Abstract
Ionotropic glutamate receptors (iGluRs) are ligand gated ion channels which mediate excitatory synaptic transmission in the brain of vertebrates. A rapidly growing body of crystal structures for isolated iGluR extracellular domains, and more recently a full length AMPA receptor, combined with data from electrophysiological experiments and MD simulations, provides a framework which makes it possible to investigate the molecular basis for assembly, gating and modulation. These unprecedented advances in structural biology are constantly challenged by novel functional properties which emerge despite decades of functional analysis, and by a growing family of auxiliary proteins which modulate iGluR activity and assembly.
Introduction
In the vertebrate CNS three families of glutamate-gated ion channels (iGluRs) named AMPA, kainate and NMDA receptors mediate signal transduction at excitatory synapses. iGluRs are tetrameric proteins encoded by 18 genes, many of which undergo alternative splicing and RNA editing [1]. A rapidly emerging structural biology of iGluRs is leading towards a molecular understanding of mechanisms controlling their assembly, gating and modulation by accessory proteins and drugs [2]. The ability to break down the function of individual iGluRs into discrete, semi-autonomous domains, and to analyze the effects of subunit composition and mutations on the functional properties of receptor subtypes, within the context of structural information provided by X-ray crystallography, underlies much of this progress [3]. The crowning achievement of recent work is the crystal structure of a full length AMPA receptor [4••]. Other major advances revealed an unexpected role of the NMDA receptor amino terminal domain in regulating ion channel activity; defined the initial stages of receptor assembly; and identified new families of auxiliary subunits. But, to paraphrase Winston Churchill, these results mark only the end of the beginning and we are still far from understanding many basic aspects of the molecular biology of iGluRs.
Tetrameric structure of the GluA2 AMPA receptor
Any doubts about the stoichiometry of iGluR assembly have been cast aside by the structure of the rat GluA2 AMPA receptor ion channel tetramer (Figure 1), crystallized in its resting state in a complex with the competitive antagonist ZK200775 [4••]. This structure marks the most significant advance in the study of iGluR signal transduction since the cloning of individual iGluR subunits. The structure was solved using X-ray diffraction data to 3.6 Å resolution, which is not sufficient to build side chains without additional information, but the availability, as search probes for molecular replacement, of higher resolution structures for the isolated GluA2 amino terminal (ATD) and ligand binding (LBD) domains removes many of the problems associated with de novo model building at low resolution. Highly significant was the collection of anomalous diffraction data to verify alpha helix registers and symmetry switches in the ion channel pore for which no prior structural data was available. This was done by preparing Selenomethionine labeled protein for wild type GluA2, and four different mutants in which methionine residues were introduced into different positions in the transmembrane pore. This is notable because structural studies using prokaryotic membrane proteins labeled with heavy atoms, which are much easier to express, continue to dominate recent entries in the protein data bank.
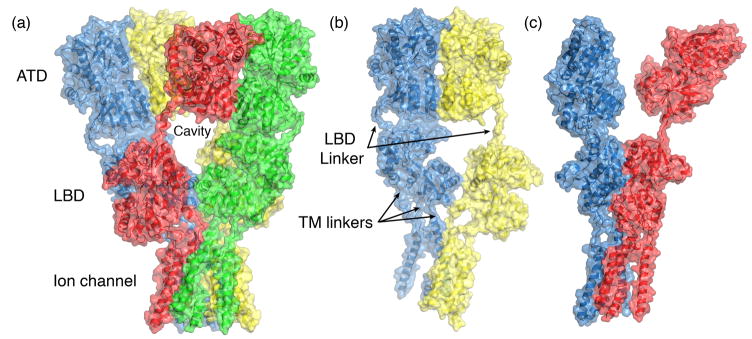
Figure 1.
Domain organization in the GluA2 ion channel crystal structure (PDB 3KG23). (a) The four subunits in the tetramer assembly are colored blue, yellow, red and green; the molecular surface representation reveals key features of the structure, including its organization in layers, the cavity at the ATD and LBD interface, and the large size of the extracellular domains compared to the ion channel. (b) ATD dimer formed by chains A and B reveals a large separation of the LBD domains in this subunit pair; linkers which connect the layered domains are indicated by black arrows which draw attention to the different conformations of the LBD linkers in the A and B subunits. (c) LBD dimer formed by chains A and D reveals a large separation of the ATD domains in this subunit pair.
Key features revealed by the GluA2 structure include an unexpected cross over of subunits forming dimer pairs in the ATD and LBD layers (Figure 1a); the multiple conformations of linkers which connect the ATD to the LBD, and the LBD to the ion channel (Figure 1b and c); the 2-fold to 4-fold switch of molecular symmetry between the ion channel and extracellular domains; and definition of the subunit interfaces in the ATD and LBD which form the dimer of dimers assembly. The loose packing of the ATD and LBD tetramer assemblies (Figure 1a), which have an enormous chalice-shaped hole on the central axis of symmetry located between the two layers of the extracellular domain, is remarkable. The GluA2 construct used for crystallization had a six amino acid deletion in the ATD to LBD linker, suggesting that coupling between these layers may be even weaker in the wildtype receptor. In retrospect, this loose packing likely underlies some of the multiple conformations observed in pioneering single molecule EM studies, but raises the question of whether some of these are native conformations, or result from interactions with the EM substrate and stains [5,6].
