Abstract
Hormones are essential regulators of many behaviors. Steroids bind either to nuclear or membrane receptors while peptides primarily act via membrane receptors. After a ligand binds, the conformational change in the receptor initiates changes in cell signaling cascades (membrane receptors) or direct alternations in DNA transcription (steroid receptors). Changes in gene transcription that result are responsible for protein production and ultimately behavioral modifications. A significant part of how hormones affect DNA transcription is via epigenetic modifications of DNA and/or the chromatin in which it is entwined. These alterations lead to transcriptional changes that ultimately define the phenotype and function of a given cell. Importantly we now know that environmental stimuli influence epigenetic marks, which in the context of neuroendocrinology can lead to behavioral changes. Importantly tracking epigenetic states and profiling the epigenome within cells requires the use of epigenetic methodologies and subsequent data analysis. Here we describe the techniques of particular importance in the mapping of DNA methylation, histone modifications and occupancy of chromatin bound effector proteins that regulate gene expression. For researchers wanting to move into these levels of analysis we discuss the application of modern sequencing technologies applied in assays such as chromatin immunoprecipitation and the bioinformatics analysis involved in the rich datasets generated.
Keywords: Epigenetics, epigenomics, bioinformatics, DNA methylation, bisulfite sequencing, chromatin Immunoprecipitation, ChIP-sequencing
Introduction
The fundamental repeating subunit of eukaryotic chromatin is the nucleosome particle. The nucleosome consists of approximately 147 bp of DNA wrapped around an octamer of two copies of histone H2A, H2B, H3 and H4. Numerous histone modifications have been described, which include acetylation, methylation, phosphorylation, ubiquitylation and sumoylation amongst others. Furthermore DNA is extensively modified by methylation. Much of today's epigenetic research is converging on the study of such covalent modifications of DNA and histone proteins and the mechanisms by which such modifications influence chromatin function. Histone modifications do not simply illicit direct structural changes in chromatin, but recruit specific effector proteins to sites of modification that transduce the epigenetic mark (Daniel et al., 2005). A number of technical approaches have been devised to identify and characterize epigenetic marks and the effector proteins that recognize them genome wide. An exciting area of research now involves characterization of epigenetic changes in behavioral disorders where any underlying genetic abnormalities are not necessarily obvious. Importantly, current and past environmental exposure and social experiences can modify chromatin in specific areas of the brain that influence gene expression and change what is termed the “epigenome” of an individual. Epigenomics refers to a more global analysis of epigenetic changes across the entire genome. The main purpose of our review is to detail and review current epigenetic and epigenomic methods including assays and bioinformatics analysis for researchers interested in exploring the epigenetic underpinnings of behavior and enabling them to select the best set of methods for their particular study.
In recent years, as chromatin modifications have been found to mediate gene-environment interactions, the study of epigenetics has been extended to focus on the epigenetic mechanisms which mediate the effects of environmental factors (social interactions, experience, stress, endocrine disrupting compounds, drugs) on brain and behavior (reviewed in this issue and see; (Crews, 2008; Fagiolini et al., 2009; Laplant and Nestler, 2010; Meaney and Szyf, 2005). During the prenatal and postnatal periods, environmental signals produce long lasting effects on developing offspring that persist into adulthood. For example, both natural variations in maternal care as well as maternal deprivation are associated with chromatin modifications (histone acetylation and DNA methylation) which impact gene transcription and subsequent adult social and parental behavior (Champagne and Curley, 2009; Curley et al., 2010; Roth and Sweatt). Further, some behavioral epigenetic modifications can be transmitted to future generations via maternal behavior (Champagne and Curley, 2009), maternal environment (Arai et al., 2009), or via an epigenetic reprogramming of the germ-line (Crews et al., 2007). In the latter case, effects of the endocrine disrupting compound vinclozolin were transmitted to future generations that were not directly exposed to the compound (Jirtle and Skinner, 2007).
In addition to the role of epigenetic modifications in mediating gene-environment interactions, emerging evidence indicates that one mechanism through which steroid hormones exert their effects on gene transcription is through alterations in the epigenome. Therefore, an important application of epigenomics to behavioral neuroendocrinology is to enhance our understanding of how steroid hormones alter gene expression. For example, recent data indicates that the masculinization of brain and behavior by testosterone during the neonatal period is dependent upon DNA methylation and the recruitment of nuclear corepressors which act as part of the steroid hormone receptor complex and might also recruit histone deacetylases (HDACs) to repress gene expression ((Auger et al.)). Further, elucidating the role of epigenetic modifications at steroid hormone receptor complexes might be an important avenue of investigation for understanding the molecular pathways involved in regulating reproductive behaviors and the consolidation of reproductive experience. In support of this idea, administration of the HDAC inhibitor sodium butyrate has been found to potentiate the effects of sexual experience in female mice (Bonthuis et al., 2010). Interestingly, the behavioral effects of HDAC inhibition on sexual behavior were dependent upon estrogen receptor alpha (ERα), because sodium butyrate failed to facilitate sexual behavior in ERαKO mice. These data are congruent with the finding that the facilitatory effects of sodium butyrate on the consolidation of memory depend on CREB-binding protein (CBP) (Vescey et al., 2007), a histone acetyltransferase (HAT) which has been shown to interact with ERα (Kim et al., 2001). However, it is presently unclear whether the effects of sodium butyrate on female sexual behavior are associated with increased histone acetylation at sites where CBP is known to act, and whether these epigenetic marks are associated with DNA sequences in the promoter region of ERα targets or the ERα gene itself. These types of questions can be explored using the techniques described herein.
Mapping of DNA methylation
In mammalian cells methylation occurs on the fifth carbon of cytosine residues of CpG dinucleotides, which occur in high density in genomic regions known as CpG islands (Goll and Bestor, 2005). Typically, DNA methylation is associated with gene repression and provides a stable and heritable mechanism of epigenetic regulation. In order for these methylation marks to remain stable and heritable, DNA methyltransferase proteins (DNMTs) catalyze the de novo formation and maintenance of 5-methylcytosine (5mC) (Bestor, 2000). Numerous techniques have been devised to study DNA methylation over the past three decades. Regardless of whether the experimental question asks where methylation marks are present at a specific gene locus, or where the methylation marks are located across the entire genome, the first step in most commonly used assays is to distinguish 5mC from unmethylated cytosine via (1) selective digestion of unmethylated DNA using restriction enzymes, (2) conversion of unmethylated cytosine by sodium bisulfite treatment or (3) selective affinity enrichment using antibodies or proteins towards 5mC (see Table 1 and (Laird, 2010) for more details as well as (Esteller, 2007; Tost, 2009). The products of these assays can then be analyzed at a single locus or throughout the entire genome. Genome scale approaches that relied on PCR amplification and gel electrophoresis of the reaction products have more recently been replaced by array hybridization and sequencing approaches (Laird, 2010). High-throughput sequencing (HTS) platforms enable the sequencing of hundreds of millions of short DNA fragments in a single run. This has enabled the rapid sequencing of whole genomes, mRNAs and other novel RNAs for gene expression analysis, DNAse I hypersensitive sites, genomic variations and DNA associated with epigenetic marks. While microarray hybridization techniques launched the era of epigenomics, current sequencing technology has enabled base-pair resolution mapping of DNA methylation. HTS techniques have recently provided more powerful DNA methylation analysis, enabling higher resolution while covering the entire genome and avoiding the need for array design and hybridization.
