Abstract
Patients with multiple sclerosis (MS) show a high prevalence of myelin-reactive CD8+ and CD4+ T-cell responses, which are the putative effectors/modulators of CNS neuropathology. Utilizing a novel combination of short-term culture, CFSE-based sorting and anchored PCR, we evaluated clonal compositions of neuroantigen-targeting T-cells from RRMS patients and controls. CDR3 region analysis of TCRβ chains revealed biased use of specific TCRBV-bearing CD4+ clones. CD8+ clones showed homology to published TCR from CNS-infiltrating T-cells in MS lesions. These studies are the first description of TCR usage of CNS-specific CD8+ T-cells and provide insights into their potential regulatory role in disease.
Keywords: Multiple Sclerosis, TCR, CD8, CD4, myelin, CDR3
1. INTRODUCTION
Multiple sclerosis (MS) is an inflammatory, demyelinating disorder of the central nervous system (CNS) that affects more than one million people worldwide. Although the etiology of MS is poorly understood, there is considerable evidence for an immune-mediated pathology, such as the presence of T-cell responses to myelin antigens (Martin et al., 1992), a strong genetic association with certain HLA Class II haplotypes, and polymorphisms in cytokine receptors (Barcellos et al., 2006; Gregory et al., 2007; Hafler et al., 2007). Evidence supporting the involvement of T-cells is based upon a vast body of work in an animal model of MS, experimental autoimmune encephalomyelitis (EAE). This well-characterized model can be induced by immunization with myelin sheath proteins such as myelin basic protein (MBP) (Einstein et al., 1962), myelin oligodendrocyte glycoprotein (MOG) (Lebar et al., 1986), or proteolipid protein (PLP) (Tuohy et al., 1988; Wekerle, 1991) or by adoptive transfer of neuroantigen-targeting T-cells which can commute disease to a new host (Pettinelli and McFarlin, 1981).
Specifically, autoreactive CD4+ Th1/Th17 T-cells are believed to be the principle mediators of both EAE and MS, based on several observations, such as: IL-2 and IFN-γ production by infiltrating CD4+ T-cells isolated from the CNS of mice with acute EAE (Renno et al., 1995), presence of Th1-inducing cytokines in inflammatory lesions and treatment of mice with Th1-inducing cytokines resulting in aggravation of autoimmune disease (Gutcher and Becher, 2007) and analysis of gene transcripts in chronic MS lesions revealing an increased level of IL-17 transcripts when compared to acute lesions or control tissues from healthy subjects (Lock et al., 2002). Additionally, CD8+ T-cells are also believed to play a role in this autoimmune disease as revealed by MS and EAE studies. For example, CD8+ and CD4+ T-cells were found in demyelinating MS lesions (Traugott et al., 1983), CD8+ T-cells predominating and exhibiting oligoclonal expansion within the lesions (Babbe et al., 2000; Monteiro et al., 1996). We have previously shown that CD4+ and CD8+ T-cells responses to neuroantigens could be detected in MS as well as healthy individuals, with slightly higher CD8+ T-cell responses to myelin-associated oligodendrocytic basic protein (MOBP) in MS (Crawford et al., 2004). Studies using transgenic and wildtype myelin-specific CD8+ T-cells have revealed a potential pathogenic or regulatory role in certain models of EAE (Huseby et al., 2001; Steinman, 2001; Sun et al., 2001; York et al., 2010). Our recent studies, both in EAE (York et al., 2010) and human MS (Baughman et al., 2011) demonstrated a novel and unexpected regulatory role for autoreactive, CNS-specific CD8+ T-cells in these diseases.
In an effort to further elucidate the dynamics of CNS-reactive CD4+ and CD8+ T-cells in MS, we evaluated the clonal composition of these neuroantigen-specific T-cells. Earlier studies have evaluated mainly CNS-specific CD4+ T-cell repertoire from both the CNS as well as the periphery, using a combination of different PCR primers to amplify T-cell receptor (TCR) Vβ segments (Gran et al., 1998; Kotzin et al., 1991; Monteiro et al., 1996; Musette et al., 1996; Wucherpfennig et al., 1992), an approach that could suffer from differing reaction efficiencies. Other studies have used antibodies to different Vβ segments (Jacobsen et al., 2002), gaining a global overview of TCR usage. In addition, CDR3 spectratyping studies have been used to measure the Gaussian distribution of CDR3 lengths, with (Matsumoto et al., 2003) or without (Muraro et al., 2006) sequencing of interesting peaks. In the current study, we utilized a novel combination of short-term culture, flow cytometric sorting and a non-biased (anchored) PCR approach, to evaluate the clonal composition of both CD4+ and CD8+ T-cells responsive to two myelin antigens. These studies are the first description of TCR usage of neuroantigen-specific CD8+ T-cells and also make interesting observations regarding TCR usage by myelin-specific CD4+ T-cells in MS, compared to healthy subjects.
2. MATERIALS AND METHODS
2.1. Subject Characteristics
MS patients and healthy controls (HC) were recruited at the UT Southwestern Medical Center and peripherial blood mononuclear cells (PBMC) were obtained by performing leukapheresis under approved IRB protocol. At the time of donation, MS patients had not received steroid treatment in the preceding three months and had never received interferon-β, glatiramer acetate or any disease-modifying immunomodulatory therapy. All MS patients were clinically defined as relapsing remitting MS (RR-MS) and were in remission at the time of leukapheresis. The age (in years) and gender distribution was as follows – MS patients: M584-41/F; M971-47/F; M210-47/F; M250-48/F and healthy controls: H267-48/F; H548-32/M; H333-53/F; H504-36/F. Ficoll-Paque Plus (Amersham Biosciences) separation was used in order to obtain PBMC, which were immediately cryopreserved on the day of collection (Karandikar et al., 2002).
2.2. Myelin Antigens
Because of the diverse specificity of MS immune responses, PBMC were stimulated in vitro using pools of serial 15-mer peptides (overlapping by 10) spanning two entire putative MS antigens (human sequences), myelin basic protein (MBP) and proteolipid protein (PLP) as described before (Crawford et al., 2004). Peptides were dissolved in dimethyl sulfoxide (DMSO), such that cultures contained less than 1 μl DMSO/ml of media.