Assembly and function of AMPA, kainate and NMDA receptors
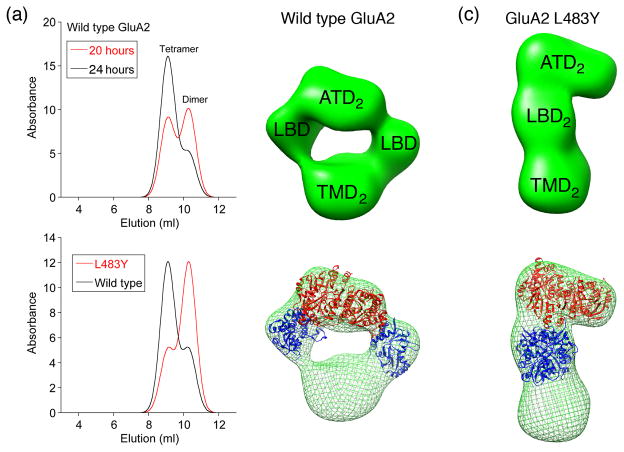
Elegant work on the initial stages of AMPA receptor assembly used synchronized induction of protein expression combined with affinity purification and single particle EM analysis to examine assembly intermediates [7•]. These studies revealed that biosynthesis and assembly for wild type GluA2 proceeds via the formation of dimers in which the ATD and TM segments are closely coupled, while the LBDs are pushed to the side and do not contact each other (Figure 2). Strikingly, the dimer assembly intermediate for the non desensitizing GluA2 L483Y mutant is an elongated structure in which both the ATD and LBD segments are closely apposed. As a consequence, the formation of tetramers is inhibited by the large energy required to separate the pair of subunits in the LBD layer of the dimer assembly, which is required to permit subunit cross over revealed by the GluA2 tetramer crystal structure. As a result, the GluA2 L483Y mutant is assembly deficient. In retrospect, for iGluRs with Cys mutants cross linking the LBD [8,9], it is not the inability to desensitize that underlies their poor cell surface expression, but rather the unexpected requirement for subunit exchange in the ATD and LBD layers during tetramer assembly.
Figure 2.
AMPA receptor assembly mechanisms revealed by synchronized protein expression and single particle EM analysis (from Ref. [•7]). (a) Schematic size exclusion chromatography profiles for elution of wild type GluA2 reveals a mixture of dimers and tetramers 20 hours after induction of expression, with an increase in tetramer and decrease in dimer populations 24 hours after induction; the lower panel shows chromatograms recorded at 24 hours for wild type GluA2 and the L483Y mutant which is trapped at the dimer stage of assembly. (b) Single particle EM analysis for wild type GluA2 dimers (top) with crystal structures for a GluA2 ATD dimer, and two GluA2 LBD monomers docked in the EM density map (bottom). (C) A similar analysis for the GluA2 L483Y dimer in which both the ATD and LBD layers contain dimer assemblies.
The four subunits in a GluA2 tetramer assembly do not have identical connections to the ion channel pore. Functional studies on kainate receptors, in which Cys mutations based on the GluA2 crystal structure were introduced to cross link subunits via different interfaces in the LBD tetramer assembly, revealed non equivalent effects on both activation and desensitization, depending on which pairs of subunits were cross linked [10•]. This suggests that in AMPA and kainate receptors, the strength of iGluR activation and desensitization varies, depending on whether subunits proximal or distal to the global axis of dimer symmetry bind glutamate. Kinetic measurements of the rate of disulfide bond formation for AMPA receptor LBD Cys mutants reveals that, in its ligand free state, GluA2 is a remarkably flexible and dynamic structure, in which interfaces between the dimer assemblies spontaneously form and break on the time scale of seconds to milliseconds [11]. Functional consequences likely to result from this include ‘silent receptors’ which are transiently unavailable for activation by glutamate [12••], and modal behavior revealed by single channel recording [13,14]. Spectroscopic measurements on GluA2 suggest that in the apo state the LBD dimers are in their dissociated state, and that the binding of glutamate triggers dimer assembly, followed by channel opening, and then desensitization [15]. There is a wealth of prior structural and mechanistic data which is at conflict with this model, including studies using positive allosteric modulators and mutants to stabilize LBD dimer assemblies [8,16,17], and crystal structures of the LBD apo and antagonist bound states which reveal an intact dimer interface [18]. This provocative hypothesis also does not address the mechanism by which binding of glutamate triggers dimer formation. For GluN2A subtype NMDA receptors, in which either the NR1 or NR2A LBD clam shells were independently locked in closed cleft conformations by engineered disulfide bonds, single channel recordings suggest that LBD opening and closing does not contribute to gating kinetics recorded at equilibrium [19]. For kainate receptors scanning mutagenesis experiments suggest that binding of fatty acids to GluK2 causes structural rearrangements in the ion channel pore [20•]
Structural and biochemical studies on isolated iGluR ATDs
As a herald to publication of the GluA2 ion channel crystal structure, the first ATD structures were reported for GluA2 and GluK2 expressed as soluble glycosylated proteins [21•,22•]. These were shortly followed by a second GluA2 crystal structure [23]; a GluN2B ATD structure in the apo and Zn2+ bound states [24•]; and most recently by structures for GluK3 and GluK5 [25]. Key features to emerge from these studies, which included measurements of affinity constants for dimer formation by analytical ultracentrifugation, was the discovery that the GluA2 and GluK2 but not GluN2B ATDs self associate at micromolar protein concentrations to form dimers, and that the GluA2, GluK2 and GluK3 but not GluN2B ATDs crystallize in a dimer of dimers assembly like that found in full length GluA2 (Figure 3a). Surprisingly, studies on the GluA2 ATD report substantial differences in the Kd for dimer assembly with dissociation constants of 0.15 and 4.3 μM; the Kd for GluA2 ATD tetramer formation was also reported to be either too weak to measure or 50 μM; these discrepancies remain to be resolved.
