticipation that we look forward to future discoveries and clinical applications.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datta J, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Lujambio A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han L, et al. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284–1288. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, et al. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu CG, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett ST, et al. Toward the 1,000 dollars human genome. Pharmacogenomics. 2005;6:373–382. doi: 10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, et al. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res. 2009;38:D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drakaki A, Iliopoulos D. MicroRNA Gene Networks in Oncogenesis. Curr Genomics. 2009;10:35–41. doi: 10.2174/138920209787581299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu A, et al. MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int J Clin Exp Pathol. 2009;2:519–527. [PMC free article] [PubMed] [Google Scholar]
- 17.Jung M, et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem. 2010;56:998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- 18.Murakami Y, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 19.Szczyrba J, et al. The MicroRNA Profile of Prostate Carcinoma Obtained by Deep Sequencing. Molecular Cancer Research. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 20.Liang Y, et al. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenfeld N, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 22.Barroso-delJesus A, et al. Embryonic stem cell-specific miR302–367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramaswamy S, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci U S A. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra R, et al. SFSSClass: an integrated approach for miRNA based tumor classification. BMC Bioinformatics. 2010;11 Suppl 1:S22. doi: 10.1186/1471-2105-11-S1-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandyopadhyay S, et al. Development of the human cancer microRNA network. Silence. 2010;1:6. doi: 10.1186/1758-907X-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y, Kwoh CK. Informative microRNA expression patterns for cancer classification. Data Mining for Biomedical Applications, Proceedings. 2006;3916:143–154. [Google Scholar]
- 27.Oien KA, Evans TR. Raising the profile of cancer of unknown primary. J Clin Oncol. 2008;26:4373–4375. doi: 10.1200/JCO.2008.17.6156. [DOI] [PubMed] [Google Scholar]
- 28.Pavlidis N. Cancer of unknown primary: biological and clinical characteristics. Ann Oncol. 2003;14 Suppl 3:iii11–iii18. doi: 10.1093/annonc/mdg742. [DOI] [PubMed] [Google Scholar]
- 29.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 30.Sorlie T, et al. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics. 2006;7:127. doi: 10.1186/1471-2164-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du L, et al. MicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCs. J Exp Clin Cancer Res. 2010;29:75. doi: 10.1186/1756-9966-29-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebanony D, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 34.Petillo D, et al. MicroRNA profiling of human kidney cancer subtypes. Int J Oncol. 2009;35:109–114. doi: 10.3892/ijo_00000318. [DOI] [PubMed] [Google Scholar]
- 35.Ueda T, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fridman E, et al. Accurate molecular classification of renal tumors using microRNA expression. J Mol Diagn. 2010;12:687–696. doi: 10.2353/jmoldx.2010.090187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ratner ES, et al. MicroRNA signatures differentiate uterine cancer tumor subtypes. Gynecol Oncol. 2010;118:251–257. doi: 10.1016/j.ygyno.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blenkiron C, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sempere LF, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 40.Mattie MD, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowery AJ, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 43.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 44.Park SM, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baffa R, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 46.Uhlmann S, et al. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010;29:4297–4306. doi: 10.1038/onc.2010.201. [DOI] [PubMed] [Google Scholar]
- 47.Sachdeva M, Mo YY. miR-145-mediated suppression of cell growth, invasion and metastasis. Am J Transl Res. 2010;2:170–180. [PMC free article] [PubMed] [Google Scholar]
- 48.Spizzo R, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2010;17:246–254. doi: 10.1038/cdd.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70:378–387. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 51.Garzon R, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jongen-Lavrencic M, et al. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 53.Garzon R, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcucci G, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 55.Dhillon AS, et al. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 56.Caramuta S, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 57.Cahill S, et al. Effect of BRAFV600E mutation on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer. 2007;6:21. doi: 10.1186/1476-4598-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee CH, et al. MicroRNA profiling of BRCA1/2 mutation-carrying and non-mutation-carrying high-grade serous carcinomas of ovary. PLoS One. 2009;4:e7314. doi: 10.1371/journal.pone.0007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Visone R, et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood. 2009;114:3872–3879. doi: 10.1182/blood-2009-06-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gutierrez NC, et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia. 2010;24:629–637. doi: 10.1038/leu.2009.274. [DOI] [PubMed] [Google Scholar]