2.3. CFSE-based flow sorting
MBP and PLP-specific CD4+ and CD8+ T-cells were sorted from CFSE-stained PBMC cultures, as described (Crawford et al., 2004; Karandikar et al., 2002). Briefly, PBMC were first suspended at 1×106/mL in phosphate-buffered saline (PBS) and incubated at 37°C for 7 mins. with 0.25 μM CFSE. Following addition of serum and two PBS washes, cells were resuspended at 2×106/mL in H5 media (RPMI 1640 supplemented with glutamine, 5% human AB serum, penicillin and streptomycin) and cultured in 15-30 ml of media in T25 or T75 flasks (BD Biosciences) with MBP or PLP peptide pools at 10 μg/mL (per 15-mer peptide). On day 7, cells were washed and stained with fluorescently tagged anti-CD4 and anti-CD8 antibodies and sorted by electronic gating into CFSE low (antigen responding) and CFSE high (non-responding), CD4+ and CD8+ T-cell populations using a BD FACSVantage SE sorter. On populations with adequate yields (>200,000 cells), a “post-sort” run was performed revealing >95% purity. Sorted cells were collected in 1.5 ml Sarstedt tubes, pelleted and frozen at −80°C in RNAlater (Ambion, Austin, TX) for subsequent molecular analyses. In previous reports, we have shown that this technique acquires a highly enriched population of antigen-specific, HLA-restricted CD4+ and CD8+ T-cells (Crawford et al., 2004).
2.4. Evaluation of TCR repertoire
As previously described, a detailed evaluation of the clonal repertoire was performed on each sorted antigen-specific T-cell population using an anchored PCR approach (Biegler et al., 2006; Douek et al., 2002) thus allowing for the characterization of endogenous levels of TCR Vβ usage. Total RNA was isolated (RNAEasy kit, Qiagen, Valencia, CA) and a portion used for anchored RT-PCR using a modified version of the Switching Mechanism at the 5′ end of RNA Transcript procedure (SMART Race cDNA Amplification Kit, BD Clonetech). A TCRβ constant region 3′ primer for the PCR was used to obtain TCRβ PCR products from the 5′ end to the start of the TCRβ constant region. The PCR product was ligated into the pGEMT Easy vector (Promega, Madison, WI) and used to transform Escherichia coli (Max Efficiency DH5α, Invitrogen). White colonies were selected, amplified by PCR with M13 primers, and sequenced using the ABI BigDye Terminator V3.1 Cycle Sequencing Kit and sequenced on an ABI 3300 sequencer (ABI, Foster City, CA). Sequences were translated and then defined using the nomenclature from the International ImMunoGeneTics information system® (IMGT, http://imgt.cines.fr; initiator and coordinator: Marie-Paule Lefranc, Montpellier, France) (Lefranc, 2001; Lefranc, 2004). A Basic Local Alignment Search Tool (BLAST) search was conducted to compare dominant clone sequences to published TCR data (http://www.ncbi.nlm.nih.gov/BLAST/).
2.5. Data Analysis
T-cell clonality was assessed by evaluating unique TCR sequences represented in the populations [representation of a single clone at ≥10% was considered significant, as described previously (Biegler et al., 2006)]. Prism 5.0c students' t- test was used to compare the overall distribution of TCR clones between the different groups. Chi-square tests were used to compare distribution across cohorts. p < 0.05 was considered significant, whereas p value between 0.05 and 0.10 was considered a “trend”.
3. RESULTS
3.1. Clonal dominance within MBP-specific CD8+ T-cells in healthy subjects but not MS patients
We evaluated the myelin-specific CD4+ and CD8+ T-cell TCR repertoire in PBMC specimens from MS patients and healthy subjects. As described in prior studies (Biegler et al., 2006; Crawford et al., 2004; Karandikar et al., 2002) we combined flow sorting and CFSE-labeled PBMC in order to obtain a high yield of antigen-specific T-cells. In conjunction with a short term in vitro culture (7 days) and myelin antigen stimulation, we were successfully able to obtain myelin-specific CD4+ and CD8+ T-cells (Crawford et al., 2004; Douek et al., 2002). For molecular analysis of the TCR repertoire, we utilized a constant pair of primers for the anchor and TCRβ constant region in order to amplify the complete TCR in a given population of cells (Biegler et al., 2006; Douek et al., 2002). This method allowed us to circumvent the use of multiple primer pairs and possible differences in amplification efficiencies, thus allowing us to more accurately assess the TCR distribution on a sorted T-cell population.
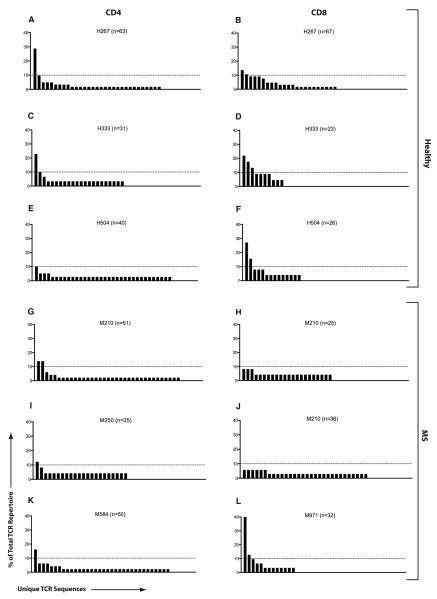
Individual TCR repertoire analysis of MBP-specific CD4+ and CD8+ T-cell responses from three healthy subjects and three treatment-naïve MS patients are depicted in Figure 1. The figure provides a visual depiction of the distribution of specific sequences within the total MBP-specific repertoire. Overall, both untreated MS patients and healthy subjects exhibited a diverse clonotypic distribution within their MBP-specific T-cell responses. MBP-specific CD4+ T-cell responses showed at least one dominant clone (>10% of the total repertoire) in every subject. Overall, clonal dominance was similar between healthy subjects [2.2% (3 prominent sequences out of 134), Figures 1A, C, E] and MS patients [3.1% (4 out of 126), Figures 1G, I, K], p =n.s.. Interestingly, healthy subjects showed a slight trend toward a more focused MBP-specific response in the CD8+ compartment as revealed by 6.0% clonal dominance (7 out of 116, Figures 1B, D, F). In contrast, MBP-specific CD8+ T-cells from MS patients were much more polytypic, similar to their CD4 responses, as revealed by a 2.1% clonal dominance (2 out of 93, Figures 1H, J, L).
Figure 1. TCR repertoire of MBP-specific T-cells in MS and healthy subjects.
PBMC from three healthy subjects (A-F) and three treatment-naïve RRMS patients (G-L) were used to obtain purified populations of MBP-specific CD4+ (left column) and CD8+ (right column) T-cells using CFSE-based flow sorting. TCRβ repertoire was evaluated within each population. The total number of sequences (n) analyzed per population is denoted in the corresponding bar graph. Each bar along the X-axis represents a unique TCRβ sequence and the height of each bar represents its contribution to the total TCR repertoire (% of total). Dotted line is drawn at the 10% level, used as a cutoff for significant contribution by a single clone to the overall response.