Table 1.
DNA methylation and histone modification analysis methods.
| Analysis | Pretreatment | Locus- specific analysis |
Genome-wide analysis | Pros and Cons | |
|---|---|---|---|---|---|
| Array-based | Sequencing- based |
||||
| DNA Methylation | Enzyme digestion | HpaII-PCR | DMH | Methyl-seq | Pros- Sensitive assay and sequence-based analysis allows for allele-specific analysis, covering more of the genome and avoiding hybridization artifacts. |
| MCAM | MCA-seq | ||||
| HELP | HELP-seq | ||||
| MethylScope | MSCC | ||||
| CHARM | |||||
| MMASS | |||||
| Cons- Some approaches can occasionally be prone to error caused by incomplete digestion that is not related to methylation and sequence library biases can occur. | |||||
| Sodium bisulfite | MethylLight | BiMP | BC-seq | Pros- Bisufite converted DNA is particularly well-suited for sequence-based approaches, giving unambiguous and allele-specific CpG identification. | |
| EpiTYPER | GoldenGate | RRBS | |||
| Pyro-Sequencing | Infinium | BSPP | |||
| WGSBS | |||||
| Cons- Incomplete bisulphate conversion and bisuphite-PCR can introduce bias. About 10% of mammalian CpGs remain refractory to genome alignment. | |||||
| Affinity enrichment | MeDIP-PCR | MeDIP | MeDIP-seq | Pros- Usually rapid and efficient genome-wide analysis | |
| mDIP | MIRA-seq | ||||
| mCIP | Cons- Does not yield single CpG resolution and requires adjustment for CpG density at different regions. | ||||
| MIRA | |||||
| Histone modification | Affinity enrichment | ChIP-PCR | ChIP-chip | ChIP-seq | Pros- Compared to ChIP-ChIP, ChIP-seq allows for nucleotide resolution with a broader dynamic range, allows for multiplexing, requires less input DNA (10–50ng) and less amplification. |
| Cons- ChIP-chip is limited by sequences on the array, is subject to saturation and requires high amounts of genomic DNA (low micrograms). However, ChIP-seq data does contain some GC bias. | |||||
Restriction enzyme digestion
Methylation sensitive restriction enzymes are inhibited by 5mC and so their digestion patterns can give a read-out of DNA methylation. The most widely used of these endonucleases are HpaII and SmaI. When used in combination with methylation-insensitive enzymes with the same sequence recognition, a methylation digestion map can be developed. Several array hybridization techniques have been developed that analyze restriction enzyme digestion products (Laird, 2010). This includes differential methylation hybridization, which generates a pool of methylation-sensitive restriction enzyme digested DNA and compares it to a mock digested pool. The DNAs are amplified and labeled with different dyes for two-color array hybridization (Yan et al., 2009). The relative signal intensities allow for the identification of loci with DNA methylation. However, using HTS for a genome-wide analysis would be even more powerful. For example, Methyl-seq (methylation/bisulfite conversion sequencing) is the term given to an approach that involves sequence based analysis of HpaII digested DNA libraries (Brunner et al., 2009). A number of similar restriction enzyme approaches have now been adapted to utilize sequencing (Table 1). Furthermore, HTS is becoming the method of choice in chromatin immunoprecipitation assays, discussed below.
Chemical conversion of unmethylated cytosine
A particularly useful tool in the mapping of DNA methylation marks has been the chemical deamination of unmethylated cytosines by sodium bisulfite (Frommer et al., 1992). In this approach unmethylated C’s undergo conversion to T’s. Originally Sanger sequencing of treated and untreated DNA PCR products gave a readout of base-pair resolution methylation patterns. Bisulfite-converted DNA has also been used in designer hybridization arrays, where the mismatches created using this approach lead to lower levels of hybridization efficiency when compared to non-methylated DNA (Laird, 2010). A further adaptation of this approach is the Illumina GoldenGate technonology. This approach involves a multiplexed methylation specific primer extension of bisulfite-converted DNA at around 1,500 CpG sites, which is compared to unmethylated sequences. Each of the primers is labeled with a different dye and products are hybridized to CpG bead arrays. The Illumina Infinium DNA methylation analysis involves hybridization of amplified bisulfite converted DNA to methylation specific oligos linked to beads. This approach analyzes more than 27,500 CpG sites (Laird, 2010). HTS platforms are now being utilized in the analysis of bisulfite converted DNA, avoiding some of the challenges of array hybridization. Although this treated DNA has low sequence complexity in relation to untreated DNA, a number of targeted capture approaches have been developed to reduce sequence redundancy (reviewed by (Laird, 2010)). However, particularly exciting is whole-genome bisulfite sequencing that has been achieved in human cells using an Illumina Genome Analyzer, generating base pair resolution maps of DNA methylation (Lister et al., 2009). Strikingly, bisulfilte sequencing revealed that nearly 25% of all methylation identified was non-CpG in embryonic stem cells (Lister et al., 2009). In addition, non-CpG methylation showed enrichment in gene bodies and depletion in transcription factor binding sites and enhancers (Lister et al., 2009). While this approach is not free from problems, it is perhaps the wave of the future as sequencing technology becomes more available and as sequencing costs drop.
Affinity Enrichment
Chromatin Immunoprecipitation (ChIP) is a technique that assays protein-DNA binding in vivo (Solomon et al., 1988). This approach is described in more detail below in the context of histone modifications. Briefly, ChIP assays utilize an antibody with specificity against a selected epigenetic mark or protein bound to chromatin and allows for the enrichment of DNA fragments that are associated with it. This method allows one to determine whether the epigenetic mark is linked to a particular target at a particular time point. The isolated DNAs can then be hybridized to microarrays (ChIP-chip). Such arrays can bear oligonucleotides that encompass the entire non-repetitive genome or select promoter regions. A recent adaptation of ChIP involves the sequencing of precipitated DNA fragments, referred to as ChIP-seq. In contrast to ChIP-chip, ChIP-seq has a number of advantages including higher resolution mapping of epigenetic marks and lower cost for mammalian genomes. Affinity enrichment of methylated DNA is achieved using antibodies for 5mC on denatured DNA (Mukhopadhyay et al., 2004) or methyl-binding proteins on native DNA, such as methyl-CpG-binding protein 2 (MECP2) or methyl-CpG-binding domain 1 (MBD1) (Cross et al., 1994; Jorgensen et al., 2006). The enriched products can then be hybridized onto microarrays (reviewed by (Laird, 2010) or sequenced by next generation sequencing (NGS) (Down et al., 2008). The approach allows rapid generation of bulk genome-wide maps of DNA methylation, yet does not yield information on individual CpG sites and may reflect bias towards the identification of genomic locations of a higher density of CpG methylation.
