Abstract
Variation in drug efficacy and toxicity remains an important clinical concern. Presently, single nucleotide polymorphisms (SNP) only explain a portion of this problem, even in situations where the pharmacological trait is clearly heritable. The Human CNV Project identified copy number variations (CNVs) across approximately 12% of the human genome, and these CNVs were considered causes of diseases. Although the contribution of CNVs to the pathogenesis of many common diseases is questionable, CNVs play a clear role in drug related genes by altering drug metabolizing and drug response. Here we provide a comprehensive review of the clinical relevance of CNVs to drug efficacy, toxicity, disease prevalence in world populations and discuss the implication of using CNVs as diagnosis in clinical intervention.
Keywords: pharmacogenetics, copy number variants, drug efficacy, drug toxicity
Pharmacogenetics
Pharmacogenetics aims to study the gene variants associated with drug metabolism enzymes, transporters, drug receptors and relieve the burden of sickness caused by interindividual differences in drug response or vulnerabilities to drug toxicity [1, 2]. Since the completion of the Human Genome Project in 2000, there has been a burst in the number of pharmacogenetic research studies published, with some of the findings now being introduced into clinical practice. To practice pharmacogenetics testing in the clinic, the United States Food and Drug Administration (FDA) approved the first DNA microarray molecular diagnostic test in 2004, the AmpliChip CYP450 (Roche Molecular Diagnostics, Alameda, CA, USA), for the analysis of single nucleotide polymorphisms (SNPs) and copy number variants (CNVs) of CYP2D6 and CYP2C19 genes which are closely related with the drug therapy of depression, gastroesophageal reflux and cardiovascular diseases. [3]. The FDA also issued “Draft Guidelines for Industry: Pharmacogenomic Data Submission”, making the submission of pharmacogenomic data mandatory for new drug applications from drug companies [4]. Since 2008, the FDA has issued a list of valid pharmacogenetic biomarkers in the context of approved drug labels that are updated every year [5], which accelerates the translation of pharmacogenetics from bench to bedside.
At present, pharmacogenetics can be applied to the treatment of patients with psychiatric illnesses, cancer, cardiovascular disease, pain, HIV infection, and microorganism infection, to enhance drug efficacy and decrease drug toxicities [6–9]. According to a survey of 1200 drug labels for the years 1945–2005, 121 drugs labels contained pharmacogenetic information [10]. Of those, 69 labels referred to human genomic biomarkers (e.g. atomoxetine, fluoxetine and codeine are labeled with CYP2D6 variants associated drug efficacy and toxicity change) [10]. Over 20 of these markers were either required or recommended by the FDA to be tested in patients before starting drug therapy [11]. For example, tramadol is an orally administered synthetic analogue of codeine and its product label warns that patients with cytochrome P450 2D6 (CYP2D6) gene (CYP2D6) duplications are at increased risk of respiratory depression [12].
What is a copy number variant?
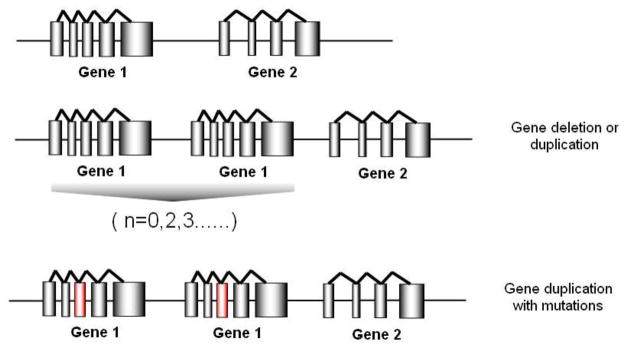
Human DNA has one copy of autosomal regions on each chromosome. However, as discovered by the Human Genome Project, many genetic regions display a variation in the number of copies (more or less than two copies totally). Alleles containing 0–13 gene copies have been reported across the human population [13]. These genetic variants are termed copy number variants (CNVs) and are defined as DNA segments ranging in size from one kilobase to several megabases among individuals due to deletion, insertion, inversion, duplication or complex recombination [14] (Figure 1). Many groups have addressed the presence of CNV in the human genome and their associations with common human diseases including neuropsychiatric, autoimmune, infectious and cardiovascular diseases[15, 16], while others found evidence that CNVs are not associated with common diseases [17]. While the contribution of CNVs to the pathogenesis of common diseases is questionable, CNVs in some pharmacogenetic genes play a clear role in drug efficacy and toxicity [18]. Inspired by the increasing interest in CNVs, here we review our current knowledge of CNVs in relation to drug efficacy and toxicity, their prevalence in ethnically diverse populations, and the potential utilization of CNV knowledge in the clinical setting.
Figure 1.
A diagram for copy number variants in human genome. If the gene recombination event occurs between two genes, a gene duplication or multiplication (n=2,3..) or a gene deletion (n=0) could happen. A duplication of gene could also carry mutations from the original copy (show in red column).
