Abstract
AIM
To investigate the impact of genetic polymorphisms in CYP2D6, CYP3A5, CYP2C9 and CYP2C19 on the pharmacokinetics of tamoxifen and its metabolites in Asian breast cancer patients.
METHODS
A total of 165 Asian breast cancer patients receiving 20 mg tamoxifen daily and 228 healthy Asian subjects (Chinese, Malay and Indian; n = 76 each) were recruited. The steady-state plasma concentrations of tamoxifen and its metabolites were quantified using high-performance liquid chromatography. The CYP2D6 polymorphisms were genotyped using the INFINITI™ CYP450 2D6I assay, while the polymorphisms in CYP3A5, CYP2C9 and CYP2C19 were determined via direct sequencing.
RESULTS
The polymorphisms, CYP2D6*5 and *10, were significantly associated with lower endoxifen and higher N-desmethyltamoxifen (NDM) concentrations. Patients who were *1/*1 carriers exhibited 2.4- to 2.6-fold higher endoxifen concentrations and 1.9- to 2.1-fold lower NDM concentrations than either *10/*10 or *5/*10 carriers (P < 0.001). Similarly, the endoxifen concentrations were found to be 1.8- to 2.6-times higher in *1/*5 or *1/*10 carriers compared with *10/*10 and *5/*10 carriers (P≤ 0.001). Similar relationships were observed between the CYP2D6 polymorphisms and metabolic ratios of tamoxifen and its metabolites. No significant associations were observed with regards to the polymorphisms in CYP3A5, CYP2C9 and CYP2C19.
CONCLUSIONS
The present study in Asian breast cancer patients showed that CYP2D6*5/*10 and *10/*10 genotypes are associated with significantly lower concentrations of the active metabolite of tamoxifen, endoxifen. Identifying such patients before the start of treatment may be useful in optimizing therapy with tamoxifen. The role of CYP3A5, CYP2C9 and CYP2C19 seem to be minor.
Keywords: CYP2C19, CYP2D6, CYP3A5, pharmacogenetics, pharmacokinetics, tamoxifen
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
Tamoxifen is metabolized to active metabolites, 4-hydroxytamoxifen and endoxifen, by multiple cytochrome P450 (CYP) enzymes including CYP2D6, CYP3A4/5, CYP2C9/19, CYP1A2 and CYP2B6.
The steady-state plasma concentrations of tamoxifen and its metabolites can be affected by variations in the activity of these enzymes.
Although CYP2D6*4 and *10 have been shown to influence the plasma concentration of endoxifen in Caucasian and Korean patients respectively, there is still a paucity of data on CYP2D6 pharmacogenetics in other Orientals such as Chinese, Malays and Indians.
WHAT THIS STUDY ADDS
Pharmacogenetic analyses of a comprehensive panel of CYP2D6 polymorphisms (*2, *2A, *3, *4, *5, *6, *7, *8, *9, *10, *12, *14, *17, *29, *41 and *xN) were performed in three distinct Asian ethnic groups and breast cancer patients with CYP2D6*5 and *10 found to be highly prevalent.
Both CYP2D6*5 and *10 were significantly associated with lower endoxifen and higher N-desmethyltamoxifen concentrations as well as a lower rate of metabolic conversion of N-desmethyltamoxifen to endoxifen.
Polymorphisms present in CYP3A5, CYP2C9 and CYP2C19 were not found to be significantly associated with plasma concentrations of analytes suggesting that these enzymes may be playing minor roles in the metabolic pathway of tamoxifen compared with CYP2D6.
Introduction
Tamoxifen is widely used in the treatment and prevention of oestrogen receptor (ER) and/or progesterone receptor (PR) positive breast cancers [1]. It undergoes metabolic activation to form two highly potent metabolites, namely 4-hydroxytamoxifen (4-OHT) formed via 4-hydroxylation of tamoxifen and 4-hydroxy-N-desmethyltamoxifen (endoxifen) resulting from the N-demethylation of 4-OHT. Both metabolites exhibit 100-fold greater affinity for ER and 30- to 100-fold greater potency than tamoxifen [2]. Up to 97% of endoxifen is formed via an alternative pathway involving sequential CYP3A4/5 mediated N-demethylation of tamoxifen to form N-desmethyltamoxifen (NDM) and CYP2D6 catalysed 4-hydroxylation of NDM [3]. Two other enzymes, CYP2C9 and CYP2C19, have been found to play minor roles in these metabolic pathways [4], although recent studies have highlighted an important role of CYP2C19 in influencing treatment response to tamoxifen [5]. There is a wide degree of interindividual and interethnic differences in the steady-state plasma concentrations of tamoxifen and its metabolites which have been shown to influence therapeutic outcome [6, 7]. This area of research has generated intense clinical interest because of its potential to select women with postmenopausal hormone receptor positive early stage breast cancer who are poor metabolizers of tamoxifen to receive alternative adjuvant endocrine therapy with aromatase inhibitors [8]. The deviations in plasma concentrations of tamoxifen and its metabolites can be due to drug–drug interactions or genetic variants present in the genes responsible for encoding the various proteins involved in regulating its disposition [3].
Among the various candidate genes, polymorphic variants in the CYP2D6 gene play an important role in affecting the catalytic activity of the CYP2D6 enzyme and hence the plasma concentrations of 4-OHT and endoxifen [3, 9–12]. Caucasian breast cancer patients homozygous for the *4 (1846G>A; rs3892097) allele are known to display lower concentrations of endoxifen compared with patients who are *1/*1 carriers [3, 9, 13, 14]. The plasma concentration of endoxifen is also affected by the presence of other null alleles such as *3 (2549delA; rs35742686), *5 (CYP2D6del) and *6 (1707delT; rs5030655) [9, 10, 13]. However, the distribution of functional CYP2D6 polymorphisms varies between breast cancer patients from different ethnic backgrounds which may influence therapeutic outcome to tamoxifen treatment [10].
Although CYP2D6*4 (1846G>A; rs3892097) is the predominant defective allele in the Caucasian population [3, 9, 13, 14], it is rare in the Asian population (1–3%). Instead, CYP2D6*5 (CYP2D6del) and CYP2D6*10 (100C>T; rs1065852) alleles [15] have been reported to be more prevalent [16] in Asian populations, occurring at frequencies of approximately 5% to 10% and more than 40%, respectively [17, 18]. A recent study in Korean breast cancer patients has shown that the steady-state plasma concentrations of 4-OHT and endoxifen were significantly lower and the median time to progression significantly shorter in CYP2D6*10 homozygotes compared with patients who were wild-type or heterozygotes [19]. In view of the greater prevalence of CYP2D6*5 (CYP2D6del) and *10 (100C>T; rs1065852) among Asian populations, these polymorphisms may be more important in influencing the pharmacokinetics of tamoxifen in Asian breast cancer patients. Apart from CYP2D6, other cytochrome P450 enzymes (CYP3A4/5, CYP2C9 and CYP2C19) also contribute to the overall metabolism of tamoxifen and its metabolites, albeit to different extents [4, 20–23].
On the basis of previous studies, we hypothesize that different functionally defective CYP2D6 alleles which are operative in the Asian population [CYP2D6*5 (CYP2D6del) and *10 (100C>T; rs1065852)] may impact on the pharmacokinetics of tamoxifen and its active metabolites (4-OHT and endoxifen) and hence affect therapeutic outcome. The objectives of the present study were to study the pharmacogenetics of CYP3A5, CYP2C9, CYP2C19 and CYP2D6 polymorphisms in healthy Asian subjects (Chinese, Malays, Indians) and to delineate the impact of known functional polymorphisms in these genes on the plasma concentrations as well as metabolic ratios of tamoxifen and its metabolites in Asian breast cancer patients.
Methods
Study subjects
The healthy subject population comprised of three ethnic groups predominant in the Asian population of Singapore, namely Chinese, Malays and Indians (n = 76 in each group). The ethnicities of all subjects were confirmed by individual screening and verified against their National Registry Identification Cards.