Recently a second type of DNA methylation, 5-Hydroxymethylcytosine (5hmC), has been reported (Kriaucionis and Heintz, 2009) arising from enzymatic oxidation of methyl cytosine by tet oncogene 1 (TET1) (Tahiliani et al., 2009). 5hmC may represent a biologically important DNA modification or an intermediate in a DNA demethylation pathway. Sodium bisulfite treatment of DNA is incapable of distinguishing between 5hmC and 5mC (Jin et al., 2010). In contrast, techniques based on ChIP with an anti-5mC antibody (referred to as MeDIP) or proteins that bind to methylated CpG sequences, such as MBD1, are specific for 5mC and do not detect 5hmC unless both modified bases occur in the same DNA fragment. Commercially developed antibodies specific to 5hmC have very recently become available and ultimately the mapping of 5hmC will require the verification of these reagents in ChIP approaches. This may help shed led light onto the relevance of 5hmC.
Mapping of histone modifications and histone variants
Mapping of histone modifications, histone variants and protein-DNA interactions is key in understanding the epigenome and its associated regulation of gene expression. Histone modifications can combine to alter the packaging of DNA, nucleosome positioning and the recruitment of effector proteins that ultimately influence transcription. The main tool for mapping the location of histone modifications is ChIP. To perform this technique histones and other DNA-binding proteins are typically crosslinked to DNA with formaldehyde and subsequently sonicated to generate small DNA fragments of 150bp or larger. An antibody specific to a particular histone modification, variant or chromatin-binding protein is used to enrich for bound DNA fragments by immunoprecipitation. The crosslink is reversed and the DNA assayed. Alternatively micrococcal nuclease digestion of DNA without crosslinking is often used to digest linker DNA between nucleosomes and is of particular use to map nucleosome positions.
Recall in our previous example that although administration of sodium butyrate has been found to facilitate female sexual behavior (Bonthuis et al., 2010), the mechanism through which HDAC inhibition modulates sex behavior is presently unknown. ChIP would be a useful tool to investigate this issue. A potential first step would be to choose an antibody to either one of the acetylated histone proteins (H2A, H2B, H3, or H4) or to choose an antibody to a more specific acetylation site that enzymes such as CBP modify (e.g. H3K14ac). In light of the finding that the facilitatory effects of sodium butyrate on female sexual behavior are dependent upon ERα, a relevant question is whether H3K14ac and CBP activity are associated ERα-dependent transcriptional changes. A locus-specific analysis of the ChIP products would answer the question of whether the mark H3K14ac is associated with known ERα bound promoters, however, another unbiased way to answer this question would be to use a genome-wide analysis in order to determine all of the sequences associated with H3K14ac and ERα across the entire genome. A genome-wide approach, such as ChIP-seq, would answer the question of whether H3K14ac is associated with the transcriptional activity of ERα, but it would also potentially determine all of the genes that are associated with H3K14ac and female sexual behavior.
A critical element in any of these approaches is the validation of the antibody to be used. For example, it is key that an antibody recognizing a given histone methylation mark, such as mono methylated H3 at lysine 4 (H3K4me3) does not crossreact with higher methylated isoforms, other methylated lysines, or other non-histone proteins that likely have different biological functions. Furthermore, the efficiency of an antibody as being ChIP grade needs to be established.
ChIP-chip and ChIP-seq
In ChIP-chip the precipitated DNA fragments obtained are identified by hybridization to a microarray (Blat and Kleckner, 1999; Ren et al., 2000). Tiling arrays allow for interrogation of the majority of the genome or can be designed to highlight selected regions of the genome, such as promoter arrays. Much of our progress in the understanding of histone modifications and their biological roles can be attributed to ChIP-chip approaches over the past decade. For example, genome-wide histone modification patterns have been described for yeast using ChIP-chip, but only partial maps had been generated for mammalian cells (Reviewed in(Park, 2009). However the rapid development of NGS technology and an increase in its availability and affordability has encouraged the current surge in the use of ChIP-seq in the mapping of histones and their modifications. ChIP followed by sequencing gives a higher resolution, larger genomic coverage and a greater dynamic range than that afforded by microarray approaches. Typically millions of sequencing reads of around 30–80bp are generated, and recent sequencing developments promise even longer reads (Metzker, 2010). While this approach presents some particular bioinformatics challenges, discussed below, of particular value in this approach is the more precise mapping of transcription factor and effector protein binding sites and histone locations. For example, maps for nucleosome positions, 20 histone methylation marks, 18 histone acetylation marks, RNA polymerase II, the histone variant H2A.Z and the transcription factor CTCF were generated in human T cells with ChIP-seq using the Illumina Solexa platform (Barski et al., 2007; Wang et al., 2008). Furthermore histone methylation maps have been generated for mouse embryonic stem cells (Mikkelsen et al., 2007), reinforcing the discovery of bivalent domains of methylation that mark developmental genes. In parallel, Roche 454 pyrosequencing technology was used in the mapping of H2A.Z in yeast and flies (Park, 2009).
In ChIP-seq, following the initial purification of histone or other protein-bound DNA, common adaptors are ligated to the ChIP DNA and amplicon libraries are generated. All templates are enzymatically extended in parallel, a process that incorporates fluorescent labels. There are several platforms which can be used to sequence these DNA samples. The Illumina Solexa Genome Analyzer performs sequencing-by-synthesis of clusters of clonal sequence fragments. The Roche 454 approach utilizes emulsion PCR, where a PCR reaction emulsion encapsulates bead-DNA complexes and generates beads containing thousands of copies of the same template. The Applied Biosystems SOLiD platform uses a DNA-ligase driven synthesis. The Helicos Heliscope platform allows for single molecule sequencing where a single DNA molecule is sequenced from an immobilized primer, avoiding the need for DNA amplification. Pacific Biosciences offers another adaptation in which the polymerase is immobilized on a solid support, a technology which allows for the sequencing of larger DNA molecules, resulting in potentially longer read length. Current sequencing technologies are reviewed in detail by Metzker (Metzker, 2010).