Figure 3.
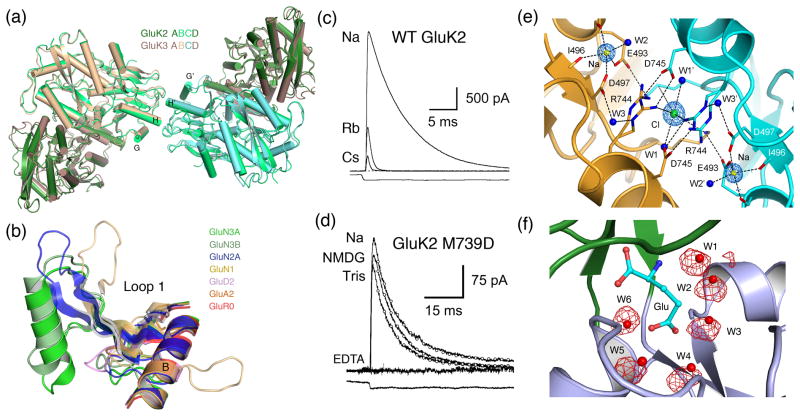
High resolution function and crystal structures for iGluR extracellular domains. (a) Tetramer assemblies of the kainate receptor GluK2 and GluK3 ATDs formed by a dimer of dimer assembly (from Ref. [25]). (b) iGluR loop 1 LBD structures superimposed using C coordinates for helix B revealing the elaborate structure in NMDA receptor subunits (from Ref. [25]). (c) Modulation of GluK1 outward current responses recorded at +60 mV by replacement of extracellular Na+ by Rb+ or Cs+. (d) The GluK2 M239D mutant retains activity in the presence of 100 μM Ca2+ and Mg2+ when Na is replaced by NMDG or Tris but is inhibited by 100 μM EDTA (from Ref. [48]). (e) Allosteric ion binding sites in the GluK1 LBD dimer assembly with alternate conformations for the Arg744 side chain and a 2:1 stoichiometry for Na+ and Cl− (from Ref. [•46]). (f) MD simulation of the GluK2 ligand binding domain docked with glutamate; red spheres indicate the position of water molecules trapped in the ligand binding site in the crystal structure; the red mesh shows MD trajectories for individual water molecules; W1, W2, W2 and W4 freely exchange with other on a ns time scale (from Ref. [64]).
In the GluA2 and GluK2 ATD crystal structures each of the subunits is in a partially closed conformation, about 10° more open than that for the ligand bound complexes of periplasmic proteins previously used as models for iGluR ATDs, but substantially more closed than for the apo state of these proteins. No ligands have been found for AMPA and kainate receptor ATDs that would trigger changes in conformation of the clam shell jaws, and the close apposition of both the upper and lower lobes of each subunit in the dimer assembly, combined with their high affinity for self assembly, suggests that the two ATDs are locked in a conformation incapable of large movements. Surprisingly, for the GluN2B ATD, the Zn2+ complex and apo structures had similar conformations, with identical extents of domain closure, but with a strikingly different relative orientation of the upper and lower lobes in the clam shell due to a substantial, around 50°, twist compared to the GluA2, GluK2 and GluK3 structures. Biochemical studies on NMDA receptor assembly suggest that the GluN1 and GluN2 ATDs likely assemble as heterodimers [26], but without crystal structures it is premature to speculate how this dimer assembly compares to that found for AMPA and kainate receptor ATD homodimers. Such knowledge will have great impact on understanding how allosteric ligands which bind to the ATD regulate NMDA receptor activity.