A total of 165 breast cancer patients were recruited prospectively. All patients were histologically diagnosed with ER and/or PR positive breast tumours and received 20 mg tamoxifen daily. Patients who had received inhibitors or inducers of CYP2D6 within 4 weeks from the time of enrolment were excluded from the study. Patients with any of the following conditions were also excluded from the study: allergy to tamoxifen, pregnancy, expected survival of less than 6 months, prior malignancies other than basal cell carcinoma of the skin or carcinoma in situ of the uterine cervix, uncontrolled intercurrent illness such as ongoing or active infection, symptomatic congestive heart failure, unstable angina pectoris, cardiac arrhythmia, or psychiatric illness/social situations that might limit compliance with the study requirements. The study was approved by the ethics review committee of the National Cancer Centre, Singapore. Written consent was obtained from both healthy subjects and patients for participation in this study.
Pharmacogenetic analyses
Purified genomic DNA was extracted from peripheral blood samples of healthy subjects and patients using the Gentra® Puregene Blood Kit (Qiagen, Inc.). A total of 228 healthy subjects and 165 breast cancer patients were screened for the following polymorphisms: CYP2D6[*2 (2850C>T; rs16947), *2A (–1584C>G), *3 (2549delA; rs35742686), *4 (1846G>A; rs3892097), *5 (CYP2D6del), *6 (1707delT; rs5030655), *7 (2935A>C; rs5030867), *8 (1758G>T), *9 (2615-2617delAAG; rs5030656), *10 (100C>T; rs1065852), *12 (124G>A; rs5030862), *14 (1758G>A), *17 (1023C>T; rs28371706), *29 (1659G>A; rs61736512), *41 (2988G>A; rs28371725) and *xN (CYP2D6dup)], CYP3A5*3 (6986A>G; rs776746), CYP2C9[*2 (430C>T; rs1799853) and *3 (1075A>C; rs1057910)] and CYP2C19[*2 (681G>A; rs4244285), *3 (636G>A; rs4986893) and *17 (–3402C>T and −806C>T; rs12248560)]. The polymorphisms in CYP2D6 were genotyped using the INFINITI™ CYP450 2D6I assay (AutoGenomics, Inc.) in accordance with the manufacturer's protocol. The polymorphic variants in CYP3A5, CYP2C9 and CYP2C19 were genotyped by amplification via polymerase chain reaction [24, 25], followed by purification by treatment with shrimp alkaline phosphatase and exonuclease I and direct sequencing on a ABI 3730 DNA Analyzer (Applied Biosystems, Inc.).
Pharmacokinetic analyses
Blood (3 ml) was collected from patients who had received a daily dose of 20 mg tamoxifen for 8 weeks or more. The samples were protected from light and immediately centrifuged at 2000 g for 10 min. Plasma was extracted and stored at −80°C until analysis.
The steady-state plasma concentrations of tamoxifen (CTAM) and its metabolites, NDM (CNDM), 4-OHT (C4-OHT) and endoxifen (CEND) were quantified by reverse-phase high-performance liquid chromatography with fluorescence detection modified from the methods developed by Lee et al. [26] and Zhu et al. [27]. Tamoxifen (≥99%), trans-4-hydroxytamoxifen (≥98%) and propranolol (99%) were purchased from Sigma-Aldrich Co., Singapore. N-Desmethyltamoxifen and trans-endoxifen were kind gifts from Dr D.A. Flockhart at Indiana University School of Medicine (Indianapolis, IN, USA) and Dr Philip Lazarus at Pennsylvania State University College of Medicine (PA, USA), respectively. The analytes were extracted from 200 µl of plasma with n-hexane : n-butanol (98:2, v : v), using propanolol as an internal standard. The chromatographic separation of the extract was performed on a C8 analytical column using a mobile phase consisting of acetonitrile and 100 mm ammonium acetate (pH 5.5) with a gradient profile of 30–80% (v : v) of acetonitrile. The limits of quantification for tamoxifen, NDM, 4-OHT and endoxifen were 10.0 ng ml−1, 10.0 ng ml−1, 0.5 ng ml−1 and 1.0 ng ml−1, respectively. The calibration curves were linear over the concentration ranges of 10.0–800.0 ng ml−1 for tamoxifen (r2= 0.9977) and NDM (r2= 0.9974), 0.5–40 ng ml−1 for 4-OHT (r2= 0.9993), and 1.0–80.0 ng ml−1 for endoxifen (r2= 0.9973). The intra-day coefficient of variation was less than 7.0%, while inter-day coefficient of variation was less than 14.0%.
The plasma concentration ratios or metabolic ratios (MRs) were calculated as follows: MRNDM-TAM=CNDM/CTAM, MR4-OHT-TAM=C4-OHT/CTAM (× 102), MREND-NDM=CEND/CNDM (× 102), and MREND-4-OHT=CEND/C4-OHT. The total plasma concentration ratios or total metabolic ratios (TMRs) were calculated as follows: TMR4-OHT=CEND/CTAM+C4-OHT (× 102) and TMRNDM=CEND/CTAM+CNDM (× 102).
Statistical analyses
The chi-squared test was employed to assess for Hardy–Weinberg equilibrium between the genotype frequencies. Genotypic–phenotypic associations were performed using the non-parametric Kruskal–Wallis test and Mann–Whitney U-test. The level of significance was set at P < 0.050. Bonferroni corrections were applied for multiple comparisons between genotype groups. The levels of significance were set at P < 0.017 and P < 0.005 for multiple pairwise comparisons across three and five groups, respectively. The statistical analyses were carried out with SPSS version 14.0 (SPSS, Inc.).
Results
Patient demographics and plasma concentrations of tamoxifen and its metabolites
The median age of the breast cancer patients was 49.3 years (range 30–74 years) with 83.0% and 17.0% of patients being premenopausal and postmenopausal, respectively. The median height and weight were 156.0 cm (range 134.0–172.3 cm) and 58.9 kg (range 38.7–91.7 kg). The majority of the patients were Chinese (83.0%) followed by Indians (8.5%) and Malays (7.3%), consistent with the racial composition in Singapore.
The inter-individual variations in plasma concentrations of tamoxifen, NDM and 4-OHT were found to vary between 11- to 20-fold, while that for endoxifen varied by more than 24-fold. The median plasma concentrations of tamoxifen, NDM, 4-OHT and endoxifen in the breast cancer patients were 205.16 ng ml−1 (range 39.26–599.91 ng ml−1), 304.05 ng ml−1 (40.82–802.98 ng ml−1), 1.96 ng ml−1 (0.47–5.33 ng ml−1) and 13.69 ng ml−1 (1.74–42.80 ng ml−1), respectively. A high degree of interindividual variation was also observed for the MR (4- to 30-fold) and the TMR (17- to 20-fold). The median MRs and TMRs of the patient population were as follows: MRNDM-TAM= 1.56 (range 0.62–2.76), MR4-OHT-TAM= 1.01 (range 0.19–2.35), MREND-NDM= 4.97 (range 0.92–27.81), MREND-4-OHT= 6.89 (range 1.99–13.18), TMR4-OHT= 6.78 (range 1.14–19.50) and TMRNDM= 2.76 (range 0.51–10.69). None of the patients was prescibed CYP2D6 inhibitors while they were receiving tamoxifen.
Pharmacogenetics of CYP2D6, CYP3A5, CYP2C and CYP2C19
The allelic frequency distributions of CYP2D6 polymorphisms in 228 healthy Asian subjects (Chinese, Malay and Indian; n = 76 each) and 165 Asian breast cancer patients are indicated in Table 1. The CYP2D6 genotypes of four patients could not be determined probably because of poor quality DNA samples.
Table 1.