In continuing with the above example, either ChIP-chip or ChIP-seq would be able to assess the location of the H3K14ac mark across the whole genome (for a good example of the advantages/disadvantages of using ChIP-chip rather than ChIP-seq using this type of data, see (Laplant and Nestler, 2010)). An obvious advantage of ChIP-seq over ChIP-chip is that whole genome analysis is not limited to probe sequences available on an array and that higher resolution can be achieved (Table 1). ChIP-seq also affords a higher dynamic range with large numbers of DNA potentially sequenced. Furthermore ChIP-chip requires 10–100ng of starting DNA, which is then PCR amplified to 2 micrograms or more per array (Reviewed in (Park, 2009). ChIP-seq requires only 10,000–100,000 cells for whole genome interrogation studies, which represents two or three orders of magnitude fewer cells than ChIP-chip. Therefore, this technique might be helpful when examining small regions of the brain where it might be difficult to get 10–100 ng of starting DNA. Also fewer rounds of PCR amplification required in the generation of the sequencing libraries reduce concerns of PCR sampling bias.
Most ChIP-seq studies to date have used input DNA as a control in sequencing reactions to account for any differences in genomic DNA shearing, solubility and amplification. A particularly valuable control when considering the mapping of histone modifications is to map bulk histones. For example, knowing the genome wide location of histone H3 methylated at lysine 4 is only fully relevant when the genomic location of all H3 histones within nucleosomes is known. Regions of nucleosome depletion or enrichment may otherwise skew interpretation of results for a given histone modification. Thus a meaningful ratio can be drawn between the fraction of nucleosomes that carry a particular modification relative to those that do not.
Another useful technique is multiplexing which allows for the simultaneous sequencing of multiple samples in a single run. Independently prepared samples are ligated to different barcode adaptors (short set of oligonucleotide base pairs) that carry a few unique nucleotides that enable their identification in a common sequencing analysis (Lefrancois et al., 2009). This approach will hopefully offset the costs of multiple sequencing runs in the future.
Analysis of Epigenetic Data
The methods outlined above offer the advantage of rapidly sequencing the entire genome, however, sequencing the entire genome generates massive amounts of data, which need to be processed and analyzed. For example, one lane of ChIP-seq data (i.e., one sample) generates ~gigabyte of sequence data. Therefore, when planning these experiments, it is important to consider how these massive amounts of data will be analyzed. Facilities that offer sequencing analysis often include some nominal statistical assessments, but this is only the tip of the iceberg and to justify the time and costs involved it is necessary to mine the data. For this, the recruitment of a bioinformatician and the use of appropriate hardware and software will be essential.
In what follows, we have outlined the process by which ChIP-seq data is analyzed using one of the available HTS platforms, the Illumina Genome Sequencer. Although we review methylation-based assays above, we will restrict our discussion to ChIP-seq analysis as the tools are more mature for this application. We focus on the bioinformatic analysis of ChIP-seq as opposed to ChIP-chip because, as described above, ChIP-seq affords many advantages including cost, improved dynamic range and ability to identify binding sites at higher resolution. It is important to note, however, that many of these approaches are either directly applicable or generalizable to methylation-based HTS assays (e.g., see (Down et al., 2008) for MeDIP-seq and MeDIP-chip analysis tools). Further, all the biologically meaningful data mining or high-level analysis steps are the same once significant sites are identified. Finally, while we focus on the Illumina Genome Analyzer, as it is the most popular HTS platform, note that the basic analysis steps are similar for the other HTS platforms.
The Illumina Genome analyzer uses a sequence-by-synthesis method in order to sequence ~10 gigabases of DNA in a few days. Once assayed, ChIP DNA samples are nebulized to ~150bp fragments and ligated to adapters that bind to linker molecules on the surface of a flow cell. Once bound, the DNA fragments are then amplified through a bridge amplification step, which generates clusters of sequence clones. The Illumina’s sequence-by-synthesis process consists of 30–80 sequencing cycles depending on the specific instrument (i.e., resulting in 30–80 nucleotide read length). Completion of the sequencing cycles constitutes an Illumina “run”. Each flow cell consists of 8 lanes, which allow 8 independent samples to be sequenced. Each lane is further subdivided into hundreds of tiles. Four image files—one for each of the four base dyes—are generated for each tile. These image files constitute the raw data from which the sequence read and sequencing error rate data are derived using Illumina’s analysis pipeline software.
There are three main stages of the analysis pipeline (each referred to by name; see Figure 1): image analysis (Firecrest), base calling (Bustard), and sequence analysis (Gerald/Eland). The first stage of analysis, Firecrest, involves direct assessment of fluorescent signals from the Illumina Genome Analyzer. During the Firecrest stage, the Illumina software calculates intensity values from the images for each raw fluorescent signal. These signal intensities are then used to determine (“call”) the bases. The second stage of the analysis, Bustard, determines the sequence of the bases. Bustard also assigns a quality score for each base. The final stage of the pipeline is executed by Gerald, during which the mapping program, Eland, aligns the nucleotide reads to a reference genome (i.e. yeast, mouse, etc.). Eland classifies all reads according to (1) whether they map to repetitive or unique regions of a genome and (2) the number of mismatches between the read and the reference genome.
Figure 1.
Summary of tools for low-level analysis (both Illumina software and 3rd party tools) as they apply to the Illumina Genome Analyzer Pipeline.
The analysis of the raw data generated from the Illumina Genome Analysis pipeline software can be broadly viewed as two-stage process. The first step consists of low-level analyses: quality assessment, calculation of enrichment profiles across the genome, and identify significantly enriched peaks/loci. High-level analysis of the data involves quantification and visualization of the data, which allows for interpretation of the data in a biologically meaningful way. The Illumina Genome Analyzer is equipped with two software packages that perform low (analysis pipeline) and high levels of analysis (BeadStudio or GenomeStudio), in addition, other open source tools are also available for these types of analyses as well.
Low-Level Analysis of Epigenomic ChIP-seq Data
Quality Assessment of Illumina Genome Analyzer Data
The analysis pipeline software assesses the quality of the data at each stage of the pipeline (Figure 1). Bustard assigns a quality score to each of the possible nucleotide calls (i.e., Q-score), which range from −40 to 40. Ideally, one nucleotide receives a score of 40 and the remaining three nucleotides receive scores of −40. The Q-score is based on the probability of an error in base calling. For example, if the probability of error is 1 in 10, the Q-score would be 10, indicating that a particular base call is 90% accurate. Q-scores of 40, therefore, indicate that a base call is 99.99% accurate. Many calls are less than ideal in which case the nucleotide with the highest Q-score is called. An aggregate quality score, QAG—defined as the maximum Q-score minus the sum of the remaining three Q-scores— is also calculated. Gerald filters out low quality reads, that would be highly ambiguous, trims the sequences by excluding low quality ends, and maps the reads to the selected reference genome. Gerald also generates a summary.html file, which tabulates a number of statistics for each lane of a run including: (1) number of clusters (i.e., raw and filtered by the Illumina pipeline), (2) average of four intensities (i.e. one per base type) for filtered clusters at the first cycle, (3) the ratio of the average of four intensities at the twentieth cycle over that at the first represented as a percentage, (4) percentage of clusters that pass Illumina’s quality filters, (5) percentage of filtered reads that were uniquely mapped to the reference genome, (6) the average filtered read alignment score and (7) the percentage of called bases that do not match the reference sequence calculated from mapped reads. This is the first file that should be viewed after a run is completed. It allows an investigator to assess the quality of each lane of a run. Tools are also emerging that perform additional quality control analysis on Illumina Genome Analyzer data, which assess data quality and help diagnose problems that may have caused a failed sequencing run.