Allosteric regulation by NMDA receptor ATDs
Accompanying the new ATD structures, electrophysiological experiments revealed a novel regulatory role of NMDA receptor ATDs [27••,28••]. The GluN2C and GluN2D subunits have an unusually low open probability, about 50-fold less than for GluN2A. By swapping both the ATD and the linkers connecting the ATD to the LBD in chimeric receptors it was possible to decrease the open probability for NR2A, and to increase the open probability for NR2D. These results are of great interest in light of experiments suggesting that domain closure underlies the negative allosteric action of drugs and ions which bind to NMDA receptor, and related to this, substantial effort has been devoted to understanding the molecular mechanisms by which the ATD controls NMDA receptor activity [29], and the basis of the selective modulation of the GluN2B by ifenprodil with the goal of developing new drugs [30,31].
Structural studies on isolated iGluR LBDs
Of the 18 iGluR genes, crystal structures have now been reported for the isolated LBDs for 10 subunits. The most recently solved novel structures are for GluN3A and GluN3B [32], GluA3 [33] and GluA4 [34,35]. The GluN3 structures have similar folds to the other iGluR LBDS, with the exception of a remarkable loop structure which forms a distinct, disulfide linked subdomain, the size and structure of which differs in the GluN1, GluN2 and GluN3 subunits (Figure 3b). By contrast, the GluA3 and GluA4 structures are essentially identical to those previously solved for GluA2, for which more than 80 ligand complexes have been solved [36•], and the data set agonist, partial agonist and antagonist structures continues to grow for the LBDs of other iGluRs [37–40]. Capitalizing on this wealth of structural data, light activated AMPA receptor irreversible antagonists have been designed [41], and photoswitched tethered ligands have been developed for kainate receptors which can be used as optical probes for triggering membrane potential depolarization [42•], and in a GluR0/GluR6 chimera, inhibition of neuronal activity [43•]. NMR experiments using Hydrogen-Deuterium exchange [44] and isotopically labeled side chain methyl groups have given unprecedented insight into motions of the LBD and the kinetics of conformational changed induced by ligands [45]. Substantial progress has been made in identifying binding sites for allosteric ions in the GluK1 and GluK2 LBDs (Figure 3e); in identifying the mechanism of action of Na+ and Cl− on kainate receptors; in measuring the effects of Na and Cl on dimer affinity by analytical ultracentrifugation; and in engineering a high affinity Ca2+ binding site [12••,46•–48]. In related work it was established that Ca2+ ions act via a similar allosteric mechanism to stabilize the dimer assembly of GluD2 LBDs [46•,49]. A large body of functional analysis for kainate receptors, combined with multiple crystal structures and AUC measurements for LBD mutants, has established beyond doubt that as in AMPA receptors the stability of LBD dimer assembly regulates the rate and extent of desensitization [17,50].
The LBD of AMPA receptors is the site of action of positive allosteric modulators, one class of which slows the decay of excitatory synaptic currents, while another class attenuates desensitization. Crystal structures for multiple GluA2 modulator complexes have recently been solved [51,52]. Several drug companies have launched medicinal chemistry programs targeting AMPA receptor positive allosteric modulators, and the 1st fruits of structurally based drug design have now emerged from this work [53–55]. Unfortunately, clinical trails have yet to convincingly demonstrate efficacy of any of these compounds, and none have entered Phase III trials [56]. All of these studies focused on flip splice variants but recent work has revealed the mechanism of binding of flop splice variant selective allosteric modulators [57]. It is anticipated that within a few years similar approaches will lead to discovery of ligands for other iGluR subtypes, and reveal definitively whether allosteric modulators which act at the ligand binding domain have clinical potential.
Auxiliary subunits
To date there is no structural data available on any of the iGluR auxiliary subunits, and ideas about how they modulate receptor function are still speculative. Key advances include the discovery of a new family of kainate receptor modulatory proteins of which NETO2 is the best characterized [58•]. When coexpressed with GluK2 NETO2 increases open channel probability and burst length, slows the decay of responses to brief applications of glutamate, and increases the duration of miniature EPSCs in cerebellar granule cells transfected with GluK1 or GluK2. For AMPA receptors two new families of modulatory proteins have been discovered. The first is CKAMP44 which unlike other iGluR auxiliary subunits down regulates the activity of GluA1 and GluA2 both in heterologous expression systems, and in vivo [59•]. The second family, named cornichons, have similar functional effects to TARPS, and up regulate AMPA receptor activity in heterologous expression systems [60•]. However, in vivo there is conflicting evidence about the role of cornichons. A recent study suggests that AMPA receptors only coassemble with cornichons during the process of biogenesis and that cornichons are not present in AMPA receptor complexes located at synapses and in the plasma membrane [61•]. By contrast, a comprehensive functional and biochemical analysis suggests that the TARP gamma-8 and the cornichon CNIH-2 are both present in hippocampal AMPA receptor complexes [62•].
