Abstract
Epithelial-mesenchymal transition (EMT) plays pivotal roles during embryonic development and carcinoma progression. Members of the Snail family of zinc finger transcription factors are central mediators of EMT and induce EMT in part by directly repressing epithelial markers such as E-cadherin, a gatekeeper of the epithelial phenotype and a suppressor of tumor invasion. However, the molecular mechanism underlying Snai1 (Snail)-mediated transcriptional repression remains incompletely understood. Here we show that Snai1 physically interacts with and recruits the histone demethylase LSD1 (KDM1A) to epithelial gene promoters. LSD1 removes dimethylation of lysine 4 on histone H3 (H3K4m2), a covalent histone modification associated with active chromatin. Importantly, LSD1 is essential for Snai1-mediated transcriptional repression and for maintenance of the silenced state of Snai1 target genes in invasive cancer cells. In the absence of LSD1, Snai1 fails to repress E-cadherin. In cancer cells in which E-cadherin is silenced, depletion of LSD1 results in partial de-repression of epithelial genes and elevated H3K4m2 levels at the E-cadherin promoter. These results underline the critical role of LSD1 in Snai1-dependent transcriptional repression of epithelial markers and suggest that the LSD1 complex may be a potential therapeutic target for prevention of tumor invasion.
Keywords: EMT, E-cadherin, Snail, histone modifications, LSD1, bivalent
Introduction
Epithelial and mesenchymal cells exhibit distinct phenotypic and functional characteristics, yet the two cell types possess substantial plasticity and are interconvertible under certain circumstances (Thiery and Sleeman JP, 2006; Polyak and Weinberg, 2009). Epithelial cells can be reprogrammed into mesenchymal cells, a process known as epithelial–mesenchymal transition (EMT) (Hay, 1995). During EMT, epithelial cells downregulate epithelial markers, disassemble cell-cell adhesions, lose cell polarity, and concomitantly acquire enhanced migratory and invasive properties. EMT is fundamental to embryonic morphogenesis (Yang and Weinberg, 2008; Thiery et al, 2009). Mesoderm formation and neural crest development represent two prototypic developmental EMT events: During gastrulation, EMT transforms epiblast epithelium into migratory mesenchymal cells that constitute the mesoderm; Through EMT, neural crest progenitor cells delaminate and emigrate extensively from the neural tube to colonize distant embryonic territories. In human cancer, EMT is an important process conducive to tumor invasion and dissemination (Thiery, 2002). The most prevalent human malignancies are carcinomas, which originate from epithelial cells. EMT facilitates single carcinoma cells to escape from the rigid structural constraints imposed by intercellular epithelial junctions and to spread to distal sites. Consistently, EMT is frequently detectable at the invasive front of carcinomas where tumor cells exhibit more mesenchymal features.
A hallmark of EMT is the loss of E-cadherin, a defining marker of epithelial cells. E-cadherin regulates the establishment of the adherens junctions during epithelial morphogenesis and is important for the maintenance of tissue architecture (Hay, 1995). E-cadherin has been considered a suppressor of tumor invasion (Perl et al, 1998). Because E-cadherin is a key caretaker of the epithelial state, understanding the regulation of E-cadherin may help develop anti-invasion cancer therapy by preventing its downregulation or restoring its expression.
The Snail family of zinc finger transcription factors Snai1 (also known as Snail) and Snai2 (also known as Slug) have been identified as direct repressors of E-cadherin transcription and central mediators of EMT (Batlle et al, 2000; Cano et al, 2000; Hajra et al, 2002; Nieto, 2002). Snai1 and Snai2 critically regulate developmental EMTs such as gastrulation movement and migration of neural crest cells (Nieto, 2002), and their aberrant expression contributes to the onset of an invasive phenotype in a wide variety of human cancers (Peinado et al, 2007). Expression levels of the E-cadherin and Snai1 genes are inversely correlated at the leading edge of carcinomas (Francí et al, 2006).