3.2. Distinct TCRBV usage in MBP-reactive CD4+ T-cells in MS
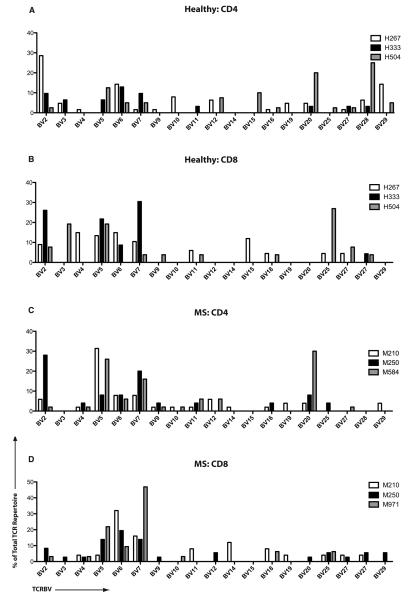
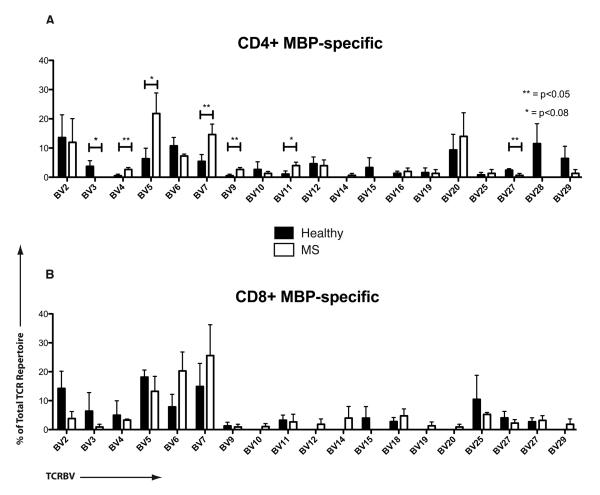
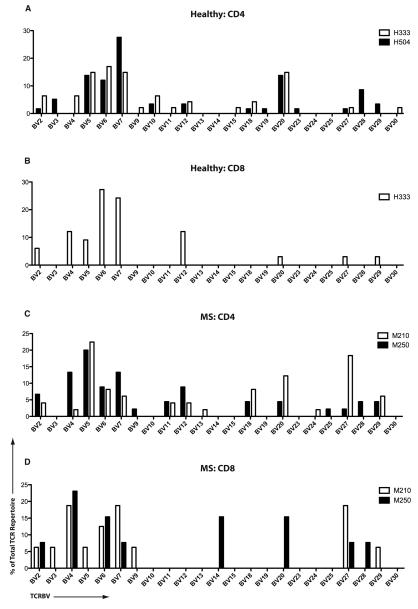
We next evaluated the specific TCR sequences of the antigen-specific clones. Tables 1-4 demonstrate the individual CDR3 sequences, showing clones that were represented at least two times within a total repertoire. Individual sequences of MBP-specific CD4+ and CD8+ TCR were distinct across different MS patients and healthy controls. To gauge the overall variable region β chain usage in MS patients vs. healthy controls, we plotted the contribution of each TCRBV region within the MBP-specific CD4 and CD8 repertoires. Figure 2 demonstrates the prominent TCRBV5 usage in patients M210 and M584. Figure 3 depicts these data in a cumulative manner to compare MS patients and healthy subjects. Interestingly, while there were clonal dominance differences in the MBP-specific CD8+ T-cells (Figure 1), TCRBV usage differences were prominent in the MBP-specific CD4+ T-cell repertoires (Figure 3). Thus, MS patients showed a significantly higher usage of TCRBV4, TCRBV5, TCRBV7, TCRBV9 and TCRBV11 and significantly lower usage of TCRBV3 and TCRBV27 (Figure 3A). Cumulatively, TCRBV usage appeared to show similar distribution within the CD8+ T-cell repertoires of healthy subjects and MS patients (Figure 3B), with only marginal differences.
Table 1.
TCR repertoire of healthy MBP-specific CD4+ T-cells
| Subject | BV locus |
CDR3 | BJ locus |
#a | Total | Freq (%) |
|---|---|---|---|---|---|---|
| H333 | 2 | CASRVGTGAQPQH | 1-5 | 7 | 31 | 22.58 |
| 2 | CASGQGAGDQPQH | 1-5 | 3 | 31 | 9.68 | |
| 6-5 | CASSYSRRGRDTGPYNEQF | 2-1 | 2 | 31 | 6.45 | |
| H267 | 2 | CASISPGSYEQY | 2-7 | 18 | 63 | 28.57 |
| 29-1 | CSVSSYNEQFF | 2-1 | 6 | 63 | 9.52 | |
| 3-1 | CASSQGGGVVTEAF | 1-1 | 3 | 63 | 4.76 | |
| 10-2 | CASSAAARGVGTDTQY | 2-3 | 3 | 63 | 4.76 | |
| 9 | CASSAPDSFYNSPLHF | 1-6 | 3 | 63 | 4.76 | |
| 10-1 | CASSLDGNNEQF | 2-1 | 2 | 63 | 3.17 | |
| 19 | CASKRGMSYNEQF | 2-1 | 2 | 63 | 3.17 | |
| 29-1 | CSVDVGGTDTQYF | 2-3 | 2 | 63 | 3.17 | |
| 28 | CASRESLDTQYF | 2-3 | 2 | 63 | 3.17 | |
| H504 | 28 | CASSLASIYEQY | 2-7 | 4 | 40 | 10.00 |
| 28 | CASSLPARSYNEQF | 2-1 | 2 | 40 | 5.00 | |
| 15 | CATSREAGYYGYT | 1-2 | 2 | 40 | 5.00 | |
| 7-2 | CASSQGRLNTEAF | 1-1 | 2 | 40 | 5.00 |
Data is restricted to those sequences that appeared at least twice within a sorted population.
Indicates the number of instances this sequence was found within the total sequence population.
Table 4.
TCR repertoire of MS MBP-specific CD8+ T-cells
| Subject | BV locus |
CDR3 | BJ locus |
# | Total | Freq (%) |
|---|---|---|---|---|---|---|
| M210 | 6-5 | CASSYDSATGELF | 2-2 | 2 | 25 | 8.00 |
| 18 | CASSPPVIANYGYT | 1-2 | 2 | 25 | 8.00 | |
| 20-1 | CSASYGGFGYT | 1-2 | 2 | 25 | 8.00 | |
| M250 | 2 | CASSEFRG | 1-3 | 2 | 36 | 5.56 |
| 5-1 | CASSHGLAEP | 2-7 | 2 | 36 | 5.56 | |
| 6-2 | CASRTHPGQL | 2-7 | 2 | 36 | 5.56 | |
| 7-9 | CASSLALTVM | 1-1 | 2 | 36 | 5.56 | |
| 16 | CASSSAFNSYNQF | 2-1 | 2 | 36 | 5.56 | |
| 30 | CAWGTGADYGYT | 1-4 | 2 | 36 | 5.56 | |
| M971 | 7-2 | CASSLEGISTDTQY | 2-1 | 13 | 32 | 40.63 |
| 5-1 | CASSLAGQGVNTEAF | 1-1 | 4 | 32 | 12.50 | |
| 6-2 | CASSLTRTYEQY | 2-7 | 3 | 32 | 9.38 | |
| 5-1 | CASSTRTGSGTEAF | 1-1 | 2 | 32 | 6.25 | |
| 27 | CASSYLEIQGLKNIQY | 2-4 | 2 | 32 | 6.25 |
Figure 2. TCRBV region gene usage in MBP-specific T-cells.