Molecular dynamics simulations
Computational studies on iGluR LBDs have made significant progress. Measurements of the free energy landscape of the GluA2 ligand binding domain in the apo and glutamate bound states reveals multiple apo conformations of similar energy [63•], consistent with the emerging picture of a highly flexible and dynamic molecule. Equilibrium MD simulations have been used to look at events leading to the spontaneous closing and opening of the LBD, and reveal that a salt bridge network around the hinge region of the clam shell likely acts as a sensor of ligand binding, triggering domain closure on a ns time scale [64]. The same study also examined the binding of water to the GluA2, GluK1, GluK2 and GluN2A LBDs, for which crystal structures reveal numerous water molecules acting as surrogate ligand atoms. The MD simulations accurately reproduce this solvent network, and reveal that all but one of the water molecules exchanges with bulk solvent via transient solvent channels not revealed in the crystal structures (Figure 3f). MD simulations have also been used to examine the binding of allosteric anions and cations to kainate receptors, and give insight into how the sites are coupled via protein electric fields [47].
Conclusion
Structure and mechanism are intimate partners in all areas of biology, but this is especially true for the study of signal transduction. For iGluRs, clues about mechanism have been obtained in the past from well designed electrophysiological experiments. However, interpretation at the molecular level becomes possible only when the results are painted onto the scaffold of a crystal structure. A third experimental approach, calculations performed using molecular dynamics, is now becoming increasingly important. This triumvirate is necessary because, like all proteins, iGluRs are dynamic molecules which are likely to be almost impossible to crystallize in the full cycle of conformational states which underlies their biological function. Despite their atomic detail, crystal structures give little insight into the thermodynamic mechanisms underlying how the binding of agonists, ions, modulators and auxiliary proteins shape the kinetics of glutamate receptor activity at synapses. Finally, even with a growing ensemble of crystal structures, NMR studies and MD simulations, functional studies continue to reveal new and unexpected aspects of receptor activity which have escaped attention despite decades of patch clamp analysis. Our understanding of these key signal transduction molecules is still far from complete, and years of interesting work and discoveries lies ahead.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Hollmann M. Structure of ionotropic glutamate receptors. In: Jonas P, Monyer H, editors. Ionotropic glutamate receptors in the CNS. Springer-Verlag; 1999. pp. 3–98. [Google Scholar]; Handbook of experimental pharmacology. Vol. 141 [Google Scholar]
- 2.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- 4••.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechamism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. The 1st full length crystal structure of an iGluR tetramer, which took more than 5 years to solve, confirmed many predictions made from the structures of soluble iGluR domains, but also revealed many unexpected features which have no counterpart in other neurotransmitter receptors including symmetry mismatch, large cavities, and subunit cross over between domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 6.Midgett CR, Madden DR. The quaternary structure of a calcium-permeable AMPA receptor: conservation of shape and symmetry across functionally distinct subunit assemblies. J Mol Biol. 2008;382:578–584. doi: 10.1016/j.jmb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 7•.Shanks NF, Maruo T, Farina AN, Ellisman MH, Nakagawa T. Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci. 2010;30:2728–2740. doi: 10.1523/JNEUROSCI.5146-09.2010. Using sophisticated cell biology and single particle EM, assembly intermediates are identified for AMPA receptors and the binding of TARPs to GluA2 tetramers but not dimer assembly intermediates is established. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nature Structural and Molecular Biology. 2006;13:1120–1127. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- 9.Priel A, Selak S, Lerma J, Stern-Bach Y. Block of kainate receptor desensitization uncovers a key trafficking checkpoint. Neuron. 2006;52:1037–1046. doi: 10.1016/j.neuron.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 10•.Das U, Kumar J, Mayer ML, Plested AJ. Domain organization and function in GluK2 subtype kainate receptors. Proc Natl Acad Sci USA. 2010;107:8463–8468. doi: 10.1073/pnas.1000838107. Cross linking for the kainate receptor GluK2 experiments test the assembly principles revealed by the GluA2 ion channel crystal structure and reveal subunit non equivalence in the activation and desensitization mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plested AJ, Mayer ML. AMPA receptor ligand binding domain mobility revealed by functional cross linking. J Neurosci. 