Additional tools for quality assessment
TileQC (Dolan and Denver, 2008) provides functions that allow tile-based quality control. The package contains a number of functions that generate image plots of various Illumina quality control metrics including the 8 read map categories generated by Eland and QAG. The metrics are coded according to color and size and placed in the physical locations of the reads within a specified tile. Companion histograms of the 8 read map categories appearing in a selected tile are also generated. A user can generate these images for a specified subset of cycles. In addition, tileQC contains functions that generate plots of Illumina read map categories and QAG as a function of sequence cycle and tile. These utilities allow assessment across tiles and cycles that can isolate artifacts (Dolan and Denver, 2008) or a particular sequencing cycle that had a high failure rate. This allows investigators to filter out particularly bad tiles or be cautious in interpreting mismatches that occur at low quality cycles as single nucleotide polymorphisms (SNPs).
A complementary quality control tool, PIQA (Martinez-Alcantara et al., 2009), generates graphical representations of cluster densities of sequence clones on the flow cell, base quality scores and nucleotide frequencies across tiles and cycles. It also provides a statistical summary of these quality metrics for a user-selected lane of a flow cell. One of the most important indicators of a successful run is the cluster density. Cluster densities that are either too high or low can result in 1–2 orders of magnitude reduction in the number of mappable reads (i.e., far below the 20–40 million (M) expected); hence a failed lane. PIQA generates a plot of the number of clusters across tiles to assess this important quality metric. In addition, PIQA generates a number of plots of base call quality metrics as a function of tile or cycle including: (a) proportion of base calls per cycle and (b) average base calls per tile or cycle. Because each lane contains randomly fragmented genomic DNA, the proportion of nucleotides observed in each tile, lane and cycle should be nearly identical. Significant deviations from this could indicate a sample preparation problem (Martinez-Alcantara et al., 2009) or a technical problem with the sequencing run. Indeed, in one case, Martinez-Alcantara highlight a sample where too many adapter sequences were introduced during sample preparation resulting in sequencing of adaptors rather than sample DNA. This is a common problem with library construction that can lead to low quality sequence data including orders of magnitude less mappable sequence reads than expected; hence, a failed lane or run.
Additional tools for sequence mapping
Although the Illumina pipeline runs Eland which maps sequence reads to a reference genome, users may want to explore alternative mapping options (e.g., allowing more than 2 mismatches; Figure 1). There are many considerations regarding sensitive and specific mapping of reads to a genome including number of allowed mismatches, degree to which reads are trimmed at the more error-prone 3’ end, number of allowed locations to which a given read can map and alignment scoring. Given that even for one lane, ~40M reads must be mapped to 3 billion base pairs of the human or mouse genome, there are also important performance criteria to consider including program speed and memory usage.
Fortunately there are many read-mapping tools emerging that are flexible (i.e., include many user defined mapping options), memory efficient and fast (Trapnell and Salzberg, 2009) including Bowtie (Langmead et al., 2009), BWA (Li and Durbin, 2009), MAQ (Li et al., 2008), Mosaik, Novoalign, SOAP (Li et al., 2008) and ZOOM (see Figure 1 for resource links). Many of these allow a user to specify the number of allowed mismatches to the reference genome, perform 3’ end read trimming, number of locations to which a read can map (with unique mappings being the most informative) and probabilistic scoring of the mapping taking into account the Illumina quality score and alignment information (e.g., number and type of mismatches). These programs achieve relatively high speeds by indexing the reference genome. Indexing a genome effectively generates a look-up table similar to the index at the end of a book that allows a rapid search of the subsequences contained in the reference genome (Trapnell and Salzberg, 2009). Thus, a user first runs a program that indexes the reference genome and then runs the mapping tool against the indexed genome. However, the different programs listed above index the reference genome using different algorithms which results in significant performance differences. For example, MAQ uses a space-seeding approach (Li et al., 2008) which requires over 50 gigabytes (Trapnell and Salzberg, 2009) while BWA and Bowtie use the Burrows-Wheeler transform which in contrast, only requires two gigabytes of memory to store the human genome (Langmead et al., 2009; Li and Durbin, 2009; Trapnell and Salzberg, 2009). The Burrows-Wheeler based algorithm is considerably more complex but much faster than a space-seed based algorithm (Langmead et al., 2009; Trapnell and Salzberg, 2009). In order to get a sense of the performance of these programs, we note that using a 2.8GHz dual quad-core MacPro with 32 GB of RAM, BWA mapped reads to the human genome in 3.5 hours. For relatively high quality data, that pass the quality control filters described above, 70–75% of the ~40M reads can be mapped to the reference genome, yielding ~30M mapped reads. Regarding genomic coverage, while 48% of the human genome is non-repetitive, 80% and 89% is mappable, with 30bp and 70bp reads respectively (Park, 2009; Rozowsky et al., 2009). This is due to the fact that repetitive and unique sequences are interspersed, and even if only a portion of a read contains unique sequence, the unique segment anchors the read to a location.
Finally, given that imprinting plays an important role in development and behavior, understanding whether the maternal, paternal or both alleles contain a mapped mark at a given locus can be of great interest in a study. Marks that map to loci that contain a heterozygous SNP allow determination of whether the mark originated from one allele or both alleles. Thus, once the reads are aligned, an investigator can call and view SNPs using SAM tools (Li et al., 2009). SAM includes a consensus sequence base caller— algorithm that identifies the same SNP in overlapping independent reads—and viewer (Li et al., 2009; Trapnell and Salzberg, 2009).
Calculation of Enrichment Profiles
After the data has been assessed for quality, poor quality reads/tiles have been filtered out and the reads have been mapped, a common next step in ChIP-seq analysis is to generate read enrichment profiles and identify significantly enriched loci. Read enrichment profiles are simply the number of overlapping mapped reads at any given nucleotide position in the reference genome as a function of genomic coordinate. The Wold lab has developed a handy tool that performs this task (Figure 1). The mapped read file must first be converted to a BED file format, which represents that mapped read locations as chromosome start position and stop position, in order to input the data into the Wold lab calculator. The Wold lab enrichment calculator outputs the profile in WIG format, a file format that represents the read enrichment profile as chromosome, start and stop of a constant stretch of overlapping reads and the number of overlapping reads. The read enrichment profile is useful for both downstream/high level analysis (discussed below) and visualization of peaks across the genome. There are a number of genome browsers available for viewing read enrichment profiles in WIG format (Figure 1). These allow further assessment of the quality of the data. For example, H3K4me3 is known to mark the 5’ end of active genes. If this mark is used for ChIP-seq, the lack of peaks at the 5’ end could indicate a bad antibody or another problem with sample preparation. This is also the exciting beginning of data exploration and discovery including checking whether or not a mark is present at specific genes, identifying specific loci wherein broad domains of epigenetic marks occur, and finding intergenic loci where, if present, peaks could mark regions where enhancers bind or the start sites of poorly annotated non-coding RNAs.