TCRBV gene usage by MBP-specific CD4+ and CD8+ T-cells from three healthy subjects (A-B) and three MS patients (C-D) is depicted. The X-axis represents unique TCRBV genes while the Y-axis plots the contribution of each gene (percentage) within the antigen-specific repertoire.
Figure 3. Cumulative TCRBV usage in MBP-specific T-cells.
These graphs represent cumulative data from Figure 3, depicting overall TCR usage among MBP-specific CD4+ (A) and CD8+ (B) T-cells from healthy subjects vs. MS patients (*p<0.08; **p<0.05).
3.3. Focused clonal composition of PLP-specific CD8+, unlike CD4+ T-cells, in MS
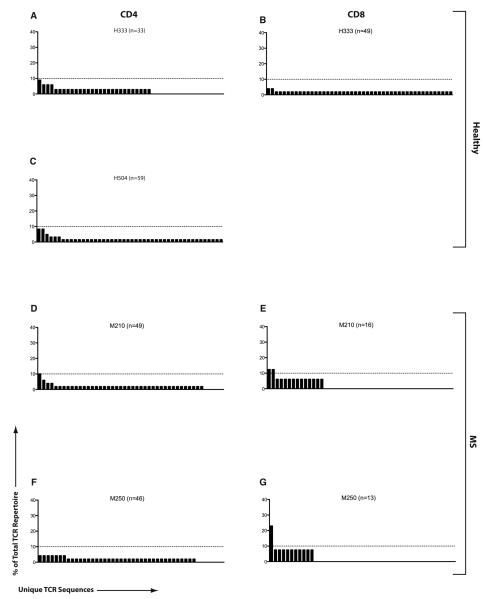
In contrast to MBP-specific T-cell responses, PLP-specific CD4+ T-cells exhibited a broadly polyclonal TCR repertoire (Figure 4 and Supplemental Tables 1-4). The PLP-specific CD4+ and CD8+ TCR repertoire possessed no unique dominating sequence in healthy subjects. MS TCR analysis revealed only one dominating CD4 sequence (1.0%* or 1 out of 95, Figure 4D), whereas PLP-CD8+ TCR repertoire revealed a significantly more focused response as revealed by a 10.3%* clonal dominance (3 out of 29), *p=0.01 (Figures 4E, G). We had difficulty getting adequate evaluable PLP-specific CD8 responses from healthy subjects and thus, comparison between healthy subjects and MS patients was not possible.
Figure 4. TCR repertoire of PLP-specific T-cells in MS and healthy subjects.
The TCRβ repertoire was evaluated in sorted populations of PLP-specific CD4+ and CD8+ T-cells from two healthy subjects and two MS patients. The figure is structured similar to Figure 1 (H504 did not have an evaluable CD8 response to PLP).
TCRBV analysis of the PLP-specific T-cell repertoire revealed broad usage across MS patients and healthy controls (Figure 5), with no striking usage differences within the CD4 repertoires. Due to limited evaluation of the PLP-specific CD8 responses within our healthy subjects, we are unable to comment on their comparative attributes.
Figure 5. TCRBV region gene usage in PLP-specific T-cells.
TCRBV gene usage by PLP-specific CD4+ and CD8+ T-cells from two healthy subjects (A-B) and two MS patients (C-D) is depicted (Figure structured similar to Figure 3).
3.4. Comparison of MBP- and PLP-specific TCR with published TCR sequences
Finally, we compared our set of CNS-specific TCR sequences to published sequences using open BLAST sequence analysis. A sequence was determined to be similar when at least four matching amino acids were located within the -NDN- hypervariable region, along with the same TCRBV and TCRJV usage.
Table 5 shows matches found against sequences derived from MS patients in our study. Interestingly, among all the published TCR sequences (not selected for any specific disease), the overwhelming majority of matches were found in sequences from other MS patients. Four positive matches were found among the published sequences from micro-dissected CNS-infiltrating CD8+ T-cells taken from active lesions of MS patients (Babbe et al., 2000). This prior study found oligoclonal expansion of CNS-infiltrating CD8+ T-cells, suggesting an important function for these cells in the MS lesion. One match was found among a set of published TCR from a study of Chronic Encephalitis of Rasmussen (Li et al., 1997). In this study, TCR sequences were analyzed among brain samples from seven patients, three MS patients, two viral encephalitis cases, four cases of medically refractory partial epilepsy and two cases with no detectable inflammatory neurologic condition. The only sequence found matching a myelin antigen-specific TCR was from one of the MS patients. The N-D-N sequence (GLAGG) also matched sequences published in other studies unrelated to MS including one of our own sequences from healthy subject H333, suggesting a more “public” CDR3 sequence (Ria et al., 2001). Of note, when TCR sequences from healthy subjects in our study were used to query the Genebank records, no matches were found that originated from any MS-related studies. Near matches were detected from other diseases such as chronic myeloid leukemia and non-Hodgkin lymphoma (data not shown), the significance of which is unclear.
Table 5.
