Abstract
Trisomy 11 (+11) as an isolated abnormality is a rare event in patients with acute myeloid leukemia (AML) and is associated with poor prognosis. We describe the clinicopathologic features of 18 AML patients with isolated +11 and their mutation status of NPM1, FLT3, NRAS ,KRAS, and KIT. Fourteen patients had de novo AML and 4 patients had a history of myelodysplastic syndrome (MDS). Fifteen patients had a progressive clinical course with refractory or relapsed disease. The median overall survival was 5 months (range, 2 to 48 months). Only 1 patient achieved complete remission after undergoing stem cell transplantation. The bone marrow median blast count was 65% (range, 22 to 86) and 14 patients had blasts >50%. The most common type of AML was AML without maturation (7 patients) classified by the World Health Organization classification system, or M1 (10 patients) by the French-American-British (FAB) system. FLT3 mutations were detected in 3 of 15 (20%) cases tested. RAS mutation was present in 1 of 16 (6%) cases and there was no evidence of NPM1 of KIT mutations (each tested in 12 cases). Our findings confirm previous reports that isolated +11 is associated with a poor prognosis in patients with AML and tends to be associated with FAB-M1 morphologic features. No evidence of NPM1 or KIT mutations were identified.
Keywords: Isolated trisomy 11, acute myeloid leukemia, gene mutations, prognosis
Introduction
Acute myeloid leukemia (AML) is a clinically and genetically heterogeneous neoplasm. In addition to the patient age, cytogenetic findings play a major role in the classification and prognosis of AML allowing for stratification into favorable, intermediate, and poor risk categories [1]. Although highly useful, approximately 40% of AML patients lack cytogenetic abnormalities. Molecular studies in this subgroup of patients have shown various gene mutations, some of which proven to have prognostic importance [1]. Some gene mutations associated with AML have been incorporated into the most recent classification system of the World Health Organization (WHO) [2].
Structural chromosomal abnormalities, such as translocations, generally constitute most of the nonrandom changes in hematopoietic neoplasms. In contrast, chromosomal gains, apart from chromosome 8, occur less frequently [3]. Unlike structural abnormalities that may lead to specific gene rearrangement, the pathogenetic significance of trisomies is unclear in most AML cases. Autosomal trisomy may occur as a secondary event in association with other cytogenetic abnormalities. However, the presence of a trisomy as part of a complex karyotype usually does not significantly influence prognosis in AML patients. The situation appears to be different for isolated autosomal trisomies as a primary cytogenetic abnormality in AML patients. Others have shown that a number of different isolated trisomies in AML are associated with an adverse outcome [4]. Of these, trisomy 11 (+11) is one of the most rare isolated abnormalities associated with AML. In a study by Heinonen and colleagues, isolated +11 was identified in 13 of 1496 adult AML patients (<1%) [5].
In this study, we focused on patients with AML associated with +11 as an isolated cytogenetic abnormality. We analyzed the clinicopathologic, immunophenotypic, and cytogenetic data in these patients and we also assessed the mutation status of the NPM1, FLT3, RAS, and KIT genes.
Materials and methods
Clinical, morphologic, and Immunophenotypic analysis
We identified AML patients with isolated +11 in the cytogenetic database of the Clinical Cytoge-netics Laboratory, in the Department of Hema-topathology, The University of Texas MD Anderson Cancer Center. These patients were treated at MD Anderson Cancer Center between 1999 and 2009. This study was approved by the Institutional Review Board.
Clinical, follow-up, and laboratory data were collected at time of diagnosis of AML from the medical records. Laboratory data included a complete blood count in all patients. All patients underwent bone marrow aspiration and biopsy and each case was classified morphologically using the 2008 WHO and the older FAB classifications. Myeloperoxidase was assessed using a standard cytochemical method on bone marrow aspirate smears of all cases.
Flow cytometry Immunophenotypic analysis was performed on peripheral blood and/or bone marrow aspirate material in a subset of patients at M.D. Anderson Cancer Center as previously described [6].The antibody panels used were highly variable, however, in all cases at least 3-color analysis was performed and myeloblasts were gated for analysis using CD45 and side scatter. CD45 was paired with peridin chlorophyll alpha protein and either fluorescein isothiocyanate or phycoerythrin conjugated antibodies that included CD13, CD33, CD34, CD117, HLA-DR, and pan-B and pan-T cell antibodies.