CNVs associated with drug efficacy in the clinic
CYP2D6
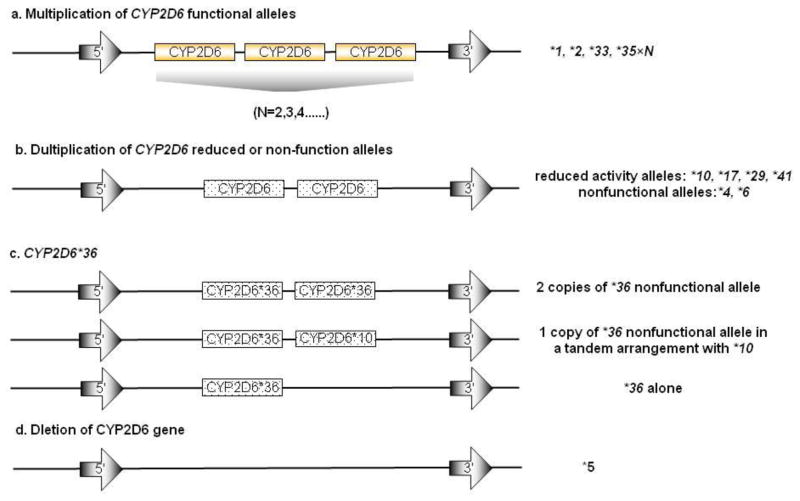
CYP2D6 is predominantly expressed in human liver and it metabolizes over 25% of drugs currently used in the clinic [19]. CYP2D6 is highly polymorphic and its variants account for the majority of the variation in enzyme activity that is observed within and between populations. To date, more than 75 CYP2D6 alleles have been documented in the Human Cytochrome P450 (CYP) Allele Nomenclature database (http://www.cypalleles.ki.se/cyp2d6.htm). Alleles are comprised of a combination of polymorphisms, including SNPs, insertion/deletions and/or gene conversions, which influence CYP2D6 enzyme activity. These alleles include normal activity alleles (*1, *2, *33, *35), reduced activity alleles (*9, *10, *17, *29, *41, *59), nonfunctional alleles, which encode enzymes with no activity or produce no enzyme at all (*3, *4, *5–*8, *11–*16, *18–*21,*36, *38, *40, *42, *44, *56, *62), and as yet functionally undetermined alleles (*22–*28, *30–*32, *34, *37, *39, *43, *45–*55, *70, *71, *73, *74) [20]. The functional polymorphisms of the nonfunctional and reduced activity alleles introduce splice defects, non-synonymous amino acid changes or frameshifts, which alter enzyme activity. In addition, CNVs of the normal, nonfunctional and reduced activity CYP2D6 alleles are observed and these can also alter in vivo CYP2D6 enzyme activity. For example, more than two copies of CYP2D6 normal alleles (e.g.*1×N, *2×N, *33×N, *35×N, 13>N>2, Figure 2) have been shown to elevate CYP2D6 enzyme activity [21]. Individuals who inherit these alleles have more rapid metabolism of CYP2D6 substrates and are classified as ultrarapid-metabolizers (UMs) [21]. CNVs are also found for nonfunctional (*4×N, *36×N) and reduced activity alleles (*10×N, *17×N), but individuals who inherit these alleles do not have increased metabolism of CYP2D6 substrates [22]. It is noteworthy that CYP2D6*36 is absent in white populations but in Asian and African American populations it exists on each chromosome in three forms: 1. CYP2D6*36×2 (2 copies of nonfunctional allele); 2. CYP2D6*36+*10 (1 copy of nonfunctional allele in a tandem arrangement with *10); and 3. CYP2D6*36 alone (Figure 2) [23]. Only the first situation is considered a CNV. In addition to gaining extra copies of the CYP2D6 gene, CYP2D6*5 is a loss of gene copy that produces no CYP2D6 protein [24]. Individuals with two nonfunctional CYP2D6 alleles, including CYP2D6*5, have no CYP2D6 activity, in turn have impaired metabolism of CYP2D6 substrates and are classified as poor-metabolizers (PMs). If two normal activity alleles are inherited, subjects have normal metabolism of CYP2D6 substrates and are classified as extensive metabolisers (EMs) [21]. CYP2D6 CNV is a significant cause of CYP2D6 phenotype diversity in the clinic.
Figure 2.
A summary of CYP2D6 CNV structure[18]. (a) Functional CYP2D6 CNVs are shown in gold. (b) Duplication of CYP2D6 reduced activity or nonfunctional alleles are shown in dotted background. (c) Three forms of CYP2D6*36 allele. The first arrangement is considered as a CNV for this allele. (d) Deletion of CYP2D6 gene.
In Caucasian and Asian populations, the frequency of duplicated normal activity alleles, CYP2D6*1×2 and *2×2, is approximately 0.5–1.5% [25], whereas in African populations, the frequency is approximately 1.6–3.3% [26, 27]. The frequency of CYP2D6*5 is similar in Caucasian, Asian and African populations (2–7%) [28]. Although the frequency of duplication of CYP2D6 normal alleles is quite low, the clinical impact of the corresponding phenotype cannot be ignored.
Nortriptyline, a tricyclic antidepressant, is prescribed for the treatment of depression and acts by inhibiting the reuptake of norepinephrine [29]. Taken orally, nortriptyline is well absorbed from the gastrointestinal tract and metabolized in the liver by CYP2D6 [30]. Dalen and colleagues found that the plasma exposure and clearance of nortriptyline was clearly related to the number of copies of the CYP2D6 gene of subjects (ranging from 0 to 13) [31]. Subjects with 13 copies of functional CYP2D6 gene displayed a 17-fold higher oral clearance and 36-fold lower plasma concentrations of nortriptyline than those carrying no copies of the functional gene [31]. Interestingly, a recent genome-wide study showed that CNVs in two intergenic regions (chromosome 1 and 10) were associated with nortriptyline response in unipolar depression patients of European ancestry [32]. However, CYP2D6 CNVs were not shown to be associated with the nortriptyline response. Further studies are required to validate the associations between CNVs and nortriptyline efficacy for CYP2D6 and new CNV candidates in the clinic.
Tamoxifen is a tissue selective estrogen receptor (ER) modulator, which serves as an antagonist of ERs in breast tissue; however, it is an agonist in endometrial tissue [33]. It is prescribed to treat and prevent hormone-dependent breast cancer in premenopausal and postmenopausal women [34]. Since the 1990s, tamoxifen has been approved by the FDA as the standard anti-estrogen treatment for metastatic breast cancer and adjuvant treatment of primary breast cancer for both female and male patients [35]. Tamoxifen is a prodrug and undergoes complex metabolism in vivo and in vitro, primarily catalysed by CYPs [36]. CYP2D6 is considered the key enzyme that converts N-desmethyltamoxifen (NDM), a primary tamoxifen metabolite, to endoxifen. Endoxifen has 10-fold higher affinity for ERs than tamoxifen, and is therefore considered the active form of tamoxifen in vivo [37]. Indeed, endoxifen/NDM plasma concentration ratios are significantly higher in UMs who carried multiple copies of CYP2D6 functional alleles than in EMs and PMs who carried two or less copies of CYP2D6 alleles [38]. It is possible that UM patients may derive more therapeutic benefit from tamoxifen than other patients; however, this has not yet been tested. The recent Italian Tamoxifen Prevention Trial genotyped 47 breast cancer patients using the AmpliChip CYP450 chip [39]. Subjects who carried more than two functional CYP2D6 alleles (*1/*1×N, *1/*2A×N and *1×N/*2A) were classified as UMs and those with one or two nonfunctional alleles (*3/*5, *41/*5 and *4A/*5) were classified as PMs. Although the small sample size limited the statistical power of the study, PMs had a higher risk of developing breast cancer than other subjects [39]. In addition, a larger German study genotyped 492 ER+ breast cancer patients who had received tamoxifen treatment. In this study patients with more than two CYP2D6 functional genes (*1/*1×N, *2/*1×N and *1/*2×N) were also classified as UMs. A Kaplan-Meier survival analyses displayed highest benefit from tamoxifen treatment in UM patients. Meanwhile, PM patients who carried no copies of CYP2D6 functional alleles had poorer outcomes than EMs (hazard ratio, 2.77; 95% confidence interval 1.31–5.89) [40]. Due to the low frequency of CYP2D6 CNV in Caucasians, small sample size is a common limitation in these studies, which contributes to low statistical power in data analysis. Clinical trials with larger sample sizes are required to further validate these findings.