Matching TCR clones with published TCR sequences
| Source clone | BV locus | CDR3 | BJ locus |
|---|---|---|---|
| CD8+ MBP from M971 (case 1) | 5-1 | SALYLCASSLAGQGVNTEAFFGQ | 1-1 |
| CD8+ active MS lesion (Babbe et al., 2000) | 5-1 | SALYLCASSLAGQGYTGELFFGE | 2-2 |
| CD8+ active MS lesion (Babbe et al., 2000) | 7-9 | SAMYLCASSLSGQGDYGYTFGS | 1-2 |
| CD8+ MBP from M971 (case 2) | 5-1 | SALYLCASSTRTGSGTEAFFGQ | 1-1 |
| CD8+ active MS lesion (Babbe et al., 2000) | 5-4 | SALYLCASSSRTGSVSYEQYFGP | 2-1 |
| CD4+ MBP from M250 | 2 | SAMYFCASRVGTGA---QPQHFGD | 1-5 |
| CD8+ active MS lesion (Babbe et al., 2000) | 6-6 | TSVYFCASRPGTGASNQPQHFGD | 1-5 |
| CD4+ PLP from M210 | 18 | SAAYFCASSPGLAGGPSYNEQFFGP | 2-1 |
| Encephalitis study control MS patient (Li et al., 1997) |
6-5 | TSVYFCASSPGLAGGPNEQYFGP | 2-1 |
| CD4+ PLP from H333 | 27 | TSLYFCASS--GLAGG-ADTQYFGP | 2-3 |
| Synovial fluid T-cell in Arthritis patient (Striebich et al., 1998) |
27 | TSLYFCASSSPGLAGGTDTQYFGP | 2-3 |
| Infiltrating Lym. in calcified aortic stenosis (Wu et al., 2007) |
27 | TSLYFCASSPGLAGGPTDTQYFGP | 2-3 |
4. DISCUSSION
In this study, we characterized the clonal composition of myelin-specific T-cells in MS patients by performing an unbiased anchored PCR assay on the TCR BV chains of sorted CD4+ and CD8+ T-cells. While there have been prior descriptions of TCR usage by myelin-specific T-cells in MS, most of these are based either on long-term T-cell lines or on analysis methods that utilize CDR3 spectratyping (length) followed by detailed sequencing only of “major” peaks. Several unique features of our approach improve upon prior analyses. First, we utilized flow-cytometric sorting of CFSE-labeled PBMC cultured for seven days with pools of myelin peptide fragments spanning the entire length of both MBP and PLP. Thus, after relatively short-term cultures, we were able to obtain antigen-specific CD4+ and CD8+ T-cells from MS patients and healthy subjects. We have shown previously that these sorted cells represent highly enriched, HLA-restricted, antigen-specific T-cell populations (Crawford et al., 2004). Second, by utilizing SMART-Race cDNA amplification in which an anchored RT-PCR is performed using a SMART switching mechanism in conjunction with a TCRβ constant region primer for the PCR we obviated the need for multiple TCR Vβ family primer sets and avoided any bias that may have arisen due to differing reaction efficiencies (Biegler et al., 2006; Douek et al., 2002). Third, the direct sequencing of the CDR3 regions allowed us to formulate the clearest picture of the clonal distribution of myelin-specific T-cells without any presumption regarding the true sequence of the CDR3 region, based simply on TCRBV usage and CDR3 length. Indeed, numerous instances can be found where the N-D-N regions are of the same length, but contain completely different amino acid sequences. Fourth, with regard to the circulating peripheral population of T-cells in MS patients, the clones of interest may not always be located in the dominant peak of a spectratype. Thus, we were able to generate a dataset containing over 500 TCR sequences from myelin-specific T-cells. Finally, our approach has allowed us to provide the first detailed descriptions of myelin antigen-specific CD8 TCR sequences from MS patients and healthy subjects.
In general, populations of CD4+ T-cells responding to MBP were polyclonal. Healthy subjects and the MS patients showed a few clones slightly dominant amongst the polyclonal response. PLP-specific CD4+ T-cells were also polyclonal with none of the clones having more than 10% representation in any MS patient or healthy subject's response. In contrast, MBP-specific CD8+ T-cells from all three healthy subjects were less polyclonal than their CD4+ counterparts, showing clear prominence of some clones. This finding correlates with previous reports showing healthy subjects with oligoclonality in their CD8+ T-cell populations (Batliwalla et al., 1996). The MBP-specific CD8+ T-cells from MS patients (especially M210 and M250) were truly polyclonal, similar to our prior finding that glatiramer acetate (GA)-reactive CD8+ T-cells from MS patients are polyclonal, distinct from those of healthy subjects (Biegler et al., 2006). This difference in clonal distribution is intriguing, especially in the context that the functional role of these cells could also be distinct between MS patients vs. healthy subjects. Prior studies from us have shown a mixed functional profile of MS CNS-specific CD8+ T cells (Crawford et al., 2004). In recent studies, we have shown that CNS-specific CD8+ T-cells appear to have an unexpected and clinically significant immune regulatory role during CNS demyelinating disease, both in human MS as well as EAE (Baughman et al., 2011; York et al., 2010). In that regard myelin-specific CD8+ T-cells behave functionally similar to GA-specific CD8+ T-cells (Tennakoon et al., 2006). Thus, the oligoclonal myelin-specific CD8+ T-cells from healthy subjects may have robust immune regulatory properties, which are lacking in the polyclonally distributed CD8+ T-cells of MS patients, especially during clinical relapses (Baughman et al., 2011). Therefore, while we did not observe specific TCRBV usage differences in the CNS-specific CD8+ T-cell responses between healthy subjects vs. MS patients, just the differences in the clonal distribution may potentially indicate the lack of an important regulatory CD8+ T-cell subset in baseline MS. While prior studies have found some global CDR3 distribution differences in CD8+ T-cells in MS patients vs. healthy subjects (Laplaud et al., 2004), this difference does not seem to represent CNS-specific CD8+ T-cells. These findings correlate with a previous study finding little or no difference in the clonal composition of myelin antigen-reactive T-cells between MS patients and healthy subjects (Hafler et al., 1996). Possibly a more detailed breakdown of the CD8+ T-cell population would provide more insights, such as one performed by Somma et al., where they found a skewed repertoire in 3 pairs of MS discordant twins when they divided the CD8+ T-cell fraction into CCR7 negative (effector memory) and CCR7 positive (central memory, naïve) fractions (Somma et al., 2007).
Reports of skewed Vβ usage in MS patients have been controversial. No difference in peripheral blood TCR Vβ chain expression was found between MS patients and healthy subjects when investigated with fluorochrome conjugated antibodies to Vβ segments (Jacobsen et al., 2002). However, two studies confirmed skewed repertoires with respect to Vβ23 by using other methods (Monteiro et al., 1996; Somma et al., 2007). Other studies have reported that Vβ5-family CDR3 gene segments were oligoclonally expanded in MS patients when compared to healthy subjects (Bourdette et al., 1994; Matsumoto et al., 2003). Another recent study performed laser micro-dissection of brain autopsy specimens from multiple sclerosis patients and found expanded Vβ5 family TCR from CD8+ T-cells in 3 of 4 patients studied (Junker et al., 2007). In our findings, MBP-stimulated CD4+ T-cells from MS patients showed high levels of clonally expanded BV5 family TCR (21.7%) when compared to their healthy counterparts (6.3%). When looking at TCRBV5 family usage among the PLP-reactive T-cells we found an increase in the CD4+ T-cells from patient M210 (21.2%) when compared to the CD4+ T-cells from healthy control H333 (14%). Thus, the higher representation of specific CD4+ T-cell clones may reflect the inability of regulatory mechanisms to control the expansion of pathogenic clones in the context of MS.
Using our newly generated TCR database, we were able to query the Genebank database for matches of interest. Although MS patients and healthy subjects possess myelin-specific T-cell responses at comparable magnitudes (Crawford et al., 2004), the TCR involved in these responses are quite distinct, in that TCR from healthy subjects did not generate any near-matches in previously published MS patients. In contrast, a number of TCR sequences from MS patients showed matches with sequences obtained from MS patients in prior studies (Babbe et al., 2000). In fact, even matches that we deemed marginal (3 consecutive amino acids inside the hyper-variable region) were not found between our healthy cohort and previous MS sequences.