Although useful, browsing is a powerful qualitative research undertaking, which does not yield a good global summary of genomic trends in the data nor provide a comprehensive ranked list of genes, pathways impacted, etc in the data. Thus, a useful next step is to identify significantly enriched loci/peaks over control (i.e., indentify the signal peaks and ignore noise peaks). Fortunately, there are a number of ChIP-seq peak finders that have recently been developed (Park, 2009).
Calculation of Significantly Enriched Loci or Sites
Currently, the most powerful ChIP-seq peak finders (Boyle et al., 2008; Jothi et al., 2008; Kharchenko et al., 2008; Park, 2009; Schmid and Bucher, 2007; Valouev et al., 2008; Zhang et al., 2008) take advantage of the fact that ChIP-seq fragments are sequenced from the 5’ to the 3’ end. Thus, when the top strand of a ~150bp ChIP-seq fragment is sequenced, a representative (i.e., 30–80bp) top-strand read is generated from the 5’ end of the fragment which, when mapped, will appear to the left of the site of a sharply peaked epigenetic mark on a genome browser. Similarly, when the bottom strand of a ~150bp ChIP-seq fragment is sequenced, a representative bottom-strand read is generated from the 5’ end of the fragment which, when mapped, will appear to the right of the site of a sharply peaked mark on a genome browser. Thus, the mapped reads should form two distributions, one to the left of the mark on the top strand and one to the right of the mark on the bottom strand. These distributions should be separated by a fixed distance. Many of these tools identify the location of the mark/peak by calculating a combined profile and shifting each distribution towards the center (Kharchenko et al., 2008; Park, 2009; Valouev et al., 2008; Zhang et al., 2008) while one (Jothi et al., 2008) directly calculates the difference in enrichment between top and bottom strand reads and identifies the crossover point where this metric reaches zero. Methods are also being developed that can identify significantly enriched broad domains of modified histones (Bernstein et al., 2005; Park, 2009). Once the combined peak is generated, it must be scored. These tools (Boyle et al., 2008; Jothi et al., 2008; Kharchenko et al., 2008; Park, 2009; Rozowsky et al., 2009; Schmid and Bucher, 2007; Valouev et al., 2008; Zhang et al., 2008) use a well-motivated null model or noise distribution (i.e., reads from sequences that were not specifically pulled down by the antibody). A reasonable null distribution for ChIP-seq data is the Poisson distribution (Park, 2009), which is appropriate for cases where the likelihood of an “event” (e.g., a read mapping in any given 150bp stretch of the genome) is rare and there are many “events” (i.e., there are 20 million 150bp stretches in the human or mouse genomes). Using the Poisson or another appropriate null distribution, p-values—defined as the fraction of the null distribution which lies above a given peak value—can be calculated. However, because ~1 million tests (i.e., in 150bp windows) are performed across a mammalian genome, one must correct for multiple hypotheses testing (Park, 2009). Briefly, if we compared control to control, we should find no significant peaks; however, we could in principle find ~50,000 peaks by applying a p-value cutoff of 0.05. This follows by the very definition of a p-value: the probability of finding a given peak by chance. Thus, most of the peak finding tools correct for multiple hypotheses testing by calculating the False Discovery Rate (FDR) (Reiner et al., 2003) which is the expected proportion of false positive sites among those identified as significant. The resultant output file from these programs is the significantly enriched sites at a user specified FDR (default is usually 5% FDR). These sites can be uploaded into genome browsers for further viewing along side the read enrichment profiles. They are also used for downstream high-level analysis.
High-Level Analysis of Epigenomic ChIP-seq Data
Assessing the activating/repressive potential of epigenetic marks
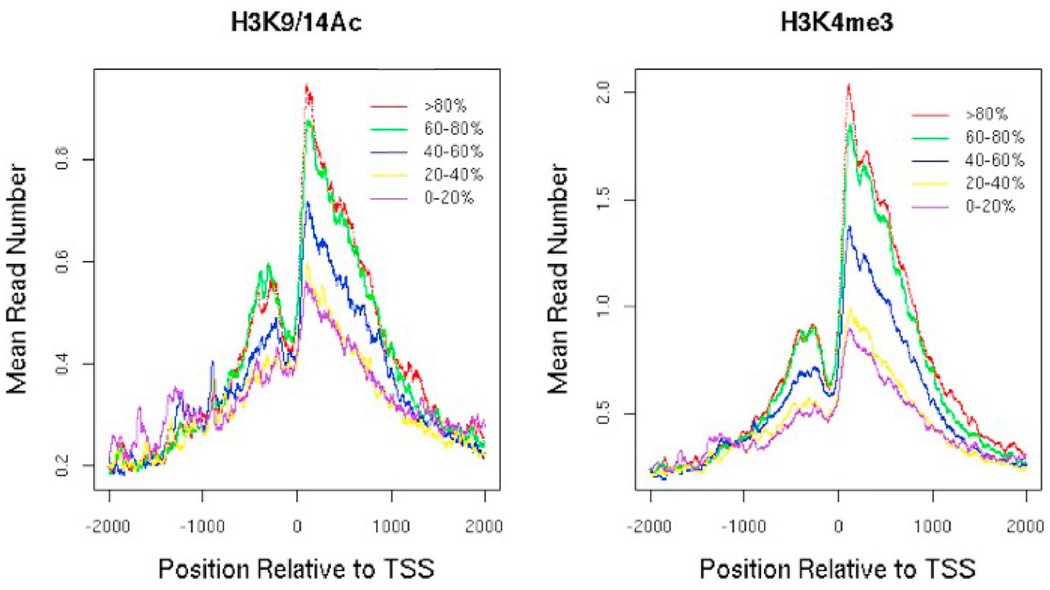
The ultimate goal of epigenetic ChIP-seq analysis in behavioral studies is to understand the molecular basis of behavior. The Illumina software package also provides a program called BeadStudio, which performs high level analysis of epigenomic data. High-level analysis involves biologically meaningful characterization including whether the mark activates or represses gene expression. Although some targets have known activating/repressive potentials, a good first step is to assess this potential regardless. For example, methylated histone 3 at lysine 4 (H3K4me3) tends to be in the promoter regions of actively transcribed genes, while methylated histone 3 at lysine 27 (H3K27me3) tends to be spread across the body of genes that are repressed. Therefore, analysis of the activating or repressive potential of these known marks serves as a quality control step to assess the quality of the antibody/sample preparation prior to sequencing. This analysis can be performed using the enrichment profile or the “sites” data together with either microarray gene expression or RNA-seq data. Using the enrichment profile data, one can stratify genes according to their gene expression levels (e.g., into 5 equal size bins or quintiles), align all gene transcription start sites (TSS) and, for each quartile of gene expression, calculate and plot the average read enrichment (y-axis) as a function of nucleotide position (x-axis) starting from the promoter region (e.g., 2kb upstream of the TSS) and ending either 2kb downstream of the TSS for 5’ associated marks or further for marks that tend to spread across the body of a gene (Barski et al., 2007) as shown in Figure 2. Read enrichment levels that rise with increasing gene expression levels/quartiles tends to be activating (Figure 2), whereas read enrichment levels that fall with increasing gene expression tend to be repressive. As can be seen from Figure 2, this analysis allows visualization of the average spatial distribution or deposition pattern of a mark across the promoter and gene and can reveal structural features (Barski et al., 2007). An alternative approach is to use the significantly enriched site data by first associating the sites with genes. For example, for a 5’ biased mark, one may require a site to overlap ±2kb of the TSS for that mark to be associated with a given gene. These rules can be easily generalized to marks that have different deposition patterns across the body of genes. One then calculates the distribution of gene expression values of genes with and without the sites (Bernstein et al., 2005). The two expression distributions can be visually represented as boxplots (i.e., boxes representing the 25th, 50th and 75th percentiles along with whiskers representing the 5th and 95th percentiles) as shown in Figure 3. Another example would be to plot two histograms. In either case, one should observe, increased expression levels for genes associated with the sites compared to those without for an activating mark and decreased expression levels for genes associated with sites for a repressive mark.