2009;29:11912–11923. doi: 10.1523/JNEUROSCI.2971-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Plested AJ, Vijayan R, Biggin PC, Mayer ML. Molecular basis of kainate receptor modulation by sodium. Neuron. 2008;58:720–735. doi: 10.1016/j.neuron.2008.04.001. A combination of X-ray crystallography, whole cell recording and MD simulations was used to identify the binding sites which mediate the positive allosteric effect of Na+ on kainate receptors. The results establish a stoichiometry of 2:1 for binding of Na+ and Cl− to kainate LBD dimer assemblies and suggest that allosteric ions stabilize dimers in their active conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prieto ML, Wollmuth LP. Gating modes in AMPA receptors. J Neurosci. 2010;30:4449–4459. doi: 10.1523/JNEUROSCI.5613-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon K, Nowak LM, Oswald RE. Characterizing single-channel behavior of GluA3 receptors. Biophys J. 2010;99:1437–1446. doi: 10.1016/j.bpj.2010.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez J, Du M, Parameshwaran K, Suppiramaniam V, Jayaraman V. Role of dimer interface in activation and desensitization in AMPA receptors. Proc Natl Acad Sci U S A. 2010;107:9891–9896. doi: 10.1073/pnas.0911854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhry C, Weston MC, Schuck P, Rosenmund C, Mayer ML. Stability of ligand-binding domain dimer assembly controls kainate receptor desensitization. EMBO J. 2009;28:1518–1530. doi: 10.1038/emboj.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 19.Kussius CL, Popescu GK. NMDA receptors with locked glutamate-binding clefts open with high efficacy. J Neurosci. 2010;30:12474–12479. doi: 10.1523/JNEUROSCI.3337-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Wilding TJ, Chen K, Huettner JE. Fatty acid modulation and polyamine block of GluK2 kainate receptors analyzed by scanning mutagenesis. J Gen Physiol. 2010;136:339–352. doi: 10.1085/jgp.201010442. Modulation of GluK2 by arachidonic acid and docosahexaenoic acid is suggested to occur via a change in structure of the ion channel pore due to reorientation of interface between M1, M2, and M3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. The 1st crystal structure for an AMPA receptor ATD dimer assembly revealed subunit interactions that are strikingly different from those found in the structurally related domains of glutamate activated GCPRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. Crystal structure for a kainate receptor ATD dimer, published at the same time as those for GluA2, revealed patterns of disulfide bonding and clam shell closure that are conserved in AMPA and kainate receptors, and which were not identified in homology models based on mGluRs or bacterial periplasmic proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clayton A, Siebold C, Gilbert RJ, Sutton GC, Harlos K, McIlhinney RA, Jones EY, Aricescu AR. Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J Mol Biol. 2009;392:1125–1132. doi: 10.1016/j.jmb.2009.07.082. [DOI] [PubMed] [Google Scholar]
- 24•.Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. Crystal structure for an NMDA receptor ATD complex with the allosteric regulator Zn2+ reveals a strikingly different conformation than found for GluA2 and GluK2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar J, Mayer ML. Crystal Structures of the Glutamate Receptor Ion Channel GluK3 and GluK5 Amino-Terminal Domains. J Mol Biol. 2010;404:680–696. doi: 10.1016/j.jmb.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuler T, Mesic I, Madry C, Bartholomaus I, Laube B. Formation of NR1/NR2 and NR1/NR3 heterodimers constitutes the initial step in N-methyl-D-aspartate receptor assembly. J Biol Chem. 2008;283:37–46. doi: 10.1074/jbc.M703539200. [DOI] [PubMed] [Google Scholar]
- 27••.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. This paper reports for the 1st time that the ATD of GluN2D subtype NMDA receptors acts as an endogenous negative allosteric modulator which greatly suppress ion channel open probability, while the GluN2A ATD acts as a positive modulator. Using a series of chimeras it is established that the linker connecting the ATD to the ligand binding domain also plays a key role in this effect, and that the gating properties of NMDA receptor subbtypes can be exchanged by transplanting the ATD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. This paper complements and extends 26•• and shows in addition that the GluN2C ATD is inhibitory, and that the ATD controls the kinetics of NMDA receptor deactivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mony L, Krzaczkowski L, Leonetti M, Le Goff A, Alarcon K, Neyton J, Bertrand HO, Acher F, Paoletti P. Structural basis of NR2B-selective antagonist recognition by NMDA receptors. Mol Pharmacol. 2009;75:60–74. doi: 10.1124/mol.108.050971. [DOI] [PubMed] [Google Scholar]