Metoprolol is a selective beta1-adrenergic antagonist, which is prescribed to decrease heart rate in patients with acute myocardial infarction (AMI), hypertension or dilated cardiomyopathy [41]. It is extensively metabolized in human liver and predominantly by CYP2D6 [42]. In a pharmacokinetics study, subjects with three copies of CYP2D6 functional alleles, classified as UMs, showed significantly lower metoprolol plasma concentrations and higher drug clearances after a single dose of 100 mg metoprolol compared with EMs and PMs (P<0.0001) [43]. For multi-doses and long-term (42 days) drug administration, metoprolol plasma concentrations (normalized for daily dose) and metoprolol/a-hydroxymetoprolol ratios at steady state were markedly lower in UMs than EMs and PMs (P<0.0001), which suggested that UMs are at potential risk of treatment failure [42]. In another study, 187 AMI patients received metoprolol and heart rate (HR) was recorded at administration and discharge. PM patients achieved pronounced HR reduction, while UM patients (*1/*1×N) failed the treatment [44]. Pre-screening for CYP2D6 CNVs may help physicians evaluate the potential benefit that patients can receive from metoprolol treatment.
In summary, associations between CYP2D6 CNVs and therapeutic response to drugs were found in wide range of drug classes including antidepressants, endocrine therapy agents, and beta-blocking agents. Pro-drugs that depend on an activation process via CYP2D6 may demonstrate a better response in individuals with multiple copies of functional CYP2D6 genes (UMs). Individuals with two copies of nonfunctional CYP2D6 alleles (PMs) may benefit from low metabolizing efficiency of drugs exempt from activation process; however, they could be at increased risk of toxicities.
CNV associations with drug toxicity in the clinic
CYP2D6
Codeine is a widely used analgesic and antitussive agent in the clinic. It is activated by CYP2D6 to a strong central nervous system (CNS) sedative, morphine. Kirchheiner and colleagues found that after a single 30 mg dose of codeine, the plasma concentrations of morphine are significantly higher in CYP2D6 UMs than EMs [45]. High plasma concentrations of morphine can lead to several fatal effects including hypotension and respiratory depression, especially in pediatric patients. A healthy 2-year-old boy recently died after elective adenotonsillectomy because of the commonly prescribed pain-killer, codeine [46]. Postmortem genotyping revealed that the boy had a duplication of a functional CYP2D6 gene, which most likely made him a UM of codeine and the subsequent elevated morphine exposure was considered the likely cause of respiratory depression and death in this tragic case [46]. There was a similar case where an infant died with high morphine concentrations in his blood. The morphine was transferred via breastmilk from a mother with a UM genotype who was taking codeine for postpartum pain [47]. Breastfed infants are at increased risk of potentially life-threatening CNS suppression in such cases [48]. Codeine and its analogues have been labeled with CYP2D6 CNVs markers by FDA.
Glutathione S-transferases (GSTs)
Glutathione S-transferases (GSTs) are a superfamily of enzymes that catalyze the detoxification of carcinogens, therapeutic chemicals and environmental toxins. GSTs catalyze the reaction of glutathione (GSH) with an acceptor molecule to form a sulfur-substituted glutathione. GST enzymes are encoded by eight distinct loci including alpha, kappa, mu, omega, pi, sigma, theta, and zeta [49]. The GST Mu-1 and Theta-1 enzymes are encoded by the GSTM1 and GSTT1 genes, respectively. Complete gene deletions (loss of functional gene copy) of GSTM1 and GSTT1 are relatively common. About 50% and 30% of Caucasians are homozygous for GSTM1 and GSTT1 gene deletions respectively, compared with >22% and >14% in Asians, and >27% and >37% in Africans [50, 51]. In addition to gene deletions, a multiplex real-time PCR method was developed to detect GSTM1 and GSTT1 gene duplications [52]. However, GSTT1 duplications have not been observed, and a GSTM1 duplication has only been detected in one of 1320 Caucasian samples. No further functional studies on GSTM1/GSTT1 gene duplications have been reported.
Consistent with the important roles that GSTM1 and GSTT1 play in the detoxification of exogenous compounds, carriers of gene deletions have elevated risks of various cancers (e.g. colorectal cancer, acute myeloid leukemia), especially in Caucasians [53, 54], and drug-related toxicities. Cho and colleagues investigated the associations between GSTM1 and GSTT1 gene deletions and toxicity from the standard chemotherapy regimen for lymphoma, R-CHOP (rituximab with cyclophosphamide, doxorubicin, vincristine and prednisone), in 94 diffuse large B-cell lymphoma (DLBCL) patients [55]. R-CHOP caused more grade III-IV toxicities, including leukocytopenia (odds ratio (OR)=3.12), fever (OR=5.27), and mucositis (OR=4.61), in patients homozygous for GSTT1 gene deletion, than other patients. Patients with a GSTM1/GSTT1 double-deletion genotype had an even higher risk of grade III-IV thrombocytopenia than other patients (OR=7.75). Loss of either or both GSTM1 and GSTT1 genes might be expected to increase toxicity along with improved efficacy of chemotherapy, however no difference was observed in this study. GSTM1 and GSTT1 gene deletions are likely to predict R-CHOP induced toxicity in DLBCL patients.