Cytogenetic analysis and fluorescence in situ hybridization (FISH)
Cytogenetic analysis and FISH analysis were performed on bone marrow aspirate specimens according to standard protocols as previously described [7]. Twenty G-banded metaphases were analyzed whenever possible, and cytogenetic diagnosis was made according to the criteria recommended by the International System for Human Cytogenetic Nomenclature [8]. Only cases with an isolated +11 as the primary change were included in the study. Fluorescence in situ hybridization using a probe SLI CEP 11 for chromosome 11 centromere (Vysis/ Abbot Molecular Inc., IL, USA) was performed to confirm the routine cytogenetic results when needed. The cutoff value for the SLI CEP11 was 1.8%.
Molecular studies
Genomic DNA was extracted from fresh bone marrow aspiration specimens for mutational analyses using the Autopure extractor (Qiagen/ Gentra, Valencia, CA, USA). FLT3 (internal tandem duplication [ITD] and codon 835) and NPM1 (exon 12) mutations were screened using polymerase chain reaction (PCR) followed by capillary electrophoresis on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA) as previously described [9]. For the FLT3 codon 835/836 tyrosine kinase domain (TKD) point mutation analysis, PCR products were digested with EcoRV before capillary electrophoresis. Mutations in NRAS and KRAS, and KIT genes were assessed by PCR followed by direct sequencing, Sanger sequencing, or py-rosequencing, according to previously described protocols [9,10].
Results
Clinical, morphologic, and immunophenotypic characteristics
We identified 18 AML patients with isolated +11 from a total of 20,029 AML patients diagnosed at MDACC between 1999 and 2009, showing a frequency of 0.09%. As shown in Table 1, there were 11 men and 7 women. The patient ages ranged from 33 to 98 years, with a median of 69 years. Twelve (67%) patients were older than 60 years. All patients presented with anemia, with a median hemoglobin of 88 g/L, range, 69-109 g/L (normal range 140-180 g/L). Sixteen (89%) patients had thrombocytopenia with a median platelet count of 65 × 109/L (normal range, 140-440 × 109/L). Fourteen (78%) patients had leukocytopenia and one patient had marked leukocytosis with a white blood cell count (WBC) of 46 × 109/L (normal range, 4-11 × 109/L). The median WBC was 2.35 ×109/L (range, 0.1-46 ×109/L).
Table 1.
Clinicopathologic findings in 18 cases of isolated trisomy 11 AML
| Case | Age/Sex | FAB Classification | WHO classification | WBC (×109/L) | Hb (g/dl) | Plt (109/L) | Blasts | OS (Months) |
|---|---|---|---|---|---|---|---|---|
| 1a | 77/F | M1 | AML with myelodysplasia-related changes | 1.4 | 7.4 | 72 | 68% | 6 |
| 2 | 76/F | M0 | AML with minimal differentiation | 5.4 | 8.1 | 95 | 50% | NA |
| 3 | 54/F | M1 | AML without maturation | 3.5 | 7.1 | 50 | 85% | 5 |
| 4 | 85/F | M1 | AML without maturation | 2.8 | 7.4 | 34 | 25% | 48 |
| 5a | 71/M | M1 | AML with myelodysplasia-related changes | 2.1 | 10.6 | 38 | 57% | 3 |
| 6 | 33/M | M2 | AML with maturation | 3.9 | 6.9 | 58 | 55% | 6 |
| 7 | 57/M | M2 | AML with maturation | 1.4 | 7.9 | 68 | 55% | NA |
| 8 | 67/M | M1 | AML without maturation | 2.6 | 8.1 | 143 | 67% | 9 |
| 9a | 68/M | M0 | AML with myelodysplasia-related changes | 2.4 | 10.9 | 94 | 22% | 8 |
| 10 | 48/M | M6 | Acute erythroid leukemia | 1.5 | 7.3 | 117 | 49%b | 4 |
| 11a | 67/M | M1 | AML with myelodysplasia-related changes | 2.3 | 9.3 | 128 | 69% | 2 |
| 12 | 70/M | M2 | AML with maturation | 5.4 | 9.6 | 242 | 33% | alive |
| 13 | 98/M | M5a | Acute monoblastic leukemia | 10.2 | 10.3 | 30 | 79% | NA |
| 14 | 74/F | M1 | AML without maturation | 46 | 9.6 | 32 | 86% | 2 |
| 15 | 52/M | M1 | AML without maturation | 0.1 | 9.7 | 11 | 68% | 42 |
| 16 | 59/F | M1 | AML without maturation | 1.5 | 9.5 | 14 | 53% | 4 |
| 17 | 80/F | M2 | AML with maturation | 0.6 | 8.3 | 63 | 68% | 5 |
| 18 | 75/M | M1 | AML without maturation | 0.9 | 10.2 | 65 | 65% | 4 |
AML, acute myeloid leukemia; FAB, French–American–British; WHO, World Health Organization; WBC, blood cell count; Hb, hemoglobin; Plt, platelet; OS, overall survival; NA, not available.