Figure 2.
Plot of H3K9/14Ac and H3K4me3 average read number relative to the TSS start site of genes stratified into 5 bins according to gene expression ranging from the lowest (0–20%) to highest (>80%): sort the genes on the Affymetrix U133 array according to their expression level and put the lowest 20% in one bin, the next 20–40% in another and continue until we have 5 bins with the same number of genes representing low to high expressed genes. For each group of genes separately, we calculate the number of mapped reads at every genomic coordinate (ranging from −2kb to +2kb) relative to the TSS of every gene in the group.
Figure 3.
Boxplots of log2 gene expression levels for genes with no modifications, H3K4me3, H3K9/14Ac and both H3K4me3 and H3K9/14Ac within 1kb of their 5' ends. We classify a gene into one of these four categories based on the present or absence of the H3K4me3 and H3K9/14Ac sites within 1kb of their 5' ends. The lower part of box is 25th percentile; the thick middle line of box is the median; the upper part of the box is 75th percentile; and the lower and upper whiskers are 5th and 95th percentile and represent outliers.
Comparison of Epigenetic Sites to Genomic Annotations and Sequences
The association of significantly enriched sites with genes also allows gene lists to be generated for every sample, which in the case of a behavior study could be different individuals. Tools have been developed that perform an overlap analysis of sites and annotations (Quinlan and Hall, 2010; Taylor et al., 2007). Venn diagram analysis can be applied to distinguish genes that contain mark(s) that are both shared among individuals and distinct to each individual. One can assess the extent to which particular pathways or functionally related genes are regulated by a given mark by performing Gene Ontology analysis (Huang da et al., 2009) and Pathway Analysis (Draghici et al., 2007) using the gene lists as input.
In the process of mapping epigenetic marks and transcription factors using ChIP-chip, that are associated with promoters (Bernstein et al., 2005; Cawley et al., 2004), or ChIP-seq (Barski et al., 2007) we have discovered that while a statistically significant fraction of the mark’s/factor’s sites map to coding gene promoter regions, a healthy fraction—if not the majority—map to noncanonical loci including introns, intergenic regions and the 3’ end of genes. Histone modifications that map to these non-canonical sites could be marking enhancer regions (Heintzman et al., 2009) that regulate genes in trans. Alternatively, these modifications could be regulating poorly annotated, non-coding RNAs (Cawley et al., 2004; Kampa et al., 2004; Kapranov et al., 2002) in cis. For example, we know that H3K4me3 tends to be at the promoter and H3K36me3—a mark that has been implicated in alternative splicing (Kolasinska-Zwierz et al., 2009)—tends to rise across the body and peaks near the 3’ end of active genes (Barski et al., 2007). In a recent study, Guttman et al. (Guttman et al., 2009) hypothesized that searching for this H3K4me3-H3K36me3 pattern in intergenic regions might reveal poorly annotated functional RNAs. They found ~1,600 large mutli-exonic RNAs which they named large intervening noncoding RNAs (lincRNAs) that were evolutionarily conserved. Thus, determining the proportion of enriched sites that are genic, intergenic, intronic and in promoters for each sample can give an indication whether the analysis should focus on genic or intergenic regions. This is a good example of why using relatively unbiased methods like sequencing are so important. In addition, the Venn diagrams of sites found in different samples can be further subdivided according to genic and non-genic categories to give an indication if epigenetic regulation of behavior is being driven by marks in non-canonical regions.
Identifying DNA Sequence Motifs/Putative Co-Factors
One of the significant advantages of ChIP-seq over ChIP-chip has been the ability of the signal peak to locate the relevant binding motif of a transcription factor. By taking advantage of the fact that top and bottom strand reads tend to be on the left and right of the transcription factor respectively, the peak locations that are calculated from the peak finding methods discussed above tend to be within 10–30bp of the binding motif (Jothi et al., 2008; Kharchenko et al., 2008; Zhang et al., 2008). In contrast, either using the overall read enrichment peaks in ChIP-seq (i.e., ignoring strand information) or the peaks called from ChIP-chip, the peaks tend to be within hundreds of base pairs from the binding motif (Jothi et al., 2008). Thus, in the case of histone modifications, the peak locations found by these programs are likely to give more precise locations of the modified tails for sharply peaked marks. In addition, motif analysis of the sequences within the significantly enriched peaks could identify co-factors including transcription factors or nuclear receptors that bind to DNA and recruit the enzymes that deposit the chemical group (i.e., methyl or acetyl) to a given histone tail residue. For example, in order to determine whether the H3K14ac is associated with ERα a motif analysis could be performed on the sequences of histone acetylation sites in order to determine if these sites yield the ERα motif (i.e., the estrogen response element). A number of powerful motif finding tools have been developed (Park, 2009; Tompa et al., 2005) including MEME (Bailey et al., 2006), MDScan (Liu et al., 2002), Weeder (Pavesi et al., 2004) and WebMOTIFS (Romer et al., 2007). Thus, using the tools and high level analyses described above, an investigator can characterize the regulation of the mapped mark, factors upstream of the mark as well as target genes and downstream pathways. In this way, ChIP-seq is a powerful tool for characterizing the epigenetic pathways regulating expression of genes that influence behavior.