Tacrine was the first drug for the treatment of mild to moderate dementia of Alzheimer’s disease [56], which is the most common form of dementia characterized for a progressive and fatal neurodegenerative disorder [57]. Tacrine-induced hepatotoxicity is primarily caused by one of its metabolites, which is de-activated by GSTM1 and GSTT1 [58]. A study in 141 Caucasian Alzheimer’s disease patients found that patients with the GSTM1 and GSTT1 double-deletion had alanine aminotransferase (ALT) levels that were three times the upper limit of normal (relative hazard: 2.84, 95% confidence interval:1.51–5.35, P=0.001), which is a marker of tacrine-induced hepatotoxicity [59]. In addition, a patient’s susceptibility to tacrine-induced hepatotoxicity could not be predicted by either GSTM1 or GSTT1 deletion alone suggesting redundancy in the enzymes [59]. Due to the high prevalence of GSTM1 and GSTT1 gene deletions in Caucasians, over 10% of patients have a GSTM1 and GSTT1 double deletion and are therefore at increased risk of tacrine-induced hepatotoxicity. A nitric oxide donor-tacrine hybrid compound may be an alternative therapy for these patients to achieve both drug efficacy and low risk of hepatotoxicity [60].
In summary, many phase I and II drug metabolism enzymes are involved in the detoxification of drugs used in clinic. CNVs can lead to the over-activation or loss of detoxification of a drug which may cause increased drug toxicities in patients. Precautions should be adopted based on individual genotyping information.
Prevalence of CNVs in drug related gene in human populations
Understanding the prevalence of major genetic variations related to drug efficacy and toxicity is critical for both health providers and patients. Both the Ministry of Health in each nation and physicians in clinics can use this knowledge to maximize benefit and minimize harm for patients prior to and during drug therapy [61], which is widely accepted as the base for personalized genomic medicine [62].
The frequency of CNVs is shown in Table 1. These data can help each population to select the most appropriate drugs and proper doses for specific treatments. For example, in the United States, over 50% and 30% of Caucasian populations carry either GSTM1 or GSTT1 gene deletions [50], which leads to increased susceptibility to colorectal cancer, acute myeloid leukemia and chemotherapy-induced toxicities [53]. Using the CNVs frequency data, physicians will be aware of the probability of achieving benefit or developing a drug-induced toxicity in his/her patients and thus precautions can be taken. Meanwhile, multi-copies of CYP2D6 functional genes are more prevalent in Europe, Saudi Arab, and Ethiopia than in Asia and the U.S. (Caucasian). In former regions, more people may respond to tamoxifen treatment, and less codeine induced CNS suppression will be observed in the latter. The data of CNVs global prevalence can help to avoid translating a regional drug problem for a specific population into a common problem for all other populations [63].
Table 1.
A summary of CNV frequencies in pharmacogenetic genes in different ethnic populations.
| CNV frequencies (%) | |||||
|---|---|---|---|---|---|
| Population | CYP2D6*1×N | CYP2D6*2×N | CYP2D6*5 | GSTM1 deletion | GSTT1 deletion |
| North America | |||||
| U.S.A. (Caucasian) | 0.2 [72] | 0.7 [72] | 6.2 [72] | 54.3 [50] | 27.6 [50] |
| U.S.A. (African-American) | 1.2 [72] | 1.6 [72] | 4.0 [72] | 23.7 [73] | 17.5 [73] |
| Mexico (Mestizo) | 1.3–2.7 [74, 75] | 33.5 [76] | 12.1 [76] | ||
| Africa | |||||
| Cameroon | 3.3 [26] | 3.3 [26] | 27.8 [51] | 46.8 [51] | |
| Ethiopia | 10–16 [21, 77] | 3.3 [77] | 43.8 [51] | 37.3 [51] | |
| Ghana | 1.6 [27] | 6 [27] | 19.3 [78] | 73.7 [78] | |
| East Asia | |||||
| China ( Han) | 2.2 [25] | 0–2 [21] | 7.2 [79] | 52 [80] | 38.7 [80] |
| Japan | 0.5 [81] | 0.5 [81] | 6.2 [82] | 50.8 [83] | 45.8 [83] |
| Korea | 0.13 [84] | 0.5 [84] | 6.1 [84] | 51.4 [85] | 51.6 [85] |
| Europe | |||||
| France | 2.0 [25] | 4.0 [25] | 49 [86] | 26 [86] | |
| Germany | 0.51 [87] | 1.34 [87] | 1.95 [87] | 51.6 [50] | 19.5 [50] |
| Middle East | |||||
| Saudi Arabia | 10.4 [88] | 1.0 [88] | 8.3 [89] | 4.2 [89] | |
Which drugs should be labeled with CNV markers?
Since 2008, the FDA has released a list of valid genomic biomarkers in the context of approved drug labels [5]. The term “valid biomarkers” has been described in the previously released “Guidance for Industry: Pharmacogenomic Data Submissions” [4] as a “biomarker that is measured in an analytical test system with well established performance characteristics and for which there is an established scientific framework or body of evidence that elucidates the physiologic, toxicological, pharmacologic, or clinical significance of the test results.” Drugs whose therapeutic response or toxicity is affected by these markers were recorded in this list including one CNV marker: CYP2D6*2×2. The package insert for codeine sulfate and other drugs containing codeine as an ingredient, such as Fiorinal® with codeine (butalbital, aspirin, caffeine, and codeine phosphate) and Fioricet® with codeine (butalbital, acetaminophen, caffeine, and codeine phosphate) warns of elevated risk of codeine-induced toxicity in individuals with more copies of CYP2D6 functional genes. However, none of the other drugs (tamoxifen, tacrine, etc) or other CNVs (CYP2D6*5, GSTM1, GSTT1 deletions) discussed in this review are included in this list yet. Meanwhile, an FDA-approved analytical test system for GSTM1 and GSTT1 deletions is not available at this time. The only approved genotyping platform for CNV markers diagnosis in clinic is the AmpliChip CYP450 chip [3], which detects CYP2D6 CNVs. There is still a gap between the knowledge of drug-related CNVs and implementation of drug label changes. The association of pharmacogenetic CNVs with drugs and clinical indications are summarized in Table 2.