In eukaryotes, post-translational covalent modifications on histone tails regulate chromatin structure and constitute important regulatory marks demarcating transcriptionally active and inactive chromatin (Jenuwein and Allis, 2001; Kouzarides, 2007; Li et al, 2007). Generally, transcriptionally active promoters are associated with hyperacetylation of core histones and methylation of histone H3 lysine 4 (H3K4), whereas inactive promoters enrich trimethylated H3K27. Previous studies have shown that Snai1 induces repressive histone modifications at the E-cadherin promoter through recruitment of histone deacetylases (HDACs) and the H3K27 methyltransferase EZH2 (Peinado et al, 2004; Herranz et al, 2008). However, the significance of H3K4 methylation in Snai1’s function remains elusive. In the present study, we show that Snai1 physically recruits the lysine-specific demethylase LSD1 (also known as KDM1A and AOF2) to reduce the level of dimethylated H3K4 (H3K4m2) at its target genes, and that LSD1 is required for Snai1-mediated transcriptional repression.
Results
Snai1 directly represses epithelial genes
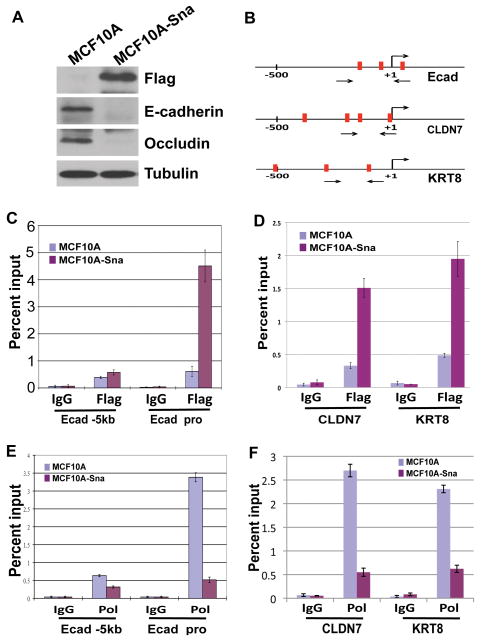
It was described previously that forced expression of Snai1 in epithelial cells drives EMT (Nieto, 2002). We decided to establish a Snai1-dependent EMT system in human mammary epithelial cells. Because amino-terminal fusions disrupted Snai1’s repressive activity (data not shown), we generated a functional Snai1-Flag construct in which the Flag tag was fused to the carboxyl terminus of Snai1. This Snai1-Flag construct was ectopically introduced into the MCF10A human mammary epithelial cell line and stable transfectants were obtained. MCF10A cells normally grow as clusters of cells with extensive cell-cell contacts. However, the Snai1-expressing cells became scattered and adopted a fibroblast-like morphology typical of mesenchymal cells. This phenotypic change was associated with loss of expression of epithelial markers E-cadherin and Occludin (Fig. 1A). These observations are indicative of an epithelial to mesenchymal morphological transition in the Snai1-expressing MCF10A cells.
Figure 1. Snai1 directly represses epithelial genes.
(A) Ectopic expression of Flag-tagged Snai1 in MCF10A cells downregulates epithelial markers E-cadherin and Occludin. Protein lysates from control and Snai1-expressing MCF10A cells were probed by Western blotting with indicated antibodies.
(B) Diagrams of the proximal promoters of E-cadherin, CLDN7, and KRT8. Vertical bars represent E-boxes. Arrows indicate primers used in ChIP assays.
(C) Snai1 is specifically enriched at the proximal promoter of E-cadherin in vivo as demonstrated by ChIP analysis. The results represent percentage of input chromatin and error bars indicate standard deviations (S.D.) from triplicate experiments.
(D) Snai1 is present at the promoters of CLDN7 and KRT8 in Snai1-expressing cells.
(E) Expression of Snai1 in MCF10A dissociates RNA polymerase II (Pol) from the E-cadherin promoter.
(F) Snai1 decreases Pol II binding to the CLDN7 and KRT8 promoters.