Genotypic and allelic frequencies of polymorphisms in CYP2D6, CYP3A5, CYP2C9 and CYP2C19 in healthy Asians (n = 76 each) and Asian breast cancer patient (n = 165) populations
| Genotypic frequencies | Allelic frequencies | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphisms | Genotypes | Chinese | Malays | Indians | Breast cancer patients | Alleles | Chinese | Malays | Indians | Breast cancer patients | Overall P value | ||||
| CYP2D6 | |||||||||||||||
| CYP2D6*2 | *1/*1 | 37 | (62.7) | 47 | (68.1) | 23 | (32.9) | 92 | (66.2) | *1 | 0.83 | 0.83 | 0.56 | 0.81 | <0.001 |
| (2850C>T; rs16947) | *1/*2 | 21 | (35.6) | 20 | (29.0) | 33 | (47.1) | 37 | (26.6) | *2 | 0.17 | 0.17§ | 0.44‡ | 0.19 | |
| *2/*2 | 1 | (1.7) | 2 | (2.9) | 14 | (20.0) | 10 | (7.2) | |||||||
| CYP2D6*2A | *1/*1 | 41 | (67.2) | 57 | (81.4) | 38 | (52.8) | 105 | (75.6) | *1 | 0.85 | 0.90 | 0.72 | 0.87 | 0.002 |
| (-1584C>G) | *1/*2A | 19 | (31.1) | 12 | (17.1) | 27 | (37.5) | 28 | (20.1) | *2A | 0.15 | 0.10§ | 0.28‡ | 0.13 | |
| *2A/*2A | 1 | (1.6) | 1 | (1.4) | 7 | (9.7) | 6 | (4.3) | |||||||
| CYP2D6*4 | *1/*1 | 50 | (98.0) | 50 | (92.6) | 57 | (89.1) | 135 | (99.3) | *1 | 0.98 | 0.96 | 0.91 | 1.00 | 0.199 |
| (1846G>A; rs3892097) | *1/*4 | 1 | (2.0) | 4 | (7.4) | 3 | (4.7) | 1 | (0.7) | *4 | 0.02 | 0.04 | 0.09 | <0.01 | |
| *4/*4 | 0 | (0.0) | 0 | (0.0) | 4 | (6.3) | 0 | (0.0) | |||||||
| CYP2D6*5 | *1/*1 | 61 | (81.3) | 70 | (92.1) | 72 | (94.7) | 139 | (84.2) | *1 | 0.91 | 0.96 | 0.97 | 0.92 | 0.018 |
| (CYP2D6 del) | *1/*5 | 14 | (18.7) | 6 | (7.9) | 4 | (5.3) | 26 | (15.8) | *5 | 0.09 | 0.04 | 0.03‡ | 0.08 | |
| *5/*5 | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | |||||||
| CYP2D6*8/*14 | *1/*1 | 50 | (92.6) | 61 | (100.0) | 69 | (100.0) | 98 | (96.1) | *1 | 0.97 | 1.00 | 1.00 | 1.00 | 0.113 |
| (1758G>T/A) | *1/*8 | 1 | (1.9) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | *8 | 0.01 | – | – | – | |
| *1/*14 | 2 | (3.7) | 0 | (0.0) | 0 | (0.0) | 4 | (3.9) | *14 | 0.02 | – | – | 0.01 | ||
| *8/*8 | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | |||||||
| *14/*14 | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | |||||||
| *8/*14 | 0 | (1.9) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | |||||||
| CYP2D6*9 | *1/*1 | 59 | (100.0) | 65 | (100) | 68 | (98.6) | 139 | (100.0) | *1 | 1.00 | 1.00 | 0.99 | 1.00 | 0.405 |
| (2615_2617delAAG; | *1/*9 | 0 | (0.0) | 0 | (0.0) | 1 | (1.4) | 0 | (0.0) | *9 | – | – | 0.01 | – | |
| rs5030656) | *9/*9 | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | ||||||
| CYP2D6*10 | *1/*1 | 14 | (23.0) | 12 | (17.4) | 56 | (77.8) | 42 | (30.2) | *1 | 0.44 | 0.43 | 0.89 | 0.49 | <0.001 |
| (100C>T; rs1065852) | *1/*10 | 29 | (47.5) | 36 | (52.2) | 15 | (20.8) | 54 | (38.9) | *10 | 0.56 | 0.57§ | 0.11‡ | 0.51 | |
| *10/*10 | 18 | (29.5) | 21 | (30.4) | 1 | (1.4) | 43 | (30.9) | |||||||
| CYP2D6*12 | *1/*1 | 61 | (100.0) | 68 | (98.6) | 72 | (100.0) | 139 | (100.0) | *1 | 1.00 | 0.99 | 1.00 | 1.00 | 0.380 |
| (124G>A; rs5030862) | *1/*12 | 0 | (0.0) | 1 | (1.4) | 0 | (0.0) | 0 | (0.0) | *12 | – | 0.01 | – | – | |
| *12/*12 | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | |||||||
| CYP2D6*41 | *1/*1 | 56 | (96.6) | 56 | (82.4) | 51 | (73.9) | 125 | (89.9) | *1 | 0.99 | 0.91 | 0.85 | 0.95 | 0.017 |
| (2988G>A; rs28371725) | *1/*41 | 2 | (3.4) | 11 | (16.2) | 17 | (24.6) | 13 | (9.4) | *41 | 0.01† | 0.09 | 0.15‡ | 0.05 | |
| *41/*41 | 0 | (0.0) | 1 | (1.5) | 1 | (1.4) | 1 | (0.7) | |||||||
| CYP2D6*xN | *1 | 61 | (100.0) | 69 | (98.6) | 71 | (98.6) | 133 | (95.7) | *1 | 1.00 | 0.99 | 0.99 | 0.98 | 0.648 |
| (multiple CYP2D6) | *1/*xN | 0 | (0.0) | 1 | (1.4) | 1 | (1.4) | 6 | (4.3) | *xN | – | 0.01 | 0.01 | 0.02 | |
| CYP3A5 | |||||||||||||||
| CYP3A5*3¶ | *1/*1 | (8.3) | (10.2) | (12.2) | 14 | (8.5) | *1 | 0.25 | 0.39 | 0.41 | 0.30 | – | |||
| (6986A>G; rs776746) | *1/*3 | (32.4) | (57.1) | (56.7) | 72 | (43.6) | *3 | 0.76 | 0.61 | 0.59 | 0.70 | ||||
| *3/*3 | (59.3) | (32.7) | (31.1) | 79 | (47.9) | ||||||||||
| CYP2C9 | |||||||||||||||
| CYP2C9*2 | *1/*1 | 76 | (100.0) | 76 | (100.0) | 72 | (94.7) | 165 | (100.0) | *1 | 1.00 | 1.00 | 0.97 | 1.00 | 0.005 |
| (430C>T; rs1799853) | *1/*2 | 0 | (0.0) | 0 | (0.0) | 4 | (5.26) | 0 | (0.0) | *2 | 0.00 | 0.00§ | 0.03‡ | 0.00 | |
| *2/*2 | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | |||||||
| CYP2C9*3 | *1/*1 | 74 | (97.4) | 69 | (90.8) | 63 | (82.9) | 151 | (92.1) | *1 | 0.99 | 0.95 | 0.91 | 0.96 | <0.001 |
| (1075A>C; rs1057910) | *1/*3 | 2 | (2.6) | 7 | (9.2) | 12 | (15.8) | 12 | (7.3) | *3 | 0.01† | 0.05 | 0.09‡ | 0.04 | |
| *3/*3 | 0 | (0.0) | 0 | (0.0) | 1 | (1.3) | 1 | (0.6) | |||||||
| CYP2C19 | |||||||||||||||
| CYP2C19*2 | *1/*1 | 38 | (50.0) | 44 | (57.9) | 28 | (36.8) | 73 | (45.1) | *1 | 0.68 | 0.77 | 0.62 | 0.65 | <0.001 |
| (681G>A; rs4244285) | *1/*2 | 28 | (36.8) | 29 | (38.2) | 38 | (50.0) | 66 | (40.7) | *2 | 0.32 | 0.23§ | 0.38 | 0.35 | |
| *2/*2 | 10 | (13.2) | 3 | (3.9) | 10 | (13.2) | 23 | (14.2) | |||||||
| CYP2C19*3 | *1/*1 | 65 | (85.5) | 65 | (85.5) | 75 | (98.7) | 149 | (90.3) | *1 | 0.93 | 0.93 | 0.99 | 0.95 | <0.001 |
| (636G>A; rs4986893) | *1/*3 | 11 | (14.5) | 11 | (14.5) | 1 | (1.3) | 15 | (9.1) | *3 | 0.07 | 0.07§ | 0.01‡ | 0.05 | |
| *3/*3 | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 1 | (0.6) | |||||||
| CYP2C19*17 | *1/*1 | 75 | (98.7) | 69 | (90.8) | 50 | (65.8) | 153 | (93.3) | *1 | 0.99 | 0.95 | 0.81 | 0.97 | <0.001 |
| (–3402C>T and | *1/*17 | 1 | (1.4) | 7 | (9.2) | 23 | (30.3) | 11 | (6.7) | *17 | 0.01† | 0.05§ | 0.19‡ | 0.03 | |
| –806C>T; rs12248560) | *17/*17 | 0 | (0.0) | 0 | (0.0) | 3 | (3.9) | 0 | (0.0) | ||||||
Chinese vs. Malay, P < 0.050.