Table 2.
Major clinical information for CNVs in pharmacogenetic genes.
| CNV type | Drug affected | Phenotype | Clinical indication | PharmGKB gene category | Reference | |
|---|---|---|---|---|---|---|
| CYP2A6*4 | Deletion | Nicotine Coumarin Tegafur |
No enzyme activity | Unknown | VIP | [90–92] |
| CYP2A6*1x2 | Duplication | Increased enzyme activity | Unknown | VIP | ||
| CYP2D6*1×N | Multiplication | Anticholinesterases Antidepressants Antipsychotics Beta Blocking Agents Opioids |
Increased enzyme activity | Alzheimer Disease Depression Hypertension Breat cancer Codeine dependence |
VIP | [19] |
| CYP2D6*2×N | Multiplication | Increased enzyme activity | VIP | |||
| CYP2D6*5 | Deletion | No enzyme activity | VIP | |||
| GSTM1*0 | Deletion | Antineoplastic agent Xenobiotics | No enzyme activity | Breast cancer Non-small cell lung cancer Leukemia Drug toxicity |
[93, 94] | |
| GSTT1*0 | Deletion | No enzyme activity | ||||
| SULT1A1 | Multiplication | Unclear clinic impact on drugs | Increased enzyme activity | Unknown | VIP | |
| SULT1A3 | Duplication | Unclear clinic impact on drugs | Unknown | Unknown | ||
| UGT2B17 | Deletion | Estradiol Testosterone |
Dcreased enzyme activity | Androgen Excretion |
[95] |
VIP = very important pharmacogene.
Concluding remarks
The translation of CNV knowledge into clinic intervention remains a big challenge, with a growing number of pharmacogenetics studies providing robust evidence for the impact of CNV on the efficacy and toxicity of drug therapy [18]. However, few comparative effectiveness studies have been conducted in this field, which has limited the expansion of genetic testing of CNVs into primary health care system. In addition, investigations on the cost versus benefits of genetic texting have been questioned. Additionally, health providers and insurance companies quite understandably are reluctant to reimburse genetic tests for low frequency variants to identify only a few patients who may benefit. That includes CYP2D6 CNVs and over 10 other clinically important pharmacogenetic variants (CYP2C19*3, CYP2C9*2,*3, CYP2D6*3,*6, DPYD*2A, TPMT*2,*3,*4, etc) that are considered rare. These variants exist in every population and may cause severe drug-induced toxicities or reduce the therapeutic benefit that a patient derives from a drug. As a result of this economic reality, diffusion of genetic tests for drug treatment has been slow. Unless we take a coordinated approach to CNV translational research, by adding policy and economics members to the investigation team, we will never see the promise of genome-guided therapy.
Box 1. Currently available methodologies for CNV detection.
Many PCR based methods have been developed for the detection of CNVs. However, the low-throughput and low reproductively rate limit the use of these methods in the clinic. Recently, new methods have been developed based on microarray hybridization technology.
Conventional methods
Long distance and multiplex PCR
This is the first method developed for the detection of CYP2D6 CNV. Long PCR is used to amplify a > 4000 bp fragment of the CYP2D6 gene. Specific primers for CYP2D6*5 have been designed. Two separate PCR reactions for either wild-type or mutated allele primers are run for each sample using the long PCR product of CYP2D6 gene. Following up with gel electrophoresis, the CYP2D6*5 can be identified [64].
TaqMan Real-Time PCR
This is a PCR quantification method. TaqMan probes emit a specific report fluorophore (usually a short-wavelength colored dye, such as green) during the elongation process of PCR reactions. The fluorophore can be detected by a corresponding detection system and compared with the signal of reference genes, such as albumin, to calculate the amount of specific genes in each sample. This is a low-through put detection method for CNVs [65].
Microarrays hybridization-based methods
Array comparative genomic hybridization (aCGH)
This is a genome-wide screening technique for CNVs [66] with higher resolution than chromosome-based comparative genomic hybridization [67], which allows detection of copy number changes of 5–10 kb of DNA sequence.
Roche AmpliChip CYP450 system
This is an oligonucleotide microarray hybridization method for genotyping 27 CYP2D6 variants (including CNVs) and two CYP2C19 variants. It has been developed by Roche based on Affymetrix microarray technology [68]. A logarithmic scale is used to predict CYP2D6 and CYP2C19 phenotypes [69]. This is the first FDA approved pharmacogenetics test for clinic use.
Genome-wide association SNP microarrays
These arrays cover 300K to more than 1 million genetic markers including thousands of probes for the detection of CNVs [70].
Affymetrix DMET plus microarray
This is a novel genotyping tool customized for pharmacogenetics research. It covers 225 essential pharmacogenetics genes and 1936 common or rare variants including five CNVs belongs to CYP2D6, CYP2A6, UGT2B17, GSTM1 and GSTT1 [71].
Acknowledgments
The authors are supported by the NIH Pharmacogenetics Research Network (U01 GM63340), the University Cancer Research Fund, and UL1RR025747 from the National Center for Research Resources. This work is part of the Pharmacogenetics for Every Nation Initiative. (www.pgeni.org).