Patient presented with previous myelodysplastic syndrome.
Of non-erythroid cells.
Morphologically, the median bone marrow blast count was 65% (range, 22-86%) (Table 1). Four patients had a history of MDS and had morphologic evidence of dysplasia associated with AML. Using the WHO classification these cases were classified as AML without maturation (n=7), AML with myelodysplasia related changes (n=4), AML with maturation (n=4), AML with minimal differentiation (n=1), acute monoblastic leukemia (n=1), and acute erythroid leukemia (n=1). Using the FAB system, 10 cases were M1, 4 M2, 2 M0, 1M5a,and 1 M6.
Cytochemistry for myeloperoxidase (MPO) was positive in 16 of 18 cases assessed. The two MPO negative cases were classified using the WHO as AML with minimal differentiation and AML with myelodysplasia-related changes. Using the FAB, both cases were M0. Flow cytomet-ric immunophenotypic data were available for 11 patients. In all cases assessed the blasts were positive for CD13, CD33, CD34, and HLA-DR. CD 117 was expressed in 10 of 11 cases. Myeloperoxidase staining was negative in the 2 patients with the FAB-M0 subtype.
Conventional cytogenetic analysis and FISH
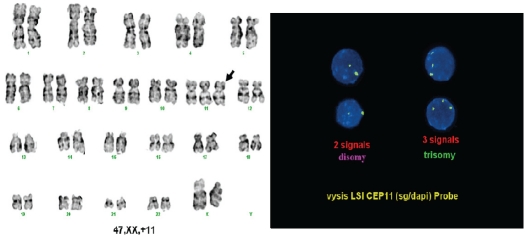
Routine cytogenetic analysis found an isolated +11 as the primary change in all 18 AML patients (Figure 1). Trisomy 11 was further confirmed by FISH in 2 cases.
Figure 1.
Female karyotype (left) and FISH (right) showing trisomy 11.
Molecular studies
Molecular testing was performed on all cases with available DNA as is summarized in Table 2. FLT3 mutations were present in 3 of 15 (20%) patient assessed. The three positive cases had an internal tandem duplication of FLT3 in 2 patients and a point mutation in D835 of the tyro-sine kinase domain in 1 patient. One of 16 (6%) cases had an NRAS mutation. There was no evidence of KRAS mutation in these 16 patients. In 12 patients assessed there was no evidence of NPM1 or KIT mutation.
Table 2.
Gene mutations in acute myeloid leukemia with isolated trisomy 11
| FLT3 % (# case positive/# cases tested) | KIT % (# case positive / # case tested) | NPM1 % (# case positive/# case tested) | RAS % (# case positive/# case tested) |
|---|---|---|---|
| 20% (3/15) | 0% (0/12) | 0% (0/12) | 6% (1/16) |
In a previous study of MDS with +11 by Wang and colleagues [11], control group of AML patients were assessed for partial tandem duplication of the MLL gene. In this study, 5 of 11 cases had PTD of the MLL gene.
Clinical Follow up
Clinical follow-up data were available for 15 patients, and the median overall survival was 5 months (range, 2 to 48 months) (Table 1). Fifteen patients had a progressive clinical course with refractory or relapsed disease. Complete remission was achieved in only 1 patient, who had undergone stem cell transplantation (SCT) (case 12).