Interestingly, patient M971 possessed MBP-reactive CD8+ T-cells that have very similar TCR sequences to CNS-infiltrating CD8+ T-cells found in micro-dissected lesions from the study by Babbe et al (Babbe et al., 2000). The clone containing the N-D-N sequence LAGQG (Table 9) was a close match for two clones from the Babbe et al., study, one of which matched the BV5.1 used and one that did not. This particular clone represented 12.5% of total clones (4/32). A second clone with N-D-N sequence TRTGSG was a close match for yet another clone from the same previous study and both used BV5 family gene segments. This clone represented 6.25% of total sequences (2/32). We believe these findings of near-matching TCR from CD8+ T-cells is both very rare and quite interesting when considered in light of other observations such as those made in a previous study by Skulina et al. in which they found clonally expanded CD8+ T-cells in MS brain lesions. These exact clones were also found in the peripheral blood and were found to persist for years (Skulina et al., 2004). The precise function of such expanded clones during the disease process (pathogenic vs. regulatory) will be of important consequence in understanding the underlying pathogenesis and devising better immunotherapeutic strategies in the future.
To summarize, we combined short term in vitro culture and flow-based sorting to isolate pure populations of myelin-specific CD4+ and CD8+ T-cells to evaluate TCR clonal composition using an unbiased approach. Our results demonstrate distinct TCR usage by MS patients in their polyclonal CNS-specific CD4+ T-cell responses. In contrast to MS patients, healthy subjects exhibit oligoclonal CD8+ TCR distribution, which may reflect important differences in overall regulatory ability between MS patients vs. healthy controls. Future studies are needed to address the functional role of these specific TCRBV myelin-specific T-cells and address the question of whether these cells are playing a part in the pathogenic process of this autoimmune disease.
Supplementary Material
Table 2.
TCR repertoire of healthy MBP-specific CD8+ T-cells.
| Subject | BV locus |
CDR3 | BJ locus |
# | Total | Freq (%) |
|---|---|---|---|---|---|---|
| H333 | 5-1 | CASSLEGRPDHEQY | 2-7 | 5 | 23 | 21.74 |
| 2 | CASSYSQGGTEAF | 1-1 | 4 | 23 | 17.39 | |
| 11 | CASSPYTDTQY | 2-1 | 3 | 23 | 13.04 | |
| 6-5 | CASSYADEQY | 2-7 | 2 | 23 | 8.70 | |
| 2 | CASKEVAGGRYTGELF | 2-2 | 2 | 23 | 8.70 | |
| 7-8 | CASSFRGDPFYGTY | 1-2 | 2 | 23 | 8.70 | |
| 7-9 | CASSLLDRGGGSTIY | 1-3 | 2 | 23 | 8.70 | |
| H267 | 5-1 | CASSLAGGYSPLH | 1-6 | 9 | 67 | 13.43 |
| 4-1 | CASSRTTSSSYNEQF | 2-1 | 7 | 67 | 10.45 | |
| 2 | CASSDRGVGTGELF | 2-2 | 6 | 67 | 8.96 | |
| 6-5 | CASSYRTSQVINSPLH | 1-6 | 6 | 67 | 8.96 | |
| 19 | CASIKRTLPGASNTEAF | 1-1 | 6 | 67 | 8.96 | |
| 7-4 | CASSLFVSGDSPLH | 1-6 | 5 | 67 | 7.46 | |
| 12-3 | CASSPGLAGYEQF | 2-1 | 3 | 67 | 4.48 | |
| 27 | CASSSWDTDSPLH | 1-6 | 3 | 67 | 4.48 | |
| 24 | CASSLQGESGPLH | 1-6 | 3 | 67 | 4.48 | |
| 28 | CASSATGVIHNEQF | 2-1 | 2 | 67 | 2.99 | |
| 6-2 | CASSYGTQGQY | 2-7 | 2 | 67 | 2.99 | |
| 20-1 | CSARDLGDSNSPLH | 1-6 | 2 | 67 | 2.99 | |
| 4-2 | CASSGYRGGNQPQH | 1-5 | 2 | 67 | 2.99 | |
| H504 | 27 | CASSSGLIRF | 2-1 | 7 | 26 | 26.92 |
| 3-1 | CASSRDSGFKDTQY | 2-5 | 4 | 26 | 15.38 | |
| 5-5 | CASSLETRGTNEQF | 2-1 | 2 | 26 | 7.69 | |
| 2 | CASRVGTGAQPQH | 1-5 | 2 | 26 | 7.69 | |
| 5-1 | CASSLAGSGYGYT | 1-2 | 2 | 26 | 7.69 |
Table 3.
TCR repertoire of MS MBP-specific CD4+ T-cells.
| Subject | BV locus |
CDR3 | BJ locus |
# | Total | Freq (%) |
|---|---|---|---|---|---|---|
| M210 | 27 | CASSLGGGGTEQFGGG | 1-1 | 7 | 51 | 13.73 |
| 5-1 | CASSPGSLSRETQY | 2-5 | 7 | 51 | 13.73 | |
| 5-1 | CASSLVLGGKNYGYT | 1-2 | 3 | 51 | 5.88 | |
| 2 | CASSEQNTQY | 2-1 | 2 | 51 | 3.92 | |
| 6-6 | CASSPGGRNEQF | 2-1 | 2 | 51 | 3.92 | |
| M250 | 2 | CASRVGTGAQPQH | 1-5 | 3 | 25 | 12.00 |
| 2 | CASGQGAQDQPQH | 1-5 | 2 | 25 | 8.00 | |
| M584 | 20-1 | CSATVGAGTYEQY | 2-7 | 8 | 50 | 16.00 |
| 20-1 | CSVSDRNNEQF | 2-1 | 3 | 50 | 6.00 | |
| 11-3 | CAPRTKIRANRAF | 1-1 | 3 | 50 | 6.00 | |
| 5-1 | CASSLAWGDTEAF | 1-1 | 3 | 50 | 6.00 | |
| 5-1 | CASSASRTGNTEAF | 1-1 | 2 | 50 | 4.00 | |
| 5-7 | CASSFYREAF | 1-1 | 2 | 50 | 4.00 | |
| 7-9 | CASSHTDRGRGNTIY | 1-3 | 2 | 50 | 4.00 |
ACKNOWLEDGEMENTS
These studies were supported, in part, by USPHS NIH grants AI053439, AI065463, AI079272, NS037513 and Grant JF2118-A-2 (Harry Weaver Neuroscience Scholar Award) from the National MS Society. The authors would like to thank Becky Price, RN, for performing the leukaphereses and Bonnie Darnell for technical assistance with cell sorting. We would also like to acknowledge Drs. Chris L. Ayers and Jason P. Mendoza for manuscript review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Deposition: Sequence data presented in the tables have been submitted to the GenBank database (Accession nos. HM236502-HM236814; HM467839-HM467908).