Footnotes
Conflicts of interest
Dr McLeod is a consultant to Medco Health Solutions, Gentris corportation, and Myriad Genetics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meyer UA. Pharmacogenetics and adverse drug reactions. Lancet. 2000;356:1667–1671. doi: 10.1016/S0140-6736(00)03167-6. [DOI] [PubMed] [Google Scholar]
- 2.Eichelbaum M, et al. Pharmacogenomics and individualized drug therapy. Annu Rev Med. 2006;57:119–137. doi: 10.1146/annurev.med.56.082103.104724. [DOI] [PubMed] [Google Scholar]
- 3.de Leon J, et al. The AmpliChip CYP450 genotyping test: Integrating a new clinical tool. Mol Diagn Ther. 2006;10:135–151. doi: 10.1007/BF03256453. [DOI] [PubMed] [Google Scholar]
- 4.U.S.FDA. Guidance for Industry: Pharmacogenomic Data Submissions. 2005. [Google Scholar]
- 5.Table of Valid Genomic Biomarkers in the Context of Approved Drug Labels. 2008. [Google Scholar]
- 6.Normanno N, et al. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6:519–527. doi: 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 7.Lonjou C, et al. A European study of HLA-B in Stevens-Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs. Pharmacogenet Genomics. 2008;18:99–107. doi: 10.1097/FPC.0b013e3282f3ef9c. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz UI, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy PD, et al. Pharmacogenomics of anti-TB drugs-related hepatotoxicity. Pharmacogenomics. 2008;9:311–321. doi: 10.2217/14622416.9.3.311. [DOI] [PubMed] [Google Scholar]
- 10.Frueh FW, et al. Pharmacogenomic biomarker information in drug labels approved by the United States food and drug administration: prevalence of related drug use. Pharmacotherapy. 2008;28:992–998. doi: 10.1592/phco.28.8.992. [DOI] [PubMed] [Google Scholar]
- 11.Administration, U.F.a.D. Table of valid genomic biomarkers in the context of approved drug labels. 2006. [Google Scholar]
- 12.Stamer UM, et al. Respiratory depression with tramadol in a patient with renal impairment and CYP2D6 gene duplication. Anesth Analg. 2008;107:926–929. doi: 10.1213/ane.0b013e31817b796e. [DOI] [PubMed] [Google Scholar]
- 13.Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348:529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 14.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoumans J, et al. Detection of chromosomal imbalances in children with idiopathic mental retardation by array based comparative genomic hybridisation (array-CGH) J Med Genet. 2005;42:699–705. doi: 10.1136/jmg.2004.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 17.Craddock N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson I, Ingelman-Sundberg M. CNVs of human genes and their implication in pharmacogenetics. Cytogenet Genome Res. 2008;123:195–204. doi: 10.1159/000184709. [DOI] [PubMed] [Google Scholar]
- 19.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinet. 2009;48:689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 20.Hoskins JM, et al. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer. 2009;9:576–586. doi: 10.1038/nrc2683. [DOI] [PubMed] [Google Scholar]
- 21.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 22.Ishiguro A, et al. Metabolic activity of dextromethorphan O-demethylation in healthy Japanese volunteers carrying duplicated CYP2D6 genes: duplicated allele of CYP2D6*10 does not increase CYP2D6 metabolic activity. Clin Chim Acta. 2004;344:201–204. doi: 10.1016/j.cccn.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Gaedigk A, et al. CYP2D6*36 gene arrangements within the cyp2d6 locus: association of CYP2D6*36 with poor metabolizer status. Drug Metab Dispos. 2006;34:563–569. doi: 10.1124/dmd.105.008292. [DOI] [PubMed] [Google Scholar]
- 24.Steen VM, et al. Detection of the poor metabolizer-associated CYP2D6(D) gene deletion allele by long-PCR technology. Pharmacogenetics. 1995;5:215–223. doi: 10.1097/00008571-199508000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Sistonen J, et al. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenet Genomics. 2007;17:93–101. doi: 10.1097/01.fpc.0000239974.69464.f2. [DOI] [PubMed] [Google Scholar]
- 26.Fuselli S, et al. Evolution of detoxifying systems: the role of environment and population history in shaping genetic diversity at human CYP2D6 locus. Pharmacogenet Genomics. 2010;20:485–499. doi: 10.1097/FPC.0b013e32833bba25. [DOI] [PubMed] [Google Scholar]
- 27.Griese EU, et al. Analysis of the CYP2D6 gene mutations and their consequences for enzyme function in a West African population. Pharmacogenetics. 1999;9:715–723. [PubMed] [Google Scholar]
- 28.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 29.Nortriptyline. 2008 http://www.ncbi.nlm.nih.gov/pubmedhealth/PMH0000732.
- 30.Yue QY, et al. Pharmacokinetics of nortriptyline and its 10-hydroxy metabolite in Chinese subjects of different CYP2D6 genotypes. Clin Pharmacol Ther. 1998;64:384–390. doi: 10.1016/S0009-9236(98)90069-8. [DOI] [PubMed] [Google Scholar]
- 31.Dalen P, et al. 10-Hydroxylation of nortriptyline in white persons with 0, 1, 2, 3, and 13 functional CYP2D6 genes. Clin Pharmacol Ther. 1998;63:444–452. doi: 10.1016/S0009-9236(98)90040-6. [DOI] [PubMed] [Google Scholar]
- 32.Uher R, et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry. 2010;167:555–564. doi: 10.1176/appi.ajp.2009.09070932. [DOI] [PubMed] [Google Scholar]
- 33.Wood CE, et al. Endometrial profile of tamoxifen and low-dose estradiol combination therapy. Clin Cancer Res. 2010;16:946–956. doi: 10.1158/1078-0432.CCR-09-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jordan VC. Tamoxifen (ICI46,474) as a targeted therapy to treat and prevent breast cancer. Br J Pharmacol. 2006;147(Suppl 1):S269–276. doi: 10.1038/sj.bjp.0706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingle JN. Pharmacogenomics of tamoxifen and aromatase inhibitors. Cancer. 2008;112:695–699. doi: 10.1002/cncr.23192. [DOI] [PubMed] [Google Scholar]