Chinese vs. Indian, P < 0.050.
Malay vs. Indian, P < 0.050.
Genotypic and allelic frequencies were from previous study by Balram et al. [23].
Wide inter-ethnic variations were observed in the allelic frequencies of CYP2D6 polymorphisms in the healthy Asian populations (Table 1). Pairwise comparisons between the three Asian ethnic groups showed that the frequencies of the various CYP2D6 variants were similar between the Chinese and Malay ethnic groups but differed significantly from the Indians with regards to selected polymorphisms (Table 1). The allelic frequency of CYP2D6*10 (100C>T; rs1065852) was approximately fivefold higher in the healthy Chinese and Malay populations compared with the Indian population (Chinese vs. Malay vs. Indians: 0.56 vs. 0.57 vs. 0.11; P < 0.001). Similarly, the allelic frequency of CYP2D6*5 (CYP2D6del) was approximately twofold to threefold higher in the Chinese compared with the Malays and Indians (Chinese vs. Malay vs. Indians: 0.09 vs. 0.04 vs. 0.03; P = 0.018; Table 1). The Indian and Malay populations were also found to harbour significantly higher frequencies of CYP2D6*41 (2988G>A; rs28371725) compared with the Chinese population (Indians vs. Malay vs. Chinese: 0.15 vs. 0.09 vs. 0.01; Table 1).
The polymorphisms in CYP2C9[*2 (430C>T; rs1799853) and *3 (1075A>C; rs1057910)] and CYP2C19[*2 (681G>A; rs4244285), *3 (636G>A; rs4986893) and *17 (–3402C>T and −806C>T; rs12248560)] were noted to display significant interethnic variations and are depicted in Table 1.The pharmacogenetics of CYP3A5*3 (6986A>G; rs776746) were previously reported in healthy Chinese, Malay and Indian populations by Balram et al. [23] and are shown in Table 1 for the purpose of comparison. The allelic and genotypic distribution profiles of the polymorphisms in the candidate genes CYP2D6, CYP3A5, CYP2C9 and CYP2C19 observed in the patient population were found to be similar to those of the healthy Chinese population (Table 1).
Influence of CYP2D6 polymorphisms on the plasma concentrations of tamoxifen and its metabolites
The median (range) plasma concentrations of tamoxifen and the three metabolites observed in patients within each genotypic group are depicted in Table 2. Although the CYP2D6*10 (100C>T; rs1065852) polymorphism was not found to be significantly associated with plasma concentrations of tamoxifen and 4-OHT, a stepwise decrease in median 4-OHT concentration was observed when comparing patients with zero, one or two copies of the CYP2D6*10 (100C>T; rs1065852) allele (Figure 1A,C, Table 2). Significant gene–dose dependent effects were, however, observed between the CYP2D6*10 (100C>T; rs1065852) allele and the plasma concentrations of NDM and endoxifen (Figure 1B,D, Table 2). The plasma concentration of NDM [median (range)] was approximately 1.6-fold and twofold higher in patients with the *1/*10 genotype [279.43 (115.41–502.13) ng ml−1] and those with *10/*10 genotype [374.41 (84.77–802.98) ng ml−1] compared with patients harbouring reference *1/*1 genotype [174.59 (40.82–448.65) ng ml−1; P < 0.001; Figure 1B, Table 2]. Conversely, the median (range) plasma endoxifen concentrations were observed to be approximately 2.5-fold higher in patients carrying the *1/*1[19.55 (4.18–39.47) ng ml−1] or the *1/*10[19.74 (7.26–33.24) ng ml−1] genotypes compared with patients who were homozygous for *10 allele [8.03 (1.74–34.68) ng ml−1; P < 0.001; Figure 1D, Table 2].
Table 2.
Effects of CYP2D6 polymorphisms on median (range) plasma concentrations of analytes and plasma metabolic ratios (n = 111)
| Parameters | Genotypes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| *1/*1 | *1/*5 | *1/*10 | *10/*10 | *5/*10 | *1/*41+*41/*41 | |||||||
| (n = 13) | (n = 9) | (n = 31) | (n = 40) | (n = 12) | (n = 6) | |||||||
| Analytes | ||||||||||||
| Tamoxifen | 161.22 | (50.06–369.89) | 216.07 | (93.56–325.56) | 194.88 | (51.80–421.16) | 217.19 | (84.27–599.91) | 169.91 | (39.26–452.29) | 166.42 | (147.72–578.17) |
| NDM | 174.59 | (40.82–448.65) | 261.07 | (206.94–464.25) | 279.43 | (115.41–502.13) | 374.41 | (84.77–802.98) | 330.67 | (78.46–757.13) | 286.30 | (159.67–511.20) |
| 4-OHT | 2.49 | (0.97–3.36) | 1.58 | (1.25–3.25) | 1.92 | (0.86–4.51) | 1.76 | (0.72–3.82) | 1.87 | (0.47–3.17) | 2.55 | (0.75–5.02) |
| Endoxifen | 19.55 | (4.18–39.47) | 14.51 | (10.73–26.04) | 19.74 | (7.26–33.24) | 8.03 | (1.74–34.68) | 7.46 | (1.79–13.77) | 16.93 | (6.37–33.99) |
| Plasma metabolic ratios (MRs) | ||||||||||||
| MREND-NDM | 10.11 | (5.63–14.86) | 5.29 | (4.48–8.00) | 6.62 | (1.65–11.65) | 2.31 | (0.92–27.81) | 2.15 | (1.06–3.30) | 6.21 | (1.94–11.85) |
| MR4-OHT-TAM | 1.34 | (0.74–1.93) | 1.04 | (0.62–1.72) | 1.08 | (0.50–2.26) | 0.83 | (0.19–1.78) | 0.94 | (0.46–1.80) | 1.00 | (0.51–2.35) |
| Total metabolic ratios (TMRs) | ||||||||||||
| TMRNDM | 5.03 | (2.94–7.45) | 3.30 | (2.61–4.56) | 3.91 | (0.98–7.24) | 1.50 | (0.51–10.69) | 1.41 | (0.65–2.21) | 3.27 | (1.35–7.37) |
| TMR4-OHT | 11.05 | (6.09–14.73) | 8.49 | (5.26–11.77) | 9.18 | (2.39–18.70) | 4.16 | (1.14–17.05) | 4.29 | (1.71–7.85) | 6.59 | (3.65–19.04) |
| Pairwise P values | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| *1/*1 vs. *1/*5 | *1/*1 vs. *1/*10 | *1/*1 vs. *10/*10 | *1/*10 vs. *10/*10 | *1/*1 vs. *5/*10 | *1/*5 vs. *5/*10 | *1/*10 vs. *5/*10 | *10/*10 vs. *5/*10 | *1/*1 vs. *1/*41+*41/*41 | |
| Analytes | |||||||||
| Tamoxifen | 0.404 | 0.177 | 0.079 | 0.578 | 0.957 | 0.477 | 0.316 | 0.171 | 0.430 |
| NDM | 0.077 | 0.006 | 0.001 | 0.011 | 0.057 | 0.434 | 0.685 | 0.409 | 0.136 |
| 4-OHT | 0.616 | 0.709 | 0.176 | 0.041 | 0.174 | 0.434 | 0.136 | 0.745 | 0.759 |
| Endoxifen | 0.243 | 0.827 | <0.001 | <0.001 | <0.001 | 0.001 | <0.001 | 0.232 | 0.792 |
| Plasma metabolic ratios (MRs) | |||||||||
| MREND-NDM | 0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.587 | 0.054 |
| MR4-OHT-TAM | 0.141 | 0.146 | <0.001 | 0.006 | 0.022 | 0.286 | 0.267 | 0.228 | 0.219 |
| Total metabolic ratios (TMRs) | |||||||||
| TMRNDM | 0.001 | 0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.811 | 0.035 |
| TMR4-OHT | 0.030 | 0.108 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.905 | 0.054 |
Figure 1.
