Abstract
Pharmacophore feature is defined by a set of chemical structure patterns having the active site of drug like molecule. Pharmacophore can be used to assist in building hypothesis about desirable chemical properties in drug molecule and hence it can be used to refine and modify drug candidates. We predicted the pharmacophoric features of 150 medicinal compounds from plants for anti-cancer, anti-carcinogenic, anti-diabetic, anti-microbial, and anti-oxidant. Estimation of pharmacophoric feature is necessary to ensure the optimal supramolecular interaction with a biological target and to trigger or block its biological response. We subsequently make this data available to open access using a database at the URL: http://www.hccbif.info/index.htm
Availability
The database is available for free at http://www.hccbif.info/index.htm
Keywords: Anti Diabetic, Anti Cancer, Anti Microbial, Pharmacophoric features, Phase
Background
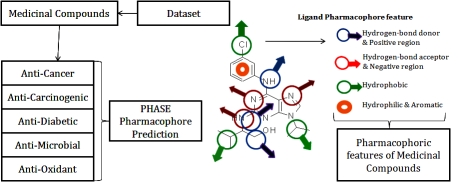
The traditional Indian medicine and use of phytocompounds against various diseases receive considerable attention in recent years. Plant-derived drugs remain an important resource, especially in developing countries to combat serious diseases. Approximately 60-80% of the world's population still relies on traditional medicines for the treatment of common illness [1–2]. Pharmacophore screening is one of the useful ways to uncover a set of features that is common to a series of active molecules responsible for the activity [3]. Perceiving a pharmacophore is the most important first step towards understanding the interaction between a receptor and a ligand. Paul Ehrlich (1900) defined a pharmacophore as “a molecular framework that carries (phoros) the essential features responsible for a drug's (pharmacon) biological activity” [4]. That definition of pharmacophore remained unperturbed for over 90 years. The current widely used definition was presented by Peter Gund (1977) refined a pharmacophore as “a set of structural features in a molecule that is recognized at a receptor site and is responsible for that molecule's biological activity” [5]. This definition is remarkably loyal to the earlier definition. A pharmacophore is an exact description of molecular features which are necessary for molecular recognition of a ligand required biological macromolecule. The IUPAC defines a pharmacophore to be an ensemble of steric and electronic features that is necessary to ensure the optimal supramolecular interactions with a specific biological target and to trigger (or block) its biological response [6]. Typical pharmacophore of a molecule consist of features like, hydrophobic, aromatic, a hydrogen bond acceptor, a hydrogen bond donor, negative and positive functional groups. Here, an effort has been taken to find the Pharmacophoric sites of certain medicinal compounds by using PHASE module. The application of database is to help researchers; particularly working with medicinal compounds. Through this database, they may get idea about ligand chemical features. Molecular level studies of pharmacophore explain about active site of drug molecule, which is important for interaction with binding target. Theoretical prediction of Pharmacophoric features of these compounds includes hydrogen bond donor, hydrogen bond acceptor, hydrophobic, aromatic, positive and negative features. These characteristics of ligand molecules are considered as pharmacophoric sites based on the differentiation (Figure 1). These pharmacophore are considered to be an essential part of a compound to become a drug, thereby these. pharmacophoric features can be an advantage for researchers to know about the chemical features and functional properties of ligand molecule.
Figure 1.
Database workflow for pharmacophoric prediction of medicinal compounds
Methodology
Data collection & Preparation
The dataset consists of 150 medicinal compounds collected from http://www.hccbif.info/ of which 89 are anti cancerous & anticarcinogenic, 43 are antidiabetic, 47 are antimicrobial and 5 are anti oxidants [7]. Data set are prepared by ligprep cleaned structures for molecular modeling environment. Prepared ligands are processed for pharmacophoric features through Pharmacophore Alignment and Scoring Engine (PHASE) module of Schrodinger suite, which is a recently developed pharmacophore modeling package, for all the above mentioned series of compounds.
Creating Pharmacophoric Sites
Phase locates features using SMARTS pattern matching, making it easy to modify existing feature definitions and to create entirely new custom features. Intuitive visualization tools and the rapid location of sites help researchers finetune feature definitions. Each compound structure is represented by a set of points in 3D space, which coincide with various chemical features that may facilitate non-covalent binding between the compound and its target receptor. PHASE provides a built-in set of six pharmacophore features, hydrogen bond acceptor(A), hydrogen bond donor(D), hydrophobic group(H), negatively ionizable (N), positively ionizable (P), and aromatic ring(R). By this methodology the pharmacophoric features are predicted and deposited in database [8–10].
Utility for biological community
A Pharmacophore is defined as an ensemble of universal chemical features that characterize a specific mode of action of a ligand in active site of the macromolecule in 3D space. Due to its importance, this method is computationally very efficient and is subsequently well suited for virtual screening of large compound libraries. This Database is open source and freely available. Applications of the work consists of user interface with dynamic web pages, more than 150 medicinal compounds having anti-carcinogenic, anticancerous, anti-diabetic, anti-microbial and anti-oxidant activity, Theoretical prediction of chemical features using commercial software's are applicable for the researchers working on medical plants. Pharmacophore predicted in this work would consider to be very useful for further development of ligand based new analog for finding a new active compound with similar Pharmacophoric features and the knowledge of the pharamacophore will result in the generation of several leads. Pharmacophore of these compounds can explain very well about the physiochemical properties required for the designing of new compounds of these series.
Future development
We plan to refine and keep updating this database and to provide more medicinal compounds with bio-physiochemical properties.
Author Contributions
Dr. Pitchai Daisy and Periyasamy Vijayalakshmi developed and maintaining the database. Dr. Sanjeev Kumar Singh and Chandrabose Selvaraj predicted the pharmacophoric features of medicinal compounds. Other authors helped in dataset and manuscript preparation. All authors read and approved the final manuscript.
Acknowledgments
This project is funded by BTIS (Biotechnology information system), DBT (Department of Biotechnology) and Ministry of Science & Technology, Government of India, India.
Footnotes
Citation:Daisy et al, Bioinformation 6(4): 167-168 (2011)
References
- 1.S Dev. Indian J Exp Biol. 2010;48:191. [PubMed] [Google Scholar]
- 2.D Schuster, G Wolber. Curr Pharm Des. 2010;16:1666. doi: 10.2174/138161210791164072. [DOI] [PubMed] [Google Scholar]
- 3.AH Khan, et al. Bioinformation. 2010;5:62. [Google Scholar]
- 4.P Ehrlich. Ber Dtsch Chem Ges. 1909;42:17. [Google Scholar]
- 5.P Gund. Prog Mol Subcell Biol. 1977;5:117. [Google Scholar]
- 6.SL Dixon, et al. Chem Biol Drug Des. 2006;67:370. doi: 10.1111/j.1747-0285.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- 7.D Pitchai, et al. Bioinformation. 2010;5:43. [Google Scholar]
- 8.S Narkhede, et al. QSAR Comb Sci. 2007;26:744. [Google Scholar]
- 9.NR Tawari, et al. J Mol Model. 2008;14:911. doi: 10.1007/s00894-008-0330-z. [DOI] [PubMed] [Google Scholar]
- 10.S Bag, et al. QSAR Comb Sci. 2009;28:296. [Google Scholar]