Discussion
Gains of whole chromosomes are frequently found in hematologic neoplasms, identified as either primary abnormalities or secondary changes in progressive or advanced stages of the disease [3]. Trisomy for chromosomes 8, 13, and 21 comprises approximately 90% of all cases with isolated trisomies, and as an isolated change is often associated with a poor outcome [4]. As an isolated abnormality, trisomy 11 (+11) in AML is a very rare abnormality. The frequency of isolated +11 at our institution, a major referral center, was 0.09%. In a recent study Wang and colleagues [11] reported a series of cases of MDS with isolated +11. These patients showed a high risk of progression to AML. Heinonen et al previously reported a series of 13 AML cases with +11 and suggested that trisomy 11 could be a marker for unfavorable prognosis [5]. Isolated +11 also has been observed uncommonly in other hematologic malignancies, including T-cell acute lymphoblas-tic leukemia (T-ALL) [12], plasma cell myeloma [13], and chronic lymphocytic leukemia [14]. A current model for AML pathogenesis suggests the importance of two categories of genetic mutation [3]. A class I mutation confers proliferation and cell survival advantages to the clone, whereas a class II mutation impairs cellular differentiation. FLT3, KIT, and RAS mutations are classified as class I mutations, whereas NPM1 mutation is considered a class II mutation. FLT3 is a type III receptor tyrosine kinase. Mutations of FLT3 result in constitutive activation of downstream molecular signaling pathways and aberrant cell growth. FLT3 mutations are seen in about 30% of AML cases and fall into two main groups: internal tandem duplication and tyrosine kinase point mutation [15]. Both of the mutations render the receptor constitutively activated. FLT3 mutations occur in approximately 20% of all cases of AML [15]. Rege-Cambrin and colleagues identified FLT-ITD mutation in 30% of isolated +11 AML cases, and their study demonstrated an association with a short survival [16]. Our data are in accord with these results. Three of 15 (20%) patients assessed had FLT3 mutation. These patients had a poor prognosis, but not apparently worse than the group as a whole, suggesting that +11 is the more important prognostic indicator.
The RAS (N- and K-) genes encode a family of membrane-associated proteins that regulate signal transduction on binding to a variety of membrane receptors. They play important roles in the regulatory processes of proliferation, differentiation, and apoptosis. In other studies, presence of a RAS mutation did not affect prognosis in AML [17,18]. In this study, an NRAS mutation was seen in a single patient (1 of 16 assessed; 6%). There were no KRAS mutations. As a group, RAS mutations are more common in AML than observed in this series of +11 AML cases [19]. Thus, RAS gene mutation does not seem to be important in pathogenesis of +11 AML.
Point mutation at exon 12 of the NPM1 gene represents one of the most frequent mutations in adult AML, in approximately 30% of unse-lected AML cases [20]. NPM1 mutation is usually associated with the FAB M4 or M5 subtypes and is more common in cytogenetically normal AML cases [20,21]. In this study, NPM1 mutations was not identified in 12 case assessed. This is consistent with previous reports in the literature as our study group all had cytogenetic abnormalities and there was a low frequency of monocytic differentiation. KIT gene is infrequently seen in AML with an overall incidence of 3%, and is most frequent in core binding factor AMLs [22]. KIT gene was not observed in all 12 patients assessed in this study.
In previous studies of AML associated with +11, mostly appearing in clinical journals, relatively little attention has been paid to the morphologic features of this disease. In addition, most studies have not used the current WHO classification system. In this study, most cases of AML associated with +11 were classified as AML without maturation using the WHO classification or as M1 using the FAB classification. The im-munophenotype of the cases assessed in this series showed evidence consistent with a stem cell phenotype with expression of CD34, CD117, and HLA-DR. These results are in accord with an earlier study by Slovak and colleagues [23].
The role of trisomy 11 in the leukemogenesis of AML remains to be defined. It has been speculated that trisomies in general lead to a gene dosage effect that results in increased gene copy number and a survival advantage [3]. Previous studies by others have shown that many +11 AML cases have partial tandem duplication of the MLL gene. In one study by Caligiuri and colleagues, up to 90% of cases of AML carrying an isolated +11 had a partial tandem duplication of MLL [24]. In this study, 5 of 11 cases assessed had partial tandem duplication of MLL [11].