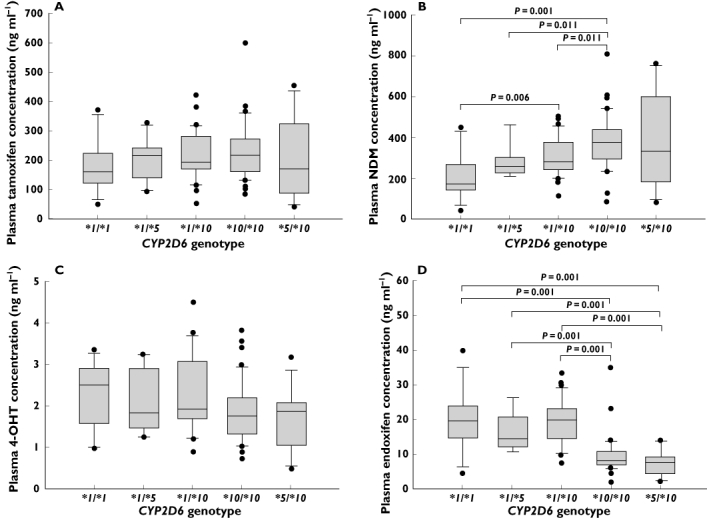
Association between CYP2D6*5- and *10-based genotypes and steady-state plasma concentrations of (A) tamoxifen, (B) NDM, (C) 4-OHT and (D) endoxifen
Although no significant associations were observed between CYP2D6*5 (CYP2D6del) polymorphism and the plasma concentrations of tamoxifen and the three metabolites (Table 2), patients harbouring the *1/*5 genotype displayed 30% to 50% higher median plasma concentrations of tamoxifen and NDM as well as 25% to 40% lower median plasma concentrations of 4-OHT and endoxifen compared with patients carrying the reference genotype (Table 2).
There were 12 compound heterozygotes (*5/*10) and their endoxifen concentrations [median (range)] were similar to patients harbouring the *10/*10 genotype [8.03 (1.74–34.68), P = 0.232] but significantly lower compared with patients homozygous [19.55 (4.18–39.47), P < 0.001] or heterozygous [*1/*5: 14.51 (10.73–26.04), P = 0.001; *1/*10: 19.74 (7.26–33.24), P < 0.001] for the reference allele (Figure 1D).
With respect to the CYP2D6*41 (2988G>A; rs28371725) allele, no significant trend in the variations of plasma concentrations of analytes was observed. However, patients who carried one or two copies of the *41 (2988G>A; rs28371725) alleles were found to have a 64.0% higher plasma NDM concentration (P = 0.136) and a 13.4% lower plasma endoxifen concentration (P = 0.792) compared with patients who were homozygous wild type (Table 2).
However, a total of seven patients carrying the CYP2D6*41 (2988G>A; rs28371725) allele were not included in this analysis as they were compound heterozygotes who also harboured other functional alleles such as *10 (100C>T; rs1065852), *14 (1758G>A) and *xN (CYP2D6dup). The presence of these variant alleles may confound the observation of the phenotypic effect of the *41 (2988G>A; rs28371725) allele. Likewise, the effect of CYP2D6 gene amplifications could not be assessed in the present study population as there was only one patient with the *1/*xN genotype. Although five other patients also carried CYP2D6 gene amplifications, the effect of this variant could not be evaluated because of the concomitant presence of *10 (100C>T; rs1065852),*14 (1758G>A) and *41 (2988G>A; rs28371725) variant alleles.
Influence of CYP2D6 polymorphisms on the metabolic ratios of tamoxifen and its metabolites
The median (range) MR4-OHT-TAM of patients carrying the *10/*10 genotype [0.83 (0.19–1.78)] was 1.25 to 1.5 times higher in patients harbouring the *1/*1[1.34 (0.74–1.93)] or *1/*10[1.08 (0.50–2.26)] genotypes (Table 2). The MR4-OHT-TAM was found to be comparable among patients harbouring the *1/*1 and *1/*10 genotypes (Figure 2B). Similarly, the median MREND-NDM was found to decrease as the number of *10 alleles increased. Patients with the *1/*1 genotype [10.11 (5.63–14.86)] were observed to have approximately 50% higher MREND-NDM than patients carrying *1/*10[6.62 (1.62–11.65)], which were in turn twice of those in patients harbouring the *10/*10 genotype [2.31 (0.92–27.81); P < 0.001; Figure 2A]. Similar genotypic–phenotypic trends were observed with regards to TMR4-OHT and TMRNDM. The TMR4-OHT median (range) was not found to differ significantly among patients who harboured *1/*1[11.05 (6.09–14.73)] and *1/*10[9.18 (2.39–18.70)] genotypes but was found to be 2.7-fold and 2.2-fold higher in patients who harboured the *10/*10 genotype [4.16 (1.14–17.05); P < 0.001; Figure 2D]. A gradual decrease in TMRNDM was observed across the different genotypes. Approximately 1.3-fold difference in TMRNDM was observed between *1/*1 and *1/*10 carriers while a 2.8-fold difference in TMRNDM was found between *1/*10 and *10/*10 carriers.
Figure 2.
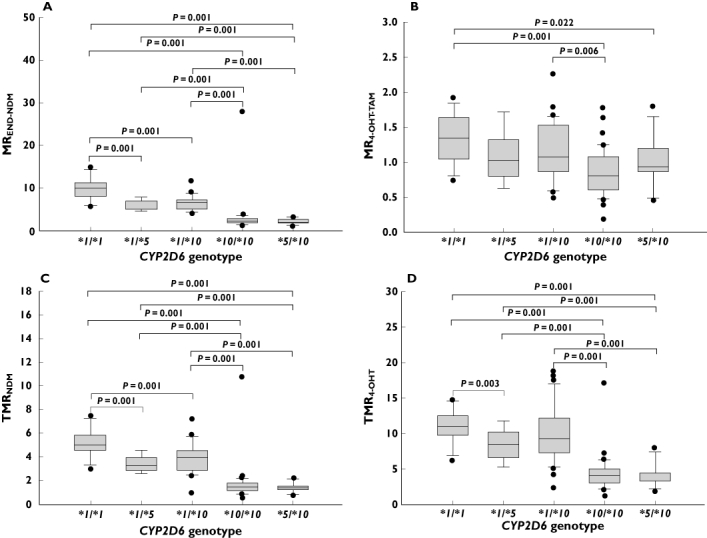
Association between CYP2D6*5- and *10-based genotypes and plasma metabolic ratios: (A) MREND-NDM, (B) MR4-OHT-TAM, (C) TMRNDM and (D) TMR4-OHT
A non-significant 20% decrease in MR4-OHT-TAM[median (range)] was observed between patients carrying the *1/*1[1.34 (0.74–1.93)] and *1/*5[10.11 (5.63–14.86)] genotypes (Figure 2B). However, the MREND-NDM[median (range)] observed in patients who were heterozygous for the *5 allele [5.29 (4.48–8.00)] was found to be half of that observed in the homozygous wild type [10.11 (5.63–14.86)] patients (Figure 2A). Likewise, a 1.3 to 1.5 times lower TMR4-OHT and TMRNDM were observed in *1/*5 carriers compared with *1/*1 carriers (Figure 2C,D)
The median MRs and TMRs were found to be similar between patients carrying *10/*10 and *5/*10 genotypes but significantly lower compared with patients who harboured *1/*1 genotypes (Figure 2A–D). The MREND-NDM[median (range)] in *5/*10[2.15 (1.06–3.30)] carriers was found to be 4.7-fold, 2.5-fold and 3.1-fold lower than that observed in *1/*1[10.11 (5.63–14.86)], *1/*5[5.29 (4.48–8.00)] and *1/*10[6.62 (1.65–11.65)] carriers, respectively (Table 2). The relationship between TMRNDM and genotypes was parallel to that observed with respect to MREND-NDM (Figure 2A,C).