- 36.Desta Z, et al. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 37.Johnson MD, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 38.Borges S, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Serrano D, et al. Efficacy of tamoxifen based on cytochrome P450 2D6, CYP2C19 and SULT1A1 genotype in the Italian Tamoxifen Prevention Trial. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.17. [DOI] [PubMed] [Google Scholar]
- 40.Schroth W, et al. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratification. Clin Cancer Res. 2010;16:4468–4477. doi: 10.1158/1078-0432.CCR-10-0478. [DOI] [PubMed] [Google Scholar]
- 41.Prakash A, Markham A. Metoprolol: a review of its use in chronic heart failure. Drugs. 2000;60:647–678. doi: 10.2165/00003495-200060030-00011. [DOI] [PubMed] [Google Scholar]
- 42.Fux R, et al. Impact of CYP2D6 genotype on adverse effects during treatment with metoprolol: a prospective clinical study. Clin Pharmacol Ther. 2005;78:378–387. doi: 10.1016/j.clpt.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Kirchheiner J, et al. Impact of the ultrarapid metabolizer genotype of cytochrome P450 2D6 on metoprolol pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 2004;76:302–312. doi: 10.1016/j.clpt.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Goryachkina K, et al. CYP2D6 is a major determinant of metoprolol disposition and effects in hospitalized Russian patients treated for acute myocardial infarction. Eur J Clin Pharmacol. 2008;64:1163–1173. doi: 10.1007/s00228-008-0525-3. [DOI] [PubMed] [Google Scholar]
- 45.Kirchheiner J, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J. 2006 doi: 10.1038/sj.tpj.6500406. [DOI] [PubMed] [Google Scholar]
- 46.Ciszkowski C, et al. Codeine, ultrarapid-metabolism genotype, and postoperative death. N Engl J Med. 2009;361:827–828. doi: 10.1056/NEJMc0904266. [DOI] [PubMed] [Google Scholar]
- 47.Madadi P, et al. Safety of codeine during breastfeeding: fatal morphine poisoning in the breastfed neonate of a mother prescribed codeine. Can Fam Physician. 2007;53:33–35. [PMC free article] [PubMed] [Google Scholar]
- 48.Madadi P, et al. Pharmacogenetics of neonatal opioid toxicity following maternal use of codeine during breastfeeding: a case-control study. Clin Pharmacol Ther. 2009;85:31–35. doi: 10.1038/clpt.2008.157. [DOI] [PubMed] [Google Scholar]
- 49.Hayes JD, et al. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 50.Garte S, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10:1239–1248. [PubMed] [Google Scholar]
- 51.Piacentini S, et al. GSTT1 and GSTM1 gene polymorphisms in European and African populations. Mol Biol Rep. 2010 doi: 10.1007/s11033-010-0221-0. [DOI] [PubMed] [Google Scholar]
- 52.Timofeeva M, et al. A multiplex real-time PCR method for detection of GSTM1 and GSTT1 copy numbers. Clin Biochem. 2009;42:500–509. doi: 10.1016/j.clinbiochem.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 53.Economopoulos KP, Sergentanis TN. GSTM1, GSTT1, GSTP1, GSTA1 and colorectal cancer risk: a comprehensive meta-analysis. Eur J Cancer. 2010;46:1617–1631. doi: 10.1016/j.ejca.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Das P, et al. Meta-analysis study of glutathione-S-transferases (GSTM1, GSTP1, and GSTT1) gene polymorphisms and risk of acute myeloid leukemia. Leuk Lymphoma. 2009;50:1345–1351. doi: 10.1080/10428190903003236. [DOI] [PubMed] [Google Scholar]
- 55.Cho HJ, et al. Glutathione-S-transferase genotypes influence the risk of chemotherapy-related toxicities and prognosis in Korean patients with diffuse large B-cell lymphoma. Cancer Genet Cytogenet. 2010;198:40–46. doi: 10.1016/j.cancergencyto.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 56.Farlow M, et al. A controlled trial of tacrine in Alzheimer’s disease. The Tacrine Study Group. Jama. 1992;268:2523–2529. [PubMed] [Google Scholar]
- 57.Kawas CH. Clinical practice. Early Alzheimer’s disease. N Engl J Med. 2003;349:1056–1063. doi: 10.1056/NEJMcp022295. [DOI] [PubMed] [Google Scholar]
- 58.Patocka J, et al. Possible role of hydroxylated metabolites of tacrine in drug toxicity and therapy of Alzheimer’s disease. Curr Drug Metab. 2008;9:332–335. doi: 10.2174/138920008784220619. [DOI] [PubMed] [Google Scholar]
- 59.Simon T, et al. Combined glutathione-S-transferase M1 and T1 genetic polymorphism and tacrine hepatotoxicity. Clin Pharmacol Ther. 2000;67:432–437. doi: 10.1067/mcp.2000.104944. [DOI] [PubMed] [Google Scholar]
- 60.Fang L, et al. NO-donating tacrine hybrid compounds improve scopolamine-induced cognition impairment and show less hepatotoxicity. J Med Chem. 2008;51:7666–7669. doi: 10.1021/jm801131a. [DOI] [PubMed] [Google Scholar]
- 61.Roederer MW, McLeod HL. Applying the genome to national drug formulary policy in the developing world. Pharmacogenomics. 2010;11:633–636. doi: 10.2217/pgs.10.55. [DOI] [PubMed] [Google Scholar]
- 62.Collins FS. The Language of Life:DNA and the Revolution in Personalized Medicine. HarperCollins Publisher; 2009. [Google Scholar]
- 63.Cavaco I, et al. CYP2C8 polymorphism frequencies among malaria patients in Zanzibar. Eur J Clin Pharmacol. 2005;61:15–18. doi: 10.1007/s00228-004-0871-8. [DOI] [PubMed] [Google Scholar]
- 64.Stuven T, et al. Rapid detection of CYP2D6 null alleles by long distance- and multiplex-polymerase chain reaction. Pharmacogenetics. 1996;6:417–421. doi: 10.1097/00008571-199610000-00005. [DOI] [PubMed] [Google Scholar]
- 65.Schaeffeler E, et al. CYP2D6 genotyping strategy based on gene copy number determination by TaqMan real-time PCR. Hum Mutat. 2003;22:476–485. doi: 10.1002/humu.10280. [DOI] [PubMed] [Google Scholar]
- 66.Shinawi M, Cheung SW. The array CGH and its clinical applications. Drug Discov Today. 2008;13:760–770. doi: 10.1016/j.drudis.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 67.Pinkel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 68.Heller T, et al. AmpliChip CYP450 GeneChip: a new gene chip that allows rapid and accurate CYP2D6 genotyping. Ther Drug Monit. 2006;28:673–677. doi: 10.1097/01.ftd.0000246764.67129.2a. [DOI] [PubMed] [Google Scholar]