- 31.Mosley CA, Myers SJ, Murray EE, Santangelo R, Tahirovic YA, Kurtkaya N, Mullasseril P, Yuan H, Lyuboslavsky P, Le P, et al. Synthesis, structural activity-relationships, and biological evaluation of novel amide-based allosteric binding site antagonists in NR1A/NR2B N-methyl-D-aspartate receptors. Bioorg Med Chem. 2009;17:6463–6480. doi: 10.1016/j.bmc.2009.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J. 2008;27:2158–2170. doi: 10.1038/emboj.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed AH, Wang Q, Sondermann H, Oswald RE. Structure of the S1S2 glutamate binding domain of GLuR3. Proteins. 2009;75:628–637. doi: 10.1002/prot.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasper C, Frydenvang K, Naur P, Gajhede M, Pickering DS, Kastrup JS. Molecular mechanism of agonist recognition by the ligand-binding core of the ionotropic glutamate receptor 4. FEBS Lett. 2008;582:4089–4094. doi: 10.1016/j.febslet.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Gill A, Birdsey-Benson A, Jones BL, Henderson LP, Madden DR. Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists. Biochemistry. 2008;47:13831–13841. doi: 10.1021/bi8013196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Pohlsgaard J, Frydenvang K, Madsen U, Kastrup JS. Lessons from more than 80 structures of the GluA2 ligand-binding domain in complex with agonists, antagonists and allosteric modulators. Neuropharmacology. 2010 doi: 10.1016/j.neuropharm.2010.08.004. This extensive review describes in detail the molecular mechanisms underlying the binding of agonists, partial agonists, antagonists and allosteric modulators to AMPA receptors. [DOI] [PubMed] [Google Scholar]
- 37.Ahmed AH, Thompson MD, Fenwick MK, Romero B, Loh AP, Jane DE, Sondermann H, Oswald RE. Mechanisms of antagonism of the GluR2 AMPA receptor: structure and dynamics of the complex of two willardiine antagonists with the glutamate binding domain. Biochemistry. 2009;48:3894–3903. doi: 10.1021/bi900107m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clausen RP, Naur P, Kristensen AS, Greenwood JR, Strange M, Brauner-Osborne H, Jensen AA, Nielsen AS, Geneser U, Ringgaard LM, et al. The glutamate receptor GluR5 agonist (S)-2-amino-3-(3-hydroxy-7,8-dihydro-6H-cyclohepta[d]isoxazol-4-yl)propion ic acid and the 8-methyl analogue: synthesis, molecular pharmacology, and biostructural characterization. J Med Chem. 2009;52:4911–4922. doi: 10.1021/jm900565c. [DOI] [PubMed] [Google Scholar]
- 39.Frydenvang K, Lash LL, Naur P, Postila PA, Pickering DS, Smith CM, Gajhede M, Sasaki M, Sakai R, Pentikainen OT, et al. Full domain closure of the ligand-binding core of the ionotropic glutamate receptor iGluR5 induced by the high affinity agonist dysiherbaine and the functional antagonist 8,9-dideoxyneodysiherbaine. J Biol Chem. 2009;284:14219–14229. doi: 10.1074/jbc.M808547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alushin GM, Jane D, Mayer ML. Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists. Neuropharmacology. 2010;60:126–134. doi: 10.1016/j.neuropharm.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cruz LA, Estebanez-Perpina E, Pfaff S, Borngraeber S, Bao N, Blethrow J, Fletterick RJ, England PM. 6-Azido-7-nitro-1,4-dihydroquinoxaline-2,3-dione (ANQX) forms an irreversible bond to the active site of the GluR2 AMPA receptor. J Med Chem. 2008;51:5856–5860. doi: 10.1021/jm701517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Numano R, Szobota S, Lau AY, Gorostiza P, Volgraf M, Roux B, Trauner D, Isacoff EY. Nanosculpting reversed wavelength sensitivity into a photoswitchable iGluR. Proc Natl Acad Sci U S A. 2009;106:6814–6819. doi: 10.1073/pnas.0811899106. Optically gated iGluRs are expected to have great utility in analyzing neuronal circuitry in brain slices and in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Janovjak H, Szobota S, Wyart C, Trauner D, Isacoff EY. A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nat Neurosci. 2010;13:1027–1032. doi: 10.1038/nn.2589. Coupled with the tools reported in 42•, the ability to be able to both silence and excite neurons by photo switching greatly increases the scope of this novel approach for network analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fenwick MK, Oswald RE. On the mechanisms of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptor binding to glutamate and kainate. J Biol Chem. 2010;285:12334–12343. doi: 10.1074/jbc.M109.086371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maltsev AS, Oswald RE. Hydrophobic side chain dynamics of a glutamate receptor ligand binding domain. J Biol Chem. 2010;285:10154–10162. doi: 10.1074/jbc.M109.088641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Chaudhry C, Plested AJ, Schuck P, Mayer ML. Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions. Proc Natl Acad Sci U S A. 2009;106:12329–12334. doi: 10.1073/pnas.0904175106. Analytical ultracentrifugation is used to show that allosteric ions directly modulate the stability of isolated LBD dimer assemblies for both kainate and delta subtype iGluRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vijayan R, Plested AJ, Mayer ML, Biggin PC. Selectivity and cooperativity of modulatory ions in a neurotransmitter receptor. Biophys J. 2009;96:1751–1760. doi: 10.1016/j.bpj.2008.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plested AJ, Mayer ML. Engineering a high-affinity allosteric binding site for divalent cations in kainate receptors. Neuropharmacology. 2009;56:114–120. doi: 10.1016/j.neuropharm.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen KB, Naur P, Kurtkaya NL, Kristensen AS, Gajhede M, Kastrup JS, Traynelis SF. Modulation of the dimer interface at ionotropic glutamate-like receptor delta2 by D-serine and extracellular calcium. J Neurosci. 