A non-significant relationship between CYP2D6*41 (2988G>A; rs28371725) and median MRs was observed. The MREND-NDM and MR4-OHT-TAM were found to be 1.6 and 1.3 times lower in patients who were heterozygous or homozygous for *41 (2988G>A; rs28371725) compared with patients who were homozygous for the reference allele, respectively (Table 2). Similarly, the plasma TMRs were observed to be lower among *41 (2988G>A; rs28371725) carriers. The TMRNDM and TMR4-OHT[median (range)] were found to be 1.53-fold and 1.68-fold lower in patients who harboured *1/*41 or *41/*41 genotypes [TMRNDM: 3.27 (1.35–7.37); TMR4-OHT: 6.59 (3.65–19.04)] compared with patients who were *1/*1[TMRNDM: 5.03 (2.94–7.45), P = 0.035; TMR4-HT: 11.05 (6.09–14.73), P = 0.054].
Influence of CYP3A5, CYP2C9 and CYP2C19 polymorphisms on the plasma concentrations of tamoxifen and its metabolites
The relationships between the polymorphic variants in CYP3A5, CYP2C9 and CYP2C19 and plasma concentrations of the analytes were not found to be statistically significant (Table 3). As there were no patients carrying the CYP2C9*2 (430C>T; rs1799853) allele, the effect of CYP2C9*2 (430C>T; rs1799853) could not be evaluated.
Table 3.
Effects of CYP2C19 polymorphisms on median (range) plasma concentrations (ng ml−1) and metabolic ratios of tamoxifen and its analytes (n = 165)
| Parameters | Genotypes | Pairwise P value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CYP2C19*2 | *1/*1 | *1/*2 | *2/*2 | *1/*1 vs.*1/*2 | *1/*1 vs.*2/*2 | *1/*2 vs.*2/*2 | |||
| (681G>A; rs4244285) | (n = 40) | (n = 44) | (n = 22) | ||||||
| Analytes | |||||||||
| Tamoxifen | 190.05 | (50.06–578.17) | 204.38 | (39.26–599.91) | 186.28 | (84.27–421.16) | 0.736 | 0.783 | 0.955 |
| NDM | 346.62 | (40.82–734.94) | 284.90 | (78.46–802.98) | 305.23 | (84.77–757.13) | 0.392 | 0.421 | 0.996 |
| 4-OHT | 1.78 | (0.70–5.02) | 2.10 | (0.47–5.33) | 2.23 | (0.87–4.51) | 0.305 | 0.480 | 0.749 |
| Endoxifen | 10.47 | (3.48–42.80) | 14.51 | (1.79–42.56) | 16.34 | (1.74–28.64) | 0.358 | 0.246 | 0.553 |
| Plasma metabolic ratios (MRs) | |||||||||
| MRNDM-TAM | 1.63 | (0.82–2.76) | 1.51 | (0.62–2.65) | 1.44 | (1.01–2.10) | 0.208 | 0.088 | 0.470 |
| MREND-4-OHT | 6.63 | (3.70–13.18) | 7.05 | (2.44–11.96) | 6.97 | (1.99–12.34) | 0.793 | 0.428 | 0.613 |
| Total metabolic ratios (TMRs) | |||||||||
| TMRNDM | 2.16 | (0.65–7.46) | 3.05 | (0.51–10.69) | 3.42 | (1.03–5.86) | 0.387 | 0.421 | 0.650 |
| TMR4-OHT | 5.96 | (1.71–19.50) | 7.28 | (1.14–18.00) | 7.34 | (2.04–15.33) | 0.408 | 0.438 | 0.836 |
| CYP2C19*3 | *1/*1 | *1/*3+*3/*3 | *1/*1 vs.*1/*3+*3/*3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (636G>A; rs4986893) | (n = 40) | (n = 8) | |||||||
| Analytes | |||||||||
| Tamoxifen | 190.05 | (50.06–578.17) | 211.17 | (164.79–364.69) | 0.138 | ||||
| NDM | 346.62 | (40.82–734.94) | 323.23 | (215.01–590.65) | 0.415 | ||||
| 4-OHT | 1.78 | (0.70–5.02) | 1.92 | (1.10–3.82) | 0.800 | ||||
| Endoxifen | 10.47 | (3.48–42.80) | 13.69 | (4.74–23.04) | 0.935 | ||||
| Plasma metabolic ratios (MRs) | |||||||||
| MRNDM-TAM | 1.63 | (0.82–2.76) | 1.53 | (1.06–1.95) | 0.328 | ||||
| MREND-4-OHT | 6.63 | (3.70–13.18) | 5.61 | (3.74–11.69) | 0.786 | ||||
| Total metabolic ratios (TMRs) | |||||||||
| TMRNDM | 2.16 | (0.65–7.46) | 1.99 | (0.75–5.52) | 0.395 | ||||
| TMR4-OHT | 5.96 | (1.71–19.50) | 4.55 | (1.83–11.74) | 0.405 | ||||
| CYP2C19*17 | *1/*1 | *1/*17 | *1/*1 vs.*1/*17 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (-3402C>T and −806C>T; rs12248560) | (n=40) | (n=6) | |||||||
| Analytes | |||||||||
| Tamoxifen | 190.05 | (50.06–578.17) | 211.65 | (100.24–369.89) | 0.887 | ||||
| NDM | 346.62 | (40.82–734.94) | 318.49 | (159.67–403.52) | 0.404 | ||||
| 4-OHT | 1.78 | (0.70–5.02) | 2.68 | (0.96–4.84) | 0.190 | ||||
| Endoxifen | 10.47 | (3.48–42.80) | 18.35 | (6.37–31.05) | 0.416 | ||||
| Plasma metabolic ratios (MRs) | |||||||||
| MRNDM-TAM | 1.63 | (0.82–2.76) | 1.64 | (0.92–1.77) | 0.422 | ||||
| MREND-4-OHT | 6.63 | (3.70–13.18) | 6.66 | (4.50–10.06) | 0.815 | ||||
| Total metabolic ratios (TMRs) | |||||||||
| TMRNDM | 2.16 | (0.65–7.46) | 2.94 | (1.67–7.37) | 0.140 | ||||
| TMR4-OHT | 5.96 | (1.71–19.50) | 6.78 | (3.65–19.04) | 0.281 | ||||
Influence of CYP3A5, CYP2C9 and CYP2C19 polymorphisms on the metabolic ratios of tamoxifen and its metabolites
The MRs were not found to differ between the different genotypes of CYP3A5*3 (6986A>G; rs776746). However, higher inter-individual variations in MRs and TMRs were observed as the number of CYP3A5*3 (6986A>G; rs776746) variant alleles increased. Similarly, the plasma metabolic ratios were not found to vary considerably between patients carrying different CYP2C9 and CYP2C19 (Table 3) variants.
Discussion
The metabolism of tamoxifen is complex and is mediated by multiple cytochrome P450 enzymes and polymorphisms in genes encoding these enzymes may influence the plasma concentrations of tamoxifen and its metabolites. In this exploratory study, we investigated the effects of previously reported polymorphisms in genes encoding the major enzymes involved in the metabolism of tamoxifen and its metabolites in Asian populations.
In the present study, the CYP2D6*10 (100C>T; rs1065852) allele was correlated with higher NDM and a lower endoxifen concentrations in the plasma while associations with tamoxifen and 4-OHT were not found to be significant. A similar trend was observed when the phenotypic effect of CYP2D6*5 (CYP2D6del) was examined in association with the *10 (100C>T; rs1065852) allele (Figure 1A–D). This is suggestive of an accumulation of NDM in the plasma when the metabolic conversion of NDM to endoxifen is impaired. This leads to the postulation that CYP2D6-mediated 4-hydroxylation of NDM is the rate-limiting step in the formation of endoxifen instead of the CYP3A4/5-catalysed N-demethylation of tamoxifen. The plasma concentrations of NDM and endoxifen were comparable between patients with *1/*5 and *1/*10 as well as *5/*10 and *10/*10 genotypes, implying that the functional impact of the *5 (CYP2D6del) allele is comparable with that of the *10 (100C>T; rs1065852) allele. These observations differed slightly from those reported by Lim et al. [19] in Koreans and Xu et al. [28] in Chinese. Lim et al. [19] demonstrated significant reductions in the steady-state plasma concentrations of both active metabolites (4-OHT and endoxifen) in patients with either *10 (100C>T; rs1065852), *5 (CYP2D6del) or both alleles compared with patients who were homozygous wild type. Likewise, Xu et al. [28] reported an association between the presence of two copies of the *10 (100C>T; rs1065852) allele and lower plasma concentrations of 4-OHT in a Chinese population. The discrepancy in the results obtained could be due to the difference in the CYP2D6 alleles screened in each study. The *5 (CYP2D6del), *10 (100C>T; rs1065852) and *2 × N (CYP2D6*2dup) alleles were screened in the Koreans while only the *10 (100C>T; rs1065852) allele was screened in the Chinese. In contrast, a comprehensive screening of 16 functionally important polymorphisms was performed using the INFINITI™ CYP450 2D6I assay in the current study. Recently, Schroth et al. [29] reported a discrepancy in the assigned phenotype status because of the difference in number of CYP2D6 polymorphisms screened. The investigators concluded that comprehensive screening of CYP2D6 polymorphisms provides more accurate prediction of CYP2D6 phenotype status.