Conclusion
Central to the field of behavioral neuroendocrinology is understanding how the external environment (maternal, social, chemical) and internal environment (fluctuating hormones) interact in order to produce long-lasting effects on development and behavior. Emerging evidence supports a role for epigenetic modifications as a mechanism through which the environment affects brain and behavior. As reviewed above, the technology available for assessing the role of epigenetic gene regulation in behavior is rapidly expanding and enabling the characterization of molecular pathways underlying reproductive and social behaviors. For example, using what are rapidly becoming standardized assays, individual labs can generate ChIP-seq data to investigate the epigenetic underpinnings not only of gene-environment interactions, but also of the molecular pathways involved in the regulation of reproductive behavior and the consolidation of reproductive experience. Moreover there are a plethora of analysis tools as detailed above. This is not to say that ChIP-seq data analysis does not require some bioinformatics skills, it does. Successful and efficient processing of these data require selection of the best software tools and algorithms, familiarity with UNIX, and writing programs/tools that reformat the data so that it can be used in multiple programs. Furthermore, many of the high level tasks are not encoded in programs that can be downloaded and require software to be written. Finally, the ability to make and test (including significance analysis and calculation of p-values) highly novel discoveries in the data requires tailored programs. Thus, a collaboration of investigators studying behavior, molecular biology and bioinformatics is highly recommended in order to truly harness the power of ChIP-seq to help define the role of epigenetics in behavior.
Acknowledgements
DS is supported by NIH T32 HD07382, and PG is supported by NIH R01 NS049065.
Abbreviations
- 5mC
5-methylcytosine
- 5-hmC
5-Hydroxymethylcytosine
- bp
base pairs
- CBP
CREB-binding protein
- ChIP
Chromatin Immunoprecipitation
- DNMTs
DNA methyltransferase proteins
- ERα
estrogen receptor alpha
- HATs
histone aceytltransferases
- HDACs
histone deacetylases
- H3K4me3
mono methylated H3 at lysine 4
- H3K14ac
acetylated H3 at lysine 14
- HTS
high-throughput sequencing
- MBD1
methyl-CpG-binding domain 1
- MECP2
methyl-CpG-binding protein 2
- Methyl-seq
methylation/bisulfite conversion sequencing
- NGS
next generation sequencing
- SNP
single nucleuotide polymorphism
- TET 1
tet oncogene 1
- TSS
transcription start site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai JA, Li S, Hartley DM, Feig LA. Transgenerational rescue of a genetic defect in long-term potentiation and memory formation by juvenile enrichment. J Neurosci. 2009;29:1496–1502. doi: 10.1523/JNEUROSCI.5057-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger AP, Jessen HM, Edelmann MN. Epigenetic organization of brain sex differences and juvenile social play behavior. Horm Behav. doi: 10.1016/j.yhbeh.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bonthuis P, Patteson JK, Rissman EF. Acquisition of sexual receptivity: roles of chromatin acetylation, estrogen receptor α, and ovarian hormones. Submitted to Endocrinology. 2010 doi: 10.1210/en.2010-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, Oyolu CB, Schroth GP, Absher DM, Baker JC, Myers RM. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci Biobehav Rev. 2009;33:593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Crews D. Epigenetics and its implications for behavioral neuroendocrinology. Front Neuroendocrinol. 2008;29:344–357. doi: 10.1016/j.yfrne.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews D, Gore AC, Hsu TS, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. Transgenerational epigenetic imprints on mate preference. Proc Natl Acad Sci U S A. 2007;104:5942–5946. doi: 10.1073/pnas.0610410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- Curley JP, Jensen CL, Mashoodh R, Champagne FA. Social influences on neurobiology and behavior: Epigenetic effects during development. Psychoneuroendocrinology. 2010 doi: 10.1016/j.psyneuen.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- Dolan PC, Denver DR. TileQC: a system for tile-based quality control of Solexa data. BMC Bioinformatics. 2008;9:250. doi: 10.1186/1471-2105-9-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavare S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Fagiolini M, Jensen CL, Champagne FA. Epigenetic influences on brain development and plasticity. Curr Opin Neurobiol. 2009;19:207–212. doi: 10.1016/j.conb.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen HF, Adie K, Chaubert P, Bird AP. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 2006;34:e96. doi: 10.1093/nar/gkl527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, Tammana H, Gingeras TR. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplant Q, Nestler EJ. CRACKing the histone code: Cocaine's effects on chromatin structure and function. Horm Behav. 2010 doi: 10.1016/j.yhbeh.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- Martinez-Alcantara A, Ballesteros E, Feng C, Rojas M, Koshinsky H, Fofanov VY, Havlak P, Fofanov Y. PIQA: pipeline for Illumina G1 genome analyzer data quality assessment. Bioinformatics. 2009;25:2438–2439. doi: 10.1093/bioinformatics/btp429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Maternal care as a model for experience-dependent chromatin plasticity? Trends Neurosci. 2005;28:456–463. doi: 10.1016/j.tins.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Yu W, Whitehead J, Xu J, Lezcano M, Pack S, Kanduri C, Kanduri M, Ginjala V, Vostrov A, Quitschke W, Chernukhin I, Klenova E, Lobanenkov V, Ohlsson R. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Romer KA, Kayombya GR, Fraenkel E. WebMOTIFS: automated discovery, filtering and scoring of DNA sequence motifs using multiple programs and Bayesian approaches. Nucleic Acids Res. 2007;35:W217–W220. doi: 10.1093/nar/gkm376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Sweatt JD. Epigenetic marking of the BDNF gene by early-life adverse experiences. Horm Behav. doi: 10.1016/j.yhbeh.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CD, Bucher P. ChIP-Seq data reveal nucleosome architecture of human promoters. Cell. 2007;131:831–832. doi: 10.1016/j.cell.2007.11.017. author reply 832-3. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics. 2007;Chapter 10(Unit 10 5) doi: 10.1002/0471250953.bi1005s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, Makeev VJ, Mironov AA, Noble WS, Pavesi G, Pesole G, Regnier M, Simonis N, Sinha S, Thijs G, van Helden J, Vandenbogaert M, Weng Z, Workman C, Ye C, Zhu Z. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Methods Mol Biol. 2009;507:3–20. doi: 10.1007/978-1-59745-522-0_1. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nat Biotechnol. 2009;27:455–457. doi: 10.1038/nbt0509-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan PS, Potter D, Deatherage DE, Huang TH, Lin S. Differential methylation hybridization: profiling DNA methylation with a high-density CpG island microarray. Methods Mol Biol. 2009;507:89–106. doi: 10.1007/978-1-59745-522-0_8. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]