In summary, AML associated with isolated trisomy 11 has a poor prognosis. In this group of 18 cases, the morphologic features most often supported classification as AML without maturation (WHO) or M1 (FAB). The immuno-phenotype usually has stem cell features. As shown by others, partial tandem duplication of the MLL gene occurs in approximately 50% of cases. FLT3 gene mutations occur in approximately 20% of cases and there is a very low frequency of mutations of the NRAS, KRAS, NPM1, and KIT genes.
Acknowledgments
This work was supported in part by funds from the Saudi Arabian Cultural Mission (SACM/ MOHE-I). We thank Sue Moreau in the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for her professional editing of our manuscript.
References
- 1.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Harris NL. 4th. Lyon: IARC; 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 3.Helm S, Mitelman F. 3rd. Mexico: Wiley-Blackwell; 2009. Cancer Cytogenetics, Chromosomal and Molecular Genetic Aberrations of Tumor Cells. [Google Scholar]
- 4.Farag SS, Archer KJ, Mrózek K, Vardiman JW, Carroll AJ, Pettenati MJ, Moore JO, Kolitz JE, Mayer RJ, Stone RM, Larson RA, Bloomfield CD. Isolated trisomy of chromosomes 8, 11, 13 and 21 is an adverse prognostic factor in adults with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B 8461. Int J Oncol. 2002;21:1041–1051. [PubMed] [Google Scholar]
- 5.Heinonen K, Mrózek K, Lawrence D, Arthur DC, Pettenati MJ, Stamberg J, Qumsiyeh MB, Verma RS, MacCallum J, Schiffer CA, Bloomfield CD. Clinical characteristics of patients with de novo acute myeloid leukaemia and isolated trisomy 11: a Cancer and Leukemia Group B study. Br J Haemaatol. 1998;101:513–520. doi: 10.1046/j.1365-2141.1998.00714.x. [DOI] [PubMed] [Google Scholar]
- 6.Yin CC, Medeiros LJ, Glassman AB, Lin P. t (8;21)(q22;q22) in blast phase of chronic mye-logenous leukemia. Am J Clin Pathol. 2004;121:836–842. doi: 10.1309/H8JH-6L09-4B9U-3HGT. [DOI] [PubMed] [Google Scholar]
- 7.Lu G, Yin CC, Medeiros LJ, Abruzzo LV. Deletion 15q as the sole abnormality in acute myeloid leukemia: report of three cases and review of the literature. Cancer Genet Cytogenet. 2009;188:118–23. doi: 10.1016/j.cancergencyto.2008.09.006. Review. [DOI] [PubMed] [Google Scholar]
- 8.Shaffer LG, Slovak ML, Campbell LJ. Basel: S. Karger; 2009. An International System for Human Cytogenetic Nomenclature (ISCN) [Google Scholar]
- 9.Bains A, Luthra R, Medeiros LJ, Zuo Z. FLT3 and NPM1 mutations in myelodysplastic syndromes: frequency and potential value for predicting progression to acute myeloid leukemia. Am J Clin Pathol. 2011;135:62–69. doi: 10.1309/AJCPEI9XU8PYBCIO. [DOI] [PubMed] [Google Scholar]
- 10.Gustafson SA, Lin P, Chen SS, Chen L, Abruzzo LV, Luthra R, Medeiros LJ, Wang SA. Therapy-related acute myeloid leukemia with t(8;21) (q22;q22) shares many features with de novo acute myeloid leukemia with t(8;21)(q22)(q22) but does not have a favorable outcome. Am J Clin Pathol. 2009;131:647–655. doi: 10.1309/AJCP5ETHDXO6NCGZ. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Jabbar K, Lu G, Chen SS, Galili N, Vega F, Jones D, Raza A, Kantarjian H, Garcia-Manero G, McDonnell TJ, Medeiros LJ. Trisomy 11 in myelodysplastic syndromes defines a unique group of disease with aggressive clinico-pathologic features. Leukemia. 2010;24:740–747. doi: 10.1038/leu.2009.289. [DOI] [PubMed] [Google Scholar]