The major active metabolite, endoxifen, can be formed from tamoxifen via two pathways involving either NDM or 4-OHT. CYP2D6 has been shown to play a major role in the 4-hydroxylation of NDM and tamoxifen to endoxifen and 4-OHT, respectively. To elucidate the influence of the CYP2D6*5 (CYP2D6del) and *10 (100C>T; rs1065852) polymorphisms on the formation of the active metabolites, the associations between the two genetic variants and the MRs were examined. The metabolic ratios, MR4-OHT-TAM and MREND-NDM were found to decrease in a stepwise manner (Figure 2A,B) as the copies of variant alleles [CYP2D6*5 (CYP2D6del) or *10 (100C>T; rs1065852)] increased. The effects were more apparent than those observed on the plasma concentrations of the analytes. These results suggest a correlation between the presence of *5 (CYP2D6del), *10 (100C>T; rs1065852) or both alleles and a significantly lower rate of catalytic conversion of tamoxifen and NDM to 4-OHT and endoxifen, respectively.
To compare the relative impact of CYP2D6 polymorphisms on the formation of endoxifen via the NDM or 4-OHT pathways, the associations between CYP2D6*5 (CYP2D6del) and *10 (100C>T; rs1065852) polymorphisms and the TMRs were studied. Consistent with the observations in MRs, both TMRs were observed to be lower in patients who were homozygous variant or compound heterozygotes with a more dominant effect being observed in TMRNDM. These observations further support that CYP2D6 plays a critical role in the biosynthesis of endoxifen although other enzymes are involved. Furthermore, the data imply that CYP2D6 polymorphisms are more influential in the 4-hydroxylation of NDM to form endoxifen.
Borges et al. [13] and Gjerde et al. [14] have previously examined the effects of CYP2D6 polymorphisms on metabolic ratios in two studies involving Caucasian populations. In both studies, patients were classified as poor metabolizers (PM), intermediate metabolizers (IM), extensive metabolizers (EM) or ultrarapid metabolizers (UM) based on their CYP2D6 genotype status. Correlations between CYP2D6 genotypes and metabolic ratios were investigated indirectly by studying the relationships between the metabolizing status and the metabolic ratios. The patients who were EM or UM exhibited higher plasma concentrations of endoxifen and MREND-NDM compared with patients who were IM or PM [13, 14]. In addition, Gjerde et al. [14] observed an association between the metabolizing status and MR4-OHT-TAM in their study population, although the effect was not as prominent compared with the association between the metabolizing status and MREND-NDM. The results of our study were consistent with those obtained by Borges et al. and Gjerde et al.
In addition, the present study evaluated the effect of CYP2D6*41 (2988G>A; rs28371725) on the disposition profile of tamoxifen which was not investigated previously. Our study showed that CYP2D6*41 (2988G>A; rs28371725) was associated with a non-significant increase in the plasma concentration of NDM and a corresponding decrease in the plasma concentration of endoxifen. The metabolic ratios except TMRNDM were also found to be lower in CYP2D6*41 (2988G>A; rs28371725) carriers suggesting lower rates of endoxifen formation in patients carrying the CYP2D6*41 (2988G>A; rs28371725) allele. However, the number of study subjects carrying the *41 (2988G>A; rs28371725) allele was only six. Although additional CYP2D6*41 (2988G>A; rs28371725) carriers were present, these patients were compound heterozygotes who harboured other functional alleles that may confound the phenotypic functional impact of the CYP2D6*41 (2988G>A; rs28371725) allele. Similarly, the effect of CYP2D6 gene amplifications in the present study population was not discernible because of its being present as compound heterozygotes in several patients. Hence the impact of *41 (2988G>A; rs28371725) as well as *xN (CYP2D6dup) should be further evaluated in a larger homogenous patient population in which these two variants are more prevalent.
Besides CYP2D6, CYP3A5 is another major enzyme involved in the metabolism of tamoxifen. It catalyses the N-demethylation of tamoxifen to NDM or 4-OHT to endoxifen and the impact of the CYP3A5*3 (6986A>G; rs776746) polymorphism on the pharmacokinetics of tamoxifen was investigated. Although Jin et al. [9] previously observed a non-significant increase in endoxifen concentrations in patients harbouring *1/*1 and *1/*3 genotypes compared with patients carrying *3/*3 genotype, the plasma concentrations of endoxifen or other analytes were not found to vary considerably across different genotypes in our study population. Correspondingly, Tucker et al. [30] also did not find any significant association between CYP3A5*3 (6986A>G; rs776746) and the plasma concentrations of tamoxifen and its metabolites. It is likely that the functional impact of CYP3A5*3 (6986A>G; rs776746) on the metabolism of tamoxifen is nullified by the presence of other enzymes (CYP3A4, CYP1A2 and CYP2C9) which catalyse the N-demethylation of tamoxifen and 4-OHT (CYP3A4). Hence, further investigations on the influence of these polygenic determinants on the pharmacokinetics of tamoxifen are warranted.
CYP2C9 and CYP2C19 polymorphisms were investigated for associations with tamoxifen pharmacokinetics. No associations between the polymorphisms in CYP2C9 and CYP2C19 and the pharmacokinetics of tamoxifen were observed, consistent with reports from previous studies. Interestingly, Schroth et al. [5] reported a positive association between CYP2C19*17 (–3402C>T and −806C>T; rs12248560) and longer relapse-free time as well as disease-free survival in patients with one copy of the CYP2D6 null allele [CYP2D6*5 (CYP2D6del) or *4 (1846G>A; rs3892097)]. The polymorphism CYP2C19*17 (–3402C>T and −806C>T; rs12248560) has been reported to cause an increase in the rate of transcription of CYP2C19 leading to an increase in enzymatic activity. No statistically significant association was found in our study between CYP2C19*17 (–3402C>T and −806C>T; rs12248560) and plasma concentrations or MRs of the analytes. This may possibly be because of the lower allelic frequency in our small patient population compared with that observed in the Caucasian population (Asian Study population vs. Caucasian population: 0.06 vs. 0.24). Sub-analysis of the impact of CYP2C19*17 (–3402C>T and −806C>T; rs12248560) in patients carrying the CYP2D6*5 (CYP2D6del) allele was not possible as only one *1/*5 carrier harboured the CYP2C19*17 (–3402C>T and −806C>T; rs12248560) allele. The frequency of the CYP2C19*17 (–3402C>T and −806C>T; rs12248560) allele observed in the Indians was found to be comparable with the Caucasians [31]. Thus, the effect of the allele should be re-examined in a larger cohort of Indian breast cancer patients.
In conclusion, the present study indicates that CYP2D6*5 (CYP2D6del) and *10 (100C>T; rs1065852) are important factors that influence the plasma concentrations of tamoxifen and its metabolites, as well as metabolic ratios of tamoxifen, in a breast cancer population which consisted predominantly of Chinese subjects. The impact of other CYP2D6 polymorphic variants needs to be evaluated further in our minor ethnic groups (Malays and Indians) owing to the high inter-ethnic variation observed. In addition to the phase I metabolic enzymes investigated in the current study, the pharmacokinetics of tamoxifen may also be influenced by uridine 5′-diphospho-glucuronyltransferases (UGT) and sulfotransferases. Therefore, future studies should elucidate the impact of polymorphisms present in other candidate genes encoding the enzymes implicated in the metabolic pathway of tamoxifen.