2009;29:907–917. doi: 10.1523/JNEUROSCI.4081-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nayeem N, Zhang Y, Schweppe DK, Madden DR, Green T. A nondesensitizing kainate receptor point mutant. Mol Pharmacol. 2009;76:534–542. doi: 10.1124/mol.109.056598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ptak CP, Ahmed AH, Oswald RE. Probing the allosteric modulator binding site of GluR2 with thiazide derivatives. Biochemistry. 2009;48:8594–8602. doi: 10.1021/bi901127s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed AH, Oswald RE. Piracetam defines a new binding site for allosteric modulators of alpha-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA) receptors. J Med Chem. 2010;53:2197–2203. doi: 10.1021/jm901905j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jamieson C, Campbell RA, Cumming IA, Gillen KJ, Gillespie J, Kazemier B, Kiczun M, Lamont Y, Lyons AJ, Maclean JK, et al. A novel series of positive modulators of the AMPA receptor: structure-based lead optimization. Bioorg Med Chem Lett. 2010;20:6072–6075. doi: 10.1016/j.bmcl.2010.08.063. [DOI] [PubMed] [Google Scholar]
- 54.Jamieson C, Basten S, Campbell RA, Cumming IA, Gillen KJ, Gillespie J, Kazemier B, Kiczun M, Lamont Y, Lyons AJ, et al. A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies. Bioorg Med Chem Lett. 2010;20:5753–5756. doi: 10.1016/j.bmcl.2010.07.138. [DOI] [PubMed] [Google Scholar]
- 55.Ward SE, Harries M, Aldegheri L, Andreotti D, Ballantine S, Bax BD, Harris AJ, Harker AJ, Lund J, Melarange R, et al. Discovery of N-[(2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H-inden-2-yl]-2-propanesulfo namide, a novel clinical AMPA receptor positive modulator. J Med Chem. 2010;53:5801–5812. doi: 10.1021/jm1005429. [DOI] [PubMed] [Google Scholar]
- 56.Ward SE, Bax BD, Harries M. Challenges for and current status of research into positive modulators of AMPA receptors. Br J Pharmacol. 2010;160:181–190. doi: 10.1111/j.1476-5381.2010.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahmed AH, Ptak CP, Oswald RE. Molecular mechanism of flop selectivity and subsite recognition for an AMPA receptor allosteric modulator: structures of GluA2 and GluA3 in complexes with PEPA. Biochemistry. 2010;49:2843–2850. doi: 10.1021/bi1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Zhang W, St-Gelais F, Grabner CP, Trinidad JC, Sumioka A, Morimoto-Tomita M, Kim KS, Straub C, Burlingame AL, Howe JR, et al. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron. 2009;61:385–396. doi: 10.1016/j.neuron.2008.12.014. Discovery of NETO2 as a kainate receptor auxiliary protein which like TARPS upregulates activity marks an import advance but raises many questions about the role of NETO2 in vivo. Based on the rate of progress made following the discovery of TARPs it will take a few years to resolve this. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.von Engelhardt J, Mack V, Sprengel R, Kavenstock N, Li KW, Stern-Bach Y, Smit AB, Seeburg PH, Monyer H. CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science. 2010;327:1518–1522. doi: 10.1126/science.1184178. The discovery of CKAMP44 as an AMPA receptor inhibitory auxiliary subunit also raises numerous questions which will take some time to resolve. [DOI] [PubMed] [Google Scholar]
- 60•.Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, et al. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. Cornichons are positive AMPA receptor modulators; see comments for •61 and •62. [DOI] [PubMed] [Google Scholar]
- 61•.Shi Y, Suh YH, Milstein AD, Isozaki K, Schmid SM, Roche KW, Nicoll RA. Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc Natl Acad Sci U S A. 2010;107:16315–16319. doi: 10.1073/pnas.1011706107. This study suggests that in neurons cornichons are not expressed at the plasma membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62•.Kato AS, Gill MB, Ho MT, Yu H, Tu Y, Siuda ER, Wang H, Qian YW, Nisenbaum ES, Tomita S, et al. Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron. 2010;68:1082–1096. doi: 10.1016/j.neuron.2010.11.026. The combined role of cornichons and TARPs in regulating AMPA receptor trafficking and function is not well understood and will require further study. In contrast to this study suggests that TARP gamma-8 and the cornichon CNIH-2 associate in postsynaptic densities in hippocampal AMPA receptor complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Lau AY, Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure. 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. Umbrella sampling was used to calculate the free energy of an isolated GluA2 ligand binding domain over a wide range of domain closure for the apo state and for complexes with glutamate and DNQX and reveals that 9–12 kcal/mol becomes available upon glutamate binding to drive receptor activation and desensitization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vijayan R, Sahai MA, Czajkowski T, Biggin PC. A comparative analysis of the role of water in the binding pockets of ionotropic glutamate receptors. Phys Chem Chem Phys. 2010 doi: 10.1039/c004336b. [DOI] [PubMed] [Google Scholar]