- 12.Cauwelier B, Cavé H, Gervais C, Lessard M, Barin C, Perot C, Van den Akker J, Mugneret F, Charrin C, Pagès MP, Grégoire MJ, Jonveaux P, Lafage-Pochitaloff M, Mozzicconacci MJ, Terré C, Luquet I, Cornillet-Lefebvre P, Laurence B, Plessis G, Lefebvre C, Leroux D, Antoine-Poirel H, Graux C, Mauvieux L, Heimann P, Chalas C, Clappier E, Verhasselt B, Benoit Y, Moerloose BD, Poppe B, Van Roy N, Keersmaecker KD, Cools J, Sigaux F, Soulier J, Hagemeijer A, Paepe AD, Dastugue N, Berger R, Speleman F. Clinical, cytogenetic and molecular characteristics of 14 T-ALL patients carrying the TCRbeta-HOXA rearrangement: a study of the Groupe Francophone de Cytogénétique Hématologique. Leukemia. 2007;21:121–128. doi: 10.1038/sj.leu.2404410. [DOI] [PubMed] [Google Scholar]
- 13.Cremer FW, Kartal M, Hose D, Bila J, Buck I, Bellos F, Raab MS, Brough M, Moebus A, Hager HD, Goldschmidt H, Moos M, Bartram CR, Jauch A. High incidence and intraclonal heterogeneity of chromosome 11 aberrations in patients with newly diagnosed multiple myeloma detected by multiprobe interphase FISH. Cancer Genet Cytogenet. 2005;161:116–124. doi: 10.1016/j.cancergencyto.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Juliusson G, Merup M. Cytogenetics in chronic lymphocytic leukemia. Semin Oncol. 1998;25(1):19–26. [PubMed] [Google Scholar]
- 15.Yanada M, Matsuo K, Suzuki T, Kiyoi H, Naoe T. Prognostic significance of FLT3 internal tandem duplication and tyrosine kinase domain mutations for acute myeloid leukemia: a meta-analysis. Leukemia. 2005;19:1345–1349. doi: 10.1038/sj.leu.2403838. [DOI] [PubMed] [Google Scholar]
- 16.Rege-Cambrin G, Giugliano E, Michaux L, Stul M, Scaravaglio P, Serra A, Saglio G, Hagemeijer A. Trisomy 11 in myeloid malignancies is associated with internal tandem duplication of both MLL and FLT3 genes. Haematologica. 2005;90:262–264. [PubMed] [Google Scholar]
- 17.Bowen D, Frew ME, Hills R, Gale RE, Wheatley K, Groves MJ, Langabeer SE, Kottaridis PD, Moorman AV, Burnett AK, Linch DC. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005;106:2113–2119. doi: 10.1182/blood-2005-03-0867. [DOI] [PubMed] [Google Scholar]
- 18.Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: a study of 2502 patients. Blood. 2006;107:3847–3853. doi: 10.1182/blood-2005-08-3522. [DOI] [PubMed] [Google Scholar]
- 19.Radich JP, Kopecky KJ, Willman CL, Weick J, Head D, Appelbaum F, Collins SJ. N-ras mutations in adult de novo acute myelogenous leukemia: prevalence and clinical significance. Blood. 1990;76(4):801–807. [PubMed] [Google Scholar]
- 20.Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, Ehninger G. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 21.Döhner K, Schlenk RF, Habdank M, Scholl C, Rücker FG, Corbacioglu A, Bullinger L, Fröhling S, Döhner H. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cyto-genetics: interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 22.Lück SC, Russ AC, Du J, Gaidzik V, Schlenk RF, Pollack JR, Döhner K, Döhner H, Bullinger L. KIT mutations confer a distinct gene expression signature in core binding factor leukaemia. Br J Haematol. 2010;148:925–937. doi: 10.1111/j.1365-2141.2009.08035.x. [DOI] [PubMed] [Google Scholar]
- 23.Slovak ML, Traweek ST, Willman CL, Head DR, Kopecky KJ, Magenis RE, Appelbaum FR, Forman SJ. Trisomy 11: an association with stem/ progenitor cell immunophenotype. Br J Haematol. 1995;90:266–273. doi: 10.1111/j.1365-2141.1995.tb05146.x. [DOI] [PubMed] [Google Scholar]
- 24.Caligiuri MA, Strout MP, Schichman SA, Mrózek K, Arthur DC, Herzig GP, Baer MR, Schiffer CA, Heinonen K, Knuutila S, Nousiainen T, Ruutu T, Block AW, Schulman P, Pedersen-Bjergaard J, Croce CM, Bloomfield CD. Partial tandem duplication of ALL1 as a recurrent molecular defect in acute myeloid leukemia with trisomy 11. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]