REFERENCES
- Babbe H, Roers A, Waisman A, Lassmann H, Goebels N, Hohlfeld R, Friese M, Schroder R, Deckert M, Schmidt S, Ravid R, Rajewsky K. Clonal expansions of CD8(+) T cells dominate the T cell infiltrate in active multiple sclerosis lesions as shown by micromanipulation and single cell polymerase chain reaction. J Exp Med. 2000;192:393–404. doi: 10.1084/jem.192.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos LF, Sawcer S, Ramsay PP, Baranzini SE, Thomson G, Briggs F, Cree BC, Begovich AB, Villoslada P, Montalban X, Uccelli A, Savettieri G, Lincoln RR, DeLoa C, Haines JL, Pericak-Vance MA, Compston A, Hauser SL, Oksenberg JR. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15:2813–2824. doi: 10.1093/hmg/ddl223. [DOI] [PubMed] [Google Scholar]
- Batliwalla F, Monteiro J, Serrano D, Gregersen PK. Oligoclonality of CD8+ T cells in health and disease: aging, infection, or immune regulation? Hum Immunol. 1996;48:68–76. doi: 10.1016/0198-8859(96)00077-8. [DOI] [PubMed] [Google Scholar]
- Baughman EJ, Mendoza JP, Ortega SB, Ayers CL, Greenberg BM, Frohman EM, Karandikar NJ. Neuroantigen-specific CD8+ regulatory T-cell function is deficient during acute exacerbation of multiple sclerosis. J Autoimmun. 2011 doi: 10.1016/j.jaut.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegler BW, Yan SX, Ortega SB, Tennakoon DK, Racke MK, Karandikar NJ. Glatiramer acetate (GA) therapy induces a focused, oligoclonal CD8+ T-cell repertoire in multiple sclerosis. J Neuroimmunol. 2006;180:159–171. doi: 10.1016/j.jneuroim.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Bourdette DN, Whitham RH, Chou YK, Morrison WJ, Atherton J, Kenny C, Liefeld D, Hashim GA, Offner H, Vandenbark AA. Immunity to TCR peptides in multiple sclerosis. I. Successful immunization of patients with synthetic V beta 5.2 and V beta 6.1 CDR2 peptides. J Immunol. 1994;152:2510–2519. [PubMed] [Google Scholar]
- Crawford MP, Yan SX, Ortega SB, Mehta RS, Hewitt RE, Price DA, Stastny P, Douek DC, Koup RA, Racke MK, Karandikar NJ. High prevalence of autoreactive, neuroantigen-specific CD8+ T cells in multiple sclerosis revealed by novel flow cytometric assay. Blood. 2004;103:4222–4231. doi: 10.1182/blood-2003-11-4025. [DOI] [PubMed] [Google Scholar]
- Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai KL, Karandikar NJ, Casazza JP, Koup RA. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- Einstein ER, Robertson DM, Dicaprio JM, Moore W. The isolation from bovine spinal cord of a homogeneous protein with encephalitogenic activity. J Neurochem. 1962;9:353–361. doi: 10.1111/j.1471-4159.1962.tb09461.x. [DOI] [PubMed] [Google Scholar]
- Gran B, Gestri D, Sottini A, Quiros Roldan E, Bettinardi A, Signorini S, Primi D, Ballerini C, Taiuti R, Amaducci L, Massacesi L. Detection of skewed T-cell receptor V-beta gene usage in the peripheral blood of patients with multiple sclerosis. J Neuroimmunol. 1998;85:22–32. doi: 10.1016/s0165-5728(97)00250-6. [DOI] [PubMed] [Google Scholar]
- Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, Caillier SJ, Ban M, Goris A, Barcellos LF, Lincoln R, McCauley JL, Sawcer SJ, Compston DA, Dubois B, Hauser SL, Garcia-Blanco MA, Pericak-Vance MA, Haines JL. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- Gutcher I, Becher B. APC-derived cytokines and T cell polarization in autoimmune inflammation. J Clin Invest. 2007;117:1119–1127. doi: 10.1172/JCI31720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- Hafler DA, Saadeh MG, Kuchroo VK, Milford E, Steinman L. TCR usage in human and experimental demyelinating disease. Immunol Today. 1996;17:152–159. doi: 10.1016/0167-5699(96)80611-6. [DOI] [PubMed] [Google Scholar]
- Huseby ES, Liggitt D, Brabb T, Schnabel B, Ohlen C, Goverman J. A pathogenic role for myelin-specific CD8(+) T cells in a model for multiple sclerosis. J Exp Med. 2001;194:669–676. doi: 10.1084/jem.194.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen M, Cepok S, Quak E, Happel M, Gaber R, Ziegler A, Schock S, Oertel WH, Sommer N, Hemmer B. Oligoclonal expansion of memory CD8+ T cells in cerebrospinal fluid from multiple sclerosis patients. Brain. 2002;125:538–550. doi: 10.1093/brain/awf059. [DOI] [PubMed] [Google Scholar]
- Junker A, Ivanidze J, Malotka J, Eiglmeier I, Lassmann H, Wekerle H, Meinl E, Hohlfeld R, Dornmair K. Multiple sclerosis: T-cell receptor expression in distinct brain regions. Brain. 2007;130:2789–2799. doi: 10.1093/brain/awm214. [DOI] [PubMed] [Google Scholar]
- Karandikar NJ, Crawford MP, Yan X, Ratts RB, Brenchley JM, Ambrozak DR, Lovett-Racke AE, Frohman EM, Stastny P, Douek DC, Koup RA, Racke MK. Glatiramer acetate (Copaxone) therapy induces CD8(+) T cell responses in patients with multiple sclerosis. J Clin Invest. 2002;109:641–649. doi: 10.1172/JCI14380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin BL, Karuturi S, Chou YK, Lafferty J, Forrester JM, Better M, Nedwin GE, Offner H, Vandenbark AA. Preferential T-cell receptor beta-chain variable gene use in myelin basic protein-reactive T-cell clones from patients with multiple sclerosis. Proc Natl Acad Sci U S A. 1991;88:9161–9165. doi: 10.1073/pnas.88.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaud DA, Ruiz C, Wiertlewski S, Brouard S, Berthelot L, Guillet M, Melchior B, Degauque N, Edan G, Brachet P, Damier P, Soulillou JP. Blood T-cell receptor beta chain transcriptome in multiple sclerosis. Characterization of the T cells with altered CDR3 length distribution. Brain. 2004;127:981–995. doi: 10.1093/brain/awh119. [DOI] [PubMed] [Google Scholar]
- Lebar R, Lubetzki C, Vincent C, Lombrail P, Boutry JM. The M2 autoantigen of central nervous system myelin, a glycoprotein present in oligodendrocyte membrane. Clin Exp Immunol. 1986;66:423–434. [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2001;29:207–209. doi: 10.1093/nar/29.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP. IMGT, The International ImMunoGeneTics Information System, http://imgt.cines.fr. Methods Mol Biol. 2004;248:27–49. doi: 10.1385/1-59259-666-5:27. http://imgt.cines.fr [DOI] [PubMed] [Google Scholar]