Acknowledgments
This study was supported by the Singapore Cancer Syndicate (SCS-PS0023R) and National Medical Research Council (NMRC/1159/2008 and NMRCB1011) grants.
Competing Interests
There are no competing interests to declare.
REFERENCES
- 1.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, Weatherman RV, Desta Z, Flockhart DA, Skaar TC. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–9. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 3.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, Hayes DF, Desta Z, Flockhart DA. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–64. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 4.Crewe HK, Ellis SW, Lennard MS, Tucker GT. Variable contribution of cytochromes P450 2D6, 2C9 and 3A4 to the 4-hydroxylation of tamoxifen by human liver microsomes. Biochem Pharmacol. 1997;53:171–8. doi: 10.1016/s0006-2952(96)00650-8. [DOI] [PubMed] [Google Scholar]
- 5.Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T, Zanger UM, Simon W, Eichelbaum M, Brauch H. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–93. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 6.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Flockhart DA, Desta Z, Perez EA, Ingle JN. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flushes. J Clin Oncol. 2005;23:9312–8. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 7.Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, Fritz P, Simon W, Suman VJ, Ames MM, Safgren SL, Kuffel MJ, Ulmer HU, Boländer J, Strick R, Beckmann MW, Koelbl H, Weinshilboum RM, Ingle JN, Eichelbaum M, Schwab M, Brauch H. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–36. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burstein HJ, Prestrud AA, Seidenfeld J, Anderson H, Buchholz TA, Davidson NE, Gelmon KE, Giordano SH, Hudis CA, Malin J, Mamounas EP, Rowden D, Solky AJ, Sowers MR, Stearns V, Winer EP, Somerfield MR, Griggs JJ. American Society of Clinical Oncology Clinical Practice Guideline: Update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. J Clin Oncol. 2010;28:3784–96. doi: 10.1200/JCO.2009.26.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, Skaar T, Storniolo AM, Li L, Araba A, Blanchard R, Nguyen A, Ullmer L, Hayden J, Lemler S, Weinshilboum RM, Rae JM, Hayes DF, Flockhart DA. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–9. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 10.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–43. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 11.Gough AC, Miles JS, Spurr NK, Moss JE, Gaedigk A, Eichelbaum M, Wolf CR. Identification of the primary gene defect at the cytochrome P450 CYP2D locus. Nature. 1990;347:773–6. doi: 10.1038/347773a0. [DOI] [PubMed] [Google Scholar]
- 12.Hanioka N, Kimura S, Meyer UA, Gonzalez FJ. The human CYP2D locus associated with a common genetic defect in drug oxidation: a G1934 – a base change in intron 3 of a mutant CYP2D6 allele results in an aberrant 3′ splice recognition site. Am J Hum Genet. 1990;47:994–1001. [PMC free article] [PubMed] [Google Scholar]
- 13.Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, Jin Y, Storniolo AM, Nikoloff DM, Wu L, Hillman G, Hayes DF, Stearns V, Flockhart DA. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Gjerde J, Hauglid M, Breilid H, Lundgren S, Varhaug JE, Kisanga ER, Mellgren G, Steen VM, Lien EA. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19:56–61. doi: 10.1093/annonc/mdm434. [DOI] [PubMed] [Google Scholar]
- 15.Sistonen J, Fuselli S, Palo JU, Chauhan N, Padh H, Sajantila A. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics. 2009;19:170–9. doi: 10.1097/FPC.0b013e32831ebb30. [DOI] [PubMed] [Google Scholar]
- 16.Gaedigk A, Blum M, Gaedigk R, Eichelbaum M, Meyer UA. Deletion of the entire cytochrome P450 CYP2D6 gene as a cause of impaired drug metabolism in poor metabolizers of the debrisoquine/sparteine polymorphism. Am J Hum Genet. 1991;48:943–50. [PMC free article] [PubMed] [Google Scholar]
- 17.Myrand SP, Sekiguchi K, Man MZ, Lin X, Tzeng RY, Teng CH, Hee B, Garrett M, Kikkawa H, Lin CY, Eddy SM, Dostalik J, Mount J, Azuma J, Fujio Y, Jang IJ, Shin SG, Bleavins MR, Williams JA, Paulauskis JD, Wilner KD. Pharmacokinetics/genotype associations for major cytochrome P450 enzymes in native and first- and third-generation Japanese populations: comparison with Korean, Chinese, and Caucasian populations. Clin Pharmacol Ther. 2008;84:347–61. doi: 10.1038/sj.clpt.6100482. [DOI] [PubMed] [Google Scholar]
- 18.Wang SL, Huang JD, Lai MD, Liu BH, Lai ML. Molecular basis of genetic variation in debrisoquine hydroxylation in Chinese subjects: polymorphism in RFLP and DNA sequence of CYP2D6. Clin Pharmacol Ther. 1993;53:410–8. doi: 10.1038/clpt.1993.44. [DOI] [PubMed] [Google Scholar]
- 19.Lim HS, Ju Lee H, Seok Lee K, Sook Lee E, Jang IJ, Ro J. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol. 2007;25:3837–45. doi: 10.1200/JCO.2007.11.4850. [DOI] [PubMed] [Google Scholar]
- 20.Crewe HK, Notley LM, Wunsch RM, Lennard MS, Gillam EMJ. Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes: formation of the 4-hydroxy, 4′-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab Dispos. 2002;30:869–74. doi: 10.1124/dmd.30.8.869. [DOI] [PubMed] [Google Scholar]
- 21.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 22.Ingelman-Sundberg M, Oscarson M, McLellan RA. Polymorphic human cytochrome P450 enzymes: an opportunity for individualized drug treatment. Trends Pharmacol Sci. 1999;20:342–9. doi: 10.1016/s0165-6147(99)01363-2. [DOI] [PubMed] [Google Scholar]
- 23.Balram C, Zhou Q, Cheung YB, Lee EJD. CYP3A5*3 and *6 single nucleotide polymorphisms in three distinct Asian populations. Eur J Clin Pharmacol. 2003;59:123–6. doi: 10.1007/s00228-003-0594-2. [DOI] [PubMed] [Google Scholar]
- 24.Roy JN, Lajoie J, Zijenah LS, Barama A, Poirier C, Ward BJ, Roger M. CYP3A5 genetic polymorphisms in different ethnic populations. Drug Metab Dispos. 2005;33:884–7. doi: 10.1124/dmd.105.003822. [DOI] [PubMed] [Google Scholar]
- 25.King BP, Leathart JBS, Mutch E, Williams FM, Daly AK. CYP3A5 phenotype-genotype correlations in a British population. Br J Clin Pharmacol. 2003;55:625–9. doi: 10.1046/j.1365-2125.2003.01798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KH, Ward BA, Desta Z, Flockhart DA, Jones DR. Quantification of tamoxifen and three metabolites in plasma by high-performance liquid chromatography with fluorescence detection: application to a clinical trial. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;791:245–53. doi: 10.1016/s1570-0232(03)00218-6. [DOI] [PubMed] [Google Scholar]
- 27.Zhu YB, Zhang Q, Zou JJ, Yu CX, Xiao DW. Optimizing high-performance liquid chromatography method with fluorescence detection for quantification of tamoxifen and two metabolites in human plasma: application to a clinical study. J Pharm Biomed Anal. 2008;46:349–55. doi: 10.1016/j.jpba.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Sun Y, Yao L, Shi L, Wu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, He L, Li P, Xie Y. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19:1423–9. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 29.Schroth W, Hamann U, Fasching PA, Dauser S, Winter S, Eichelbaum M, Schwab M, Brauch H. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratification. Clin Cancer Res. 2010;16:4468–77. doi: 10.1158/1078-0432.CCR-10-0478. [DOI] [PubMed] [Google Scholar]
- 30.Tucker AN, Tkaczuk KA, Lewis LM, Tomic D, Lim CK, Flaws JA. Polymorphisms in cytochrome P4503A5 (CYP3A5) may be associated with race and tumor characteristics, but not metabolism and side effects of tamoxifen in breast cancer patients. Cancer Lett. 2005;217:61–72. doi: 10.1016/j.canlet.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 31.Li-Wan-Po A, Girard T, Farndon P, Cooley C, Lithgow J. Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br J Clin Pharmacol. 2010;69:222–30. doi: 10.1111/j.1365-2125.2009.03578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