The E-cadherin promoter contains three “E-box” elements recognized by Snai1 (Fig. 1B) (Nieto, 2002). Two additionally selected epithelial genes claudin-7 (CLDN7) and cytokeratin-8 (KRT8), both of which were reported as direct targets of Snai1 (Ikenouchi et al, 2003; De Craene et al, 2005), also carry multiple E-boxes at their promoters (Fig. 1B). Using chromatin immunoprecipitation (ChIP) analysis, we investigated whether Snai1 bound to the promoters of these epithelial genes in vivo. Chromatin from control and Snai1-Flag-expressing MCF10A cells was precipitated with control immunoglobulin G (IgG) and anti-Flag antibodies, and quantitative PCR was performed on the recovered DNA to determine the enrichment of the proximal promoter regions of the epithelial genes as compared to a 5kb upstream region (−5kb) of E-cadherin, which serves as a negative control. Occupancy of Snai1 was detected specifically at the promoters of E-cadherin, CLDN7, and KRT8 in the Snai1-Flag cells (Fig. 1C and 1D).
To confirm that these epithelial genes are indeed inhibited by Snai1, we carried out a similar ChIP assay to monitor the binding of RNA polymerase II (Pol II) at their promoters. Consistent with E-cadherin expression, high levels of Pol II were detected at the E-cadherin promoter in MCF10A cells. By contrast, this binding was largely abolished in cells expressing Snai1 (Fig. 1E). Similar pattern was observed for the CLDN7 and KRT8 promoters (Fig. 1F). These results suggest that Snai1 directly represses epithelial markers and induces EMT in MCF10A cells.
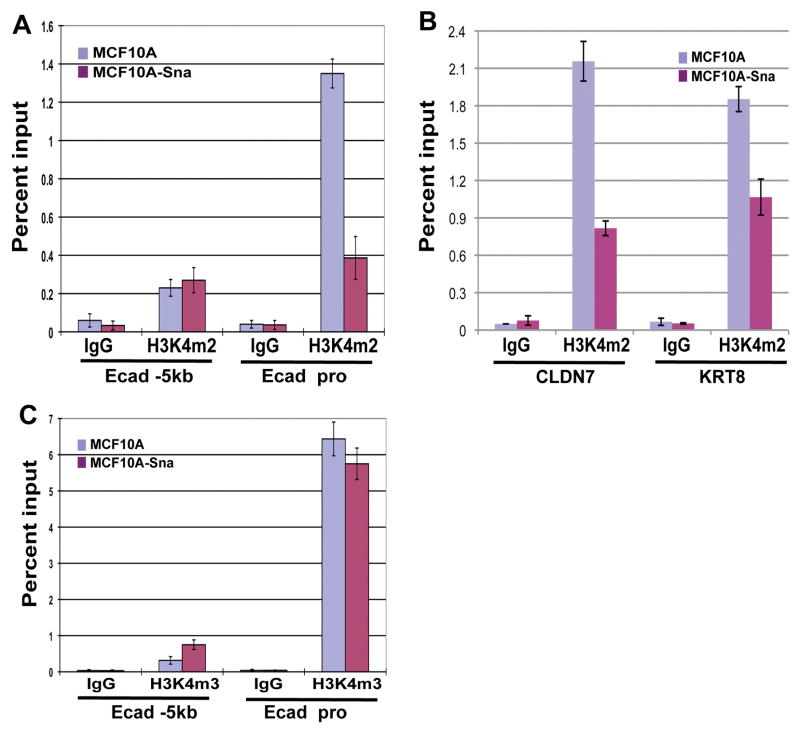
Snai1 downregulates H3K4m2 levels at epithelial gene promoters
Both di- and tri-methylated H3K4 are associated with active transcription (Kouzarides, 2007; Li et al, 2007). Having established an EMT model, we applied ChIP analysis to assess potential changes of the active H3K4 methylation marks at the epithelial gene promoters caused by ectopic Snai1 expression. Using an antibody specific for dimethyl H3K4 (H3K4m2), we detected relatively high levels of H3K4m2 at the promoter of the E-cadherin gene in MCF10A cells, and a significant decrease of this active mark specifically at the promoter region in the Snai1-expressing cells (Fig. 2A). The levels of H3K4m2 in the E-cadherin promoter thus correlate with E-cadherin transcription. Similarly, the CLDN7 and KRT8 promoters also exhibited much reduced H3K4m2 in Snai1-expressing cells (Fig. 2B). Surprisingly, when we performed ChIP assay for the abundance of trimethylated H3K4 (H3K4m3) at the E-cadherin promoter, there was no difference of this mark between control and Snai1-expressing cells (Fig. 2C). High levels of H3K4m3 were observed at the E-cadherin promoter in both groups of cells, even though E-cadherin is inactive in Snai1-expressing cells. We conclude that expression of Snai1 leads to a specific decrease of H3K4m2 at its target promoters.