- Li Y, Uccelli A, Laxer KD, Jeong MC, Vinters HV, Tourtellotte WW, Hauser SL, Oksenberg JR. Local-clonal expansion of infiltrating T lymphocytes in chronic encephalitis of Rasmussen. J Immunol. 1997;158:1428–1437. [PubMed] [Google Scholar]
- Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, Langer-Gould A, Strober S, Cannella B, Allard J, Klonowski P, Austin A, Lad N, Kaminski N, Galli SJ, Oksenberg JR, Raine CS, Heller R, Steinman L. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med. 2002;8:500–508. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- Martin R, McFarland HF, McFarlin DE. Immunological aspects of demyelinating diseases. Annu Rev Immunol. 1992;10:153–187. doi: 10.1146/annurev.iy.10.040192.001101. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Yoon WK, Jee Y, Fujihara K, Misu T, Sato S, Nakashima I, Itoyama Y. Complementarity-determining region 3 spectratyping analysis of the TCR repertoire in multiple sclerosis. J Immunol. 2003;170:4846–4853. doi: 10.4049/jimmunol.170.9.4846. [DOI] [PubMed] [Google Scholar]
- Monteiro J, Hingorani R, Peroglizzi R, Apatoff B, Gregersen PK. Oligoclonality of CD8+ T cells in multiple sclerosis. Autoimmunity. 1996;23:127–138. doi: 10.3109/08916939608995336. [DOI] [PubMed] [Google Scholar]
- Muraro PA, Cassiani-Ingoni R, Chung K, Packer AN, Sospedra M, Martin R. Clonotypic analysis of cerebrospinal fluid T cells during disease exacerbation and remission in a patient with multiple sclerosis. J Neuroimmunol. 2006;171:177–183. doi: 10.1016/j.jneuroim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Musette P, Bequet D, Delarbre C, Gachelin G, Kourilsky P, Dormont D. Expansion of a recurrent V beta 5.3+ T-cell population in newly diagnosed and untreated HLA-DR2 multiple sclerosis patients. Proc Natl Acad Sci U S A. 1996;93:12461–12466. doi: 10.1073/pnas.93.22.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettinelli CB, McFarlin DE. Adoptive transfer of experimental allergic encephalomyelitis in SJL/J mice after in vitro activation of lymph node cells by myelin basic protein: requirement for Lyt 1+ 2− T lymphocytes. J Immunol. 1981;127:1420–1423. [PubMed] [Google Scholar]
- Renno T, Krakowski M, Piccirillo C, Lin JY, Owens T. TNF-alpha expression by resident microglia and infiltrating leukocytes in the central nervous system of mice with experimental allergic encephalomyelitis. Regulation by Th1 cytokines. J Immunol. 1995;154:944–953. [PubMed] [Google Scholar]
- Ria F, van den Elzen P, Madakamutil LT, Miller JE, Maverakis E, Sercarz EE. Molecular characterization of the T cell repertoire using immunoscope analysis and its possible implementation in clinical practice. Curr Mol Med. 2001;1:297–304. doi: 10.2174/1566524013363690. [DOI] [PubMed] [Google Scholar]
- Skulina C, Schmidt S, Dornmair K, Babbe H, Roers A, Rajewsky K, Wekerle H, Hohlfeld R, Goebels N. Multiple sclerosis: brain-infiltrating CD8+ T cells persist as clonal expansions in the cerebrospinal fluid and blood. Proc Natl Acad Sci U S A. 2004;101:2428–2433. doi: 10.1073/pnas.0308689100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma P, Ristori G, Battistini L, Cannoni S, Borsellino G, Diamantini A, Salvetti M, Sorrentino R, Fiorillo MT. Characterization of CD8+ T cell repertoire in identical twins discordant and concordant for multiple sclerosis. J Leukoc Biol. 2007;81:696–710. doi: 10.1189/jlb.0906584. [DOI] [PubMed] [Google Scholar]
- Steinman L. Myelin-specific CD8 T cells in the pathogenesis of experimental allergic encephalitis and multiple sclerosis. J Exp Med. 2001;194:F27–30. doi: 10.1084/jem.194.5.f27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebich CC, Falta MT, Wang Y, Bill J, Kotzin BL. Selective accumulation of related CD4+ T cell clones in the synovial fluid of patients with rheumatoid arthritis. J Immunol. 1998;161:4428–4436. [PubMed] [Google Scholar]
- Sun D, Whitaker JN, Huang Z, Liu D, Coleclough C, Wekerle H, Raine CS. Myelin antigen-specific CD8+ T cells are encephalitogenic and produce severe disease in C57BL/6 mice. J Immunol. 2001;166:7579–7587. doi: 10.4049/jimmunol.166.12.7579. [DOI] [PubMed] [Google Scholar]
- Tennakoon DK, Mehta RS, Ortega SB, Bhoj V, Racke MK, Karandikar NJ. Therapeutic induction of regulatory, cytotoxic CD8+ T cells in multiple sclerosis. J Immunol. 2006;176:7119–7129. doi: 10.4049/jimmunol.176.11.7119. [DOI] [PubMed] [Google Scholar]
- Traugott U, Reinherz EL, Raine CS. Multiple sclerosis: distribution of T cell subsets within active chronic lesions. Science. 1983;219:308–310. doi: 10.1126/science.6217550. [DOI] [PubMed] [Google Scholar]
- Tuohy VK, Lu ZJ, Sobel RA, Laursen RA, Lees MB. A synthetic peptide from myelin proteolipid protein induces experimental allergic encephalomyelitis. J Immunol. 1988;141:1126–1130. [PubMed] [Google Scholar]
- Wekerle H. Immunopathogenesis of multiple sclerosis. Acta Neurol (Napoli) 1991;13:197–204. [PubMed] [Google Scholar]
- Wu HD, Maurer MS, Friedman RA, Marboe CC, Ruiz-Vazquez EM, Ramakrishnan R, Schwartz A, Tilson MD, Stewart AS, Winchester R. The lymphocytic infiltration in calcific aortic stenosis predominantly consists of clonally expanded T cells. J Immunol. 2007;178:5329–5339. doi: 10.4049/jimmunol.178.8.5329. [DOI] [PubMed] [Google Scholar]
- Wucherpfennig KW, Newcombe J, Li H, Keddy C, Cuzner ML, Hafler DA. T cell receptor V alpha-V beta repertoire and cytokine gene expression in active multiple sclerosis lesions. J Exp Med. 1992;175:993–1002. doi: 10.1084/jem.175.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York NR, Mendoza JP, Ortega SB, Benagh A, Tyler AF, Firan M, Karandikar NJ. Immune regulatory CNS-reactive CD8+T cells in experimental autoimmune encephalomyelitis. J Autoimmun. 2010;35:33–44. doi: 10.1016/j.jaut.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.