- 61.Chapiro E, et al. A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia. 2010;24:1362–1364. doi: 10.1038/leu.2010.93. [DOI] [PubMed] [Google Scholar]
- 62.Adams JM, et al. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A. 1983;80:1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robertus JL, et al. MiRNA profiling in B non-Hodgkin lymphoma: a MYC-related miRNA profile characterizes Burkitt lymphoma. Br J Haematol. 2010;149:896–899. doi: 10.1111/j.1365-2141.2010.08111.x. [DOI] [PubMed] [Google Scholar]
- 64.Gaur A, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 65.Marton S, et al. Small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia. 2008;22:330–338. doi: 10.1038/sj.leu.2405022. [DOI] [PubMed] [Google Scholar]
- 66.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 67.Melo SA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Kumar MS, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 69.Sevignani C, et al. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci U S A. 2007;104:8017–8022. doi: 10.1073/pnas.0702177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raveche ES, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li W, et al. Germline mutation of microRNA-125a is associated with breast cancer. J Med Genet. 2009;46:358–360. doi: 10.1136/jmg.2008.063123. [DOI] [PubMed] [Google Scholar]
- 72.Shen J, et al. Novel genetic variants in microRNA genes and familial breast cancer. Int J Cancer. 2009;124:1178–1182. doi: 10.1002/ijc.24008. [DOI] [PubMed] [Google Scholar]
- 73.Shen J, et al. Novel genetic variants in miR-191 gene and familial ovarian cancer. BMC Cancer. 2010;10:47. doi: 10.1186/1471-2407-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen J, et al. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 75.Yang R, et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res Treat. 2009;121:693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 76.Jazdzewski K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peng S, et al. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig Dis Sci. 2009;55:2288–2293. doi: 10.1007/s10620-009-1007-x. [DOI] [PubMed] [Google Scholar]
- 78.Tian T, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 79.Hu Z, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu B, et al. A functional polymorphism in Pre-miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo. Prostate. 2010;70:467–472. doi: 10.1002/pros.21080. [DOI] [PubMed] [Google Scholar]
- 81.Liu Z, et al. Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer. 2010;116:4753–4760. doi: 10.1002/cncr.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang H, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 83.Ryan BM, et al. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pelletier C, Weidhaas JB. MicroRNA binding site polymorphisms as biomarkers of cancer risk. Expert Rev Mol Diagn. 2010;10:817–829. doi: 10.1586/erm.10.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nicoloso MS, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chin LJ, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ratner E, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–6515. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Medina PP, et al. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 89.Li X, et al. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 90.Ji J, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rossi S, et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–952. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dillhoff M, et al. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou M, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496–21507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giovannetti E, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 96.Yu J, et al. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol Cancer. 2010;9:169. doi: 10.1186/1476-4598-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu JZ, Wong CW. Hunting for robust gene signature from cancer profiling data: sources of variability, different interpretations, and recent methodological developments. Cancer Lett. 2010;296:9–16. doi: 10.1016/j.canlet.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 98.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu SL, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 100.Marsit CJ, et al. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 101.Kulshreshtha R, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Catucci I, et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat. 2010;31:E1052–E1057. doi: 10.1002/humu.21141. [DOI] [PubMed] [Google Scholar]
- 103.Yazici H, et al. Investigation of the miR16-1 (C > T) + 7 Substitution in Seven Different Types of Cancer from Three Ethnic Groups. J Oncol. 2009;2009:827532. doi: 10.1155/2009/827532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brierley KL, et al. Errors in delivery of cancer genetics services: implications for practice. Conn Med. 2010;74:413–423. [PubMed] [Google Scholar]