Figure 2. Snai1 reduces H3K4 dimethylation at its target gene promoters.
(A) Snai1 expression causes a reduction in H3K4m2 levels at the E-cadherin promoter, but not in the 5kb upstream region, as determined by ChIP analysis.
(B) Snai1 reduces H3K4m2 levels at the CLDN7 and KRT8 promoters.
(C) The H3K4m3 mark at the E-cadherin promoter is not affected by Snai1.
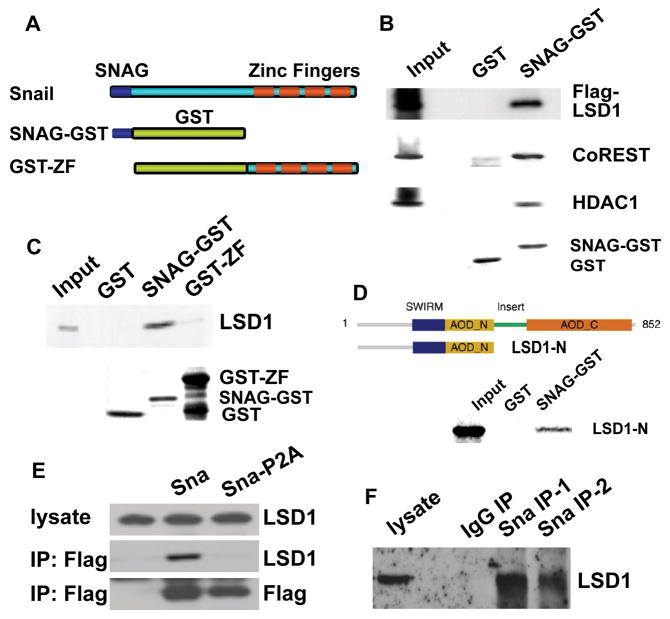
Snai1 interacts with LSD1
Decrease in H3K4me2 levels cannot be mediated directly by any known Snai1-associated histone modifying enzymes. H3K4m2 can be demethylated by two types of histone demethylases: LSD1, which belongs to the class of flavin adenine dinucleotide (FAD)-dependent monoamine oxidases, and members of the JARID1 (KDM5) family of Jumonji (JmjC) domain-containing demethylases (Shi, 2007; Klose and Zhang, 2007). LSD1 associates with co-repressors including HDAC1/2 and CoREST to form a core ternary complex, which is recruited to chromatin and can efficiently bind and modify nucleosomal substrates to repress transcription (Lan et al, 2008). Unlike JARID1, LSD1 cannot remove trimethylation of H3K4 (Shi et al, 2004). Given that Snai1 reduces the level of H3K4m2 but not H3K4m3, the LSD1 complex becomes a leading candidate to mediate Snai1’s action.
Snail family members share a carboxyl-terminal DNA binding domain composed of zinc finger motifs and a highly conserved amino-terminal repression domain designated SNAG. The SNAG domain extends to about 21 amino acids with the first 7 amino acids being nearly invariant (Grimes et al, 1996) and it is present in diverse transcriptional repressors including Gfi-1, IA-1, Gsh-1, and Ovo. The SNAG domain has been shown to be essential for repression mediated by Gfi-1 (Grimes et al, 1996) and Snai1 (Batlle et al, 2000). A recent report has demonstrated that Gfi-1 interacts with the LSD1 corepressor complex (Saleque et al, 2007). Given the well-documented transcriptional repressive activities of both the SNAG domain and the LSD1 complex, we hypothesized that the SNAG domain might be sufficient for interaction with the LSD1 complex, and hence, Snai1 might be capable of recruiting the LSD1 complex as well.
We tested this possibility using the in vitro glutathione S-transferase (GST) pull-down assays. Since the activity of SNAG is not compatible with fusions to its amino terminus, we specifically fused GST to the carboxyl terminus of the SNAG domain (Fig. 3A). GST (control) and recombinant SNAG-GST proteins were produced and affinity-purified from bacteria, and were subjected to binding assays with whole cell lysate prepared from mammalian HEK293 cells transfected with Flag-LSD1. Based on Western blotting with the anti-Flag antibody, LSD1 was found to be associated with SNAG-GST but not GST (Fig. 3B). Moreover, other components of the LSD1 core complex, the endogenous CoREST and HDAC1 specifically bound to SNAG-GST as well (Fig. 3B). The result suggests that the SNAG domain of Snai1 is sufficient for interaction with the LSD1 complex.
Figure 3. Snai1 physically interacts with LSD1 in vitro and in vivo.
(A) Schematic diagram of SNAG-GST and GST-ZF fusion proteins used in GST pull-down assays.
(B) The SNAG domain is sufficient for association with the LSD1 complex. Whole cell lysate prepared from HEK293 cells transfected with Flag-LSD1 was incubated with GST and the SNAG-GST fusion protein and followed by Western blot analysis using anti-Flag, anti-CoREST, and anti-HDAC1 antibodies. Coomassie staining shows the protein loading of GST and SNAG-GST.
(C) LSD1 directly interacts with the SNAG domain. 35S-labeled LSD1 protein was mixed with GST, SNAG-GST, or GST-ZF fusion proteins. Bound LSD1 was then detected by autoradiography after SDS/PAGE. Coomassie staining of GST proteins was shown.
(D) The amino-terminus of LSD1 interacts with SNAG.
(E) Exogenous Snai1 associates with endogenous LSD1 in intact cells. Co-immunoprecipitation of whole cell lysate from HEK293 cells overexpressing Flag-tagged wild-type Snai1 or Snai1-P2A mutant was performed with anti-Flag antibody. The presence of LSD1 in the precipitates was shown by Western blotting with an anti-LSD1 antibody. Anti-Flag Western blotting indicates expression of wild-type and mutant Snai1.
(F) Endogenous Snai1 and LSD1 form a complex in vivo. MDA-MB-231 cells were lysed and incubated with two anti-Snai1 antibodies (#1 fromSantaCruz; #2 from Cellsignaling) or control IgG, followed by Western blotting with the LSD1 antibody.
To determine whether such interactions are direct and which subunit of the LSD1 complex is responsible for physical interaction with SNAG, 35S-labeled LSD1, CoREST, and HDAC1 were produced by in vitro transcription and translation, and subjected to similar GST binding assays. Only LSD1 displayed readily detectable association with SNAG-GST (Fig. 3C), suggesting that LSD1 interacts with the SNAG domain of Snai1 in a direct manner and the other components of the core LSD1 complex (such as HDAC) are recruited to Snai1 indirectly. The specificity of the interaction was further confirmed by lack of binding between LSD1 and the carboxyl-terminal zinc finger motifs of Snai1 (Fig. 3C). Furthermore, a truncated LSD1 mutant which lacks the carboxyl-terminal half of the protein retains the ability to interact with SNAG (Fig. 3D).
We performed co-immunoprecipitation assays to examine whether Snai1 and LSD1 might form a complex in vivo. HEK293 cells were transiently transfected with a construct encoding Flag-tagged wild-type Snai1 or a P2A mutant form that carries a point mutation in the SNAG domain and fails to repress transcription (Batlle et al, 2000). Snai1 immunocomplexes were precipitated from cellular extracts with anti-Flag antibodies. The presence of endogenous LSD1 was observed in the immunoprecipitated fraction obtained only from cells expressing wild type Snai1, but not the P2A mutant (Fig. 3E). This evidence supports the specific association between Snai1 and LSD1 in vivo, and suggests that a functional SNAG domain is essential for this interaction. Finally, the ability of Snai1 to interact with LSD1 was confirmed by immunoprecipitation assays between the endogenous proteins in highly metastatic MDA-MB-231 breast tumor cells (Fig. 3F). Endogenous LSD1 was found to coimmunoprecipitate with endogenous Snai1 in the immunocomplexes obtained with two anti-Snai1 antibodies but not IgG.