- 69.Rebsamen MC, et al. The AmpliChip CYP450 test: cytochrome P450 2D6 genotype assessment and phenotype prediction. Pharmacogenomics J. 2009;9:34–41. doi: 10.1038/tpj.2008.7. [DOI] [PubMed] [Google Scholar]
- 70.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 71.Deeken J. The Affymetrix DMET platform and pharmacogenetics in drug development. Curr Opin Mol Ther. 2009;11:260–268. [PubMed] [Google Scholar]
- 72.Gaedigk A, et al. Unique CYP2D6 activity distribution and genotype-phenotype discordance in black Americans. Clin Pharmacol Ther. 2002;72:76–89. doi: 10.1067/mcp.2002.125783. [DOI] [PubMed] [Google Scholar]
- 73.Lavender NA, et al. Examination of polymorphic glutathione S-transferase (GST) genes, tobacco smoking and prostate cancer risk among men of African descent: a case-control study. BMC Cancer. 2009;9:397. doi: 10.1186/1471-2407-9-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sosa-Macias M, et al. Influence of CYP2D6 deletion, multiplication, -1584C-->G, 31G-->A and 2988G-->a gene polymorphisms on dextromethorphan metabolism among mexican tepehuanos and mestizos. Pharmacology. 2010;86:30–36. doi: 10.1159/000314334. [DOI] [PubMed] [Google Scholar]
- 75.Lopez M, et al. CYP2D6 genotype and phenotype determination in a Mexican Mestizo population. Eur J Clin Pharmacol. 2005;61:749–754. doi: 10.1007/s00228-005-0038-2. [DOI] [PubMed] [Google Scholar]
- 76.Perez-Morales R, et al. Polymorphism of CYP1A1*2C, GSTM1*0, and GSTT1*0 in a Mexican Mestizo population: a similitude analysis. Hum Biol. 2008;80:457–465. doi: 10.3378/1534-6617-80.4.457. [DOI] [PubMed] [Google Scholar]
- 77.Aklillu E, et al. Frequent distribution of ultrarapid metabolizers of debrisoquine in an ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther. 1996;278:441–446. [PubMed] [Google Scholar]
- 78.Dash B, et al. Determinants of the variability of aflatoxin-albumin adduct levels in Ghanaians. J Toxicol Environ Health A. 2007;70:58–66. doi: 10.1080/15287390600748880. [DOI] [PubMed] [Google Scholar]
- 79.Ji L, et al. Genetic polymorphisms of CYP2D6 in Chinese mainland. Chin Med J (Engl) 2002;115:1780–1784. [PubMed] [Google Scholar]
- 80.Liu L, et al. Genetic polymorphisms of glutathione S-transferase and risk of vitiligo in the Chinese population. J Invest Dermatol. 2009;129:2646–2652. doi: 10.1038/jid.2009.143. [DOI] [PubMed] [Google Scholar]
- 81.Nishida Y, et al. CYP2D6 genotypes in a Japanese population: low frequencies of CYP2D6 gene duplication but high frequency of CYP2D6*10. Pharmacogenetics. 2000;10:567–570. doi: 10.1097/00008571-200008000-00010. [DOI] [PubMed] [Google Scholar]
- 82.Kubota T, et al. Frequencies of CYP2D6 mutant alleles in a normal Japanese population and metabolic activity of dextromethorphan O-demethylation in different CYP2D6 genotypes. Br J Clin Pharmacol. 2000;50:31–34. doi: 10.1046/j.1365-2125.2000.00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fujihara J, et al. Cytochrome P450 1A1, glutathione S-transferases M1 and T1 polymorphisms in Ovambos and Mongolians. Leg Med (Tokyo) 2009;11(Suppl 1):S408–410. doi: 10.1016/j.legalmed.2009.01.073. [DOI] [PubMed] [Google Scholar]
- 84.Lee SY, et al. Sequence-based CYP2D6 genotyping in the Korean population. Ther Drug Monit. 2006;28:382–387. doi: 10.1097/01.ftd.0000211823.80854.db. [DOI] [PubMed] [Google Scholar]
- 85.Uhm YK, et al. Association of glutathione S-transferase gene polymorphisms (GSTM1 and GSTT1) of vitiligo in Korean population. Life Sci. 2007;81:223–227. doi: 10.1016/j.lfs.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 86.Abbas A, et al. GSTM1, GSTT1, GSTP1 and CYP1A1 genetic polymorphisms and susceptibility to esophageal cancer in a French population: different pattern of squamous cell carcinoma and adenocarcinoma. World J Gastroenterol. 2004;10:3389–3393. doi: 10.3748/wjg.v10.i23.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sachse C, et al. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet. 1997;60:284–295. [PMC free article] [PubMed] [Google Scholar]
- 88.McLellan RA, et al. Frequent occurrence of CYP2D6 gene duplication in Saudi Arabians. Pharmacogenetics. 1997;7:187–191. doi: 10.1097/00008571-199706000-00003. [DOI] [PubMed] [Google Scholar]
- 89.Abu-Amero KK, et al. GSTM1 and GSTT1 deletion genotypes in various spontaneous optic neuropathies in Arabs. Br J Ophthalmol. 2009;93:1101–1104. doi: 10.1136/bjo.2008.152983. [DOI] [PubMed] [Google Scholar]
- 90.Rao Y, et al. Duplications and defects in the CYP2A6 gene: identification, genotyping, and in vivo effects on smoking. Mol Pharmacol. 2000;58:747–755. doi: 10.1124/mol.58.4.747. [DOI] [PubMed] [Google Scholar]
- 91.Lerman C, et al. Genetic variation in nicotine metabolism predicts the efficacy of extended-duration transdermal nicotine therapy. Clin Pharmacol Ther. 2010;87:553–557. doi: 10.1038/clpt.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Farinola N, Piller NB. CYP2A6 polymorphisms: is there a role for pharmacogenomics in preventing coumarin-induced hepatotoxicity in lymphedema patients? Pharmacogenomics. 2007;8:151–158. doi: 10.2217/14622416.8.2.151. [DOI] [PubMed] [Google Scholar]
- 93.Yang P, et al. Role of the glutathione metabolic pathway in lung cancer treatment and prognosis: a review. J Clin Oncol. 2006;24:1761–1769. doi: 10.1200/JCO.2005.02.7110. [DOI] [PubMed] [Google Scholar]
- 94.Lamberti C, et al. Arylamine N-acetyltransferase type 2 and glutathione S-transferases M1 and T1 polymorphisms in familial adenomatous polyposis. Pharmacogenetics. 2002;12:49–54. doi: 10.1097/00008571-200201000-00007. [DOI] [PubMed] [Google Scholar]
- 95.Yang TL, et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet. 2008;83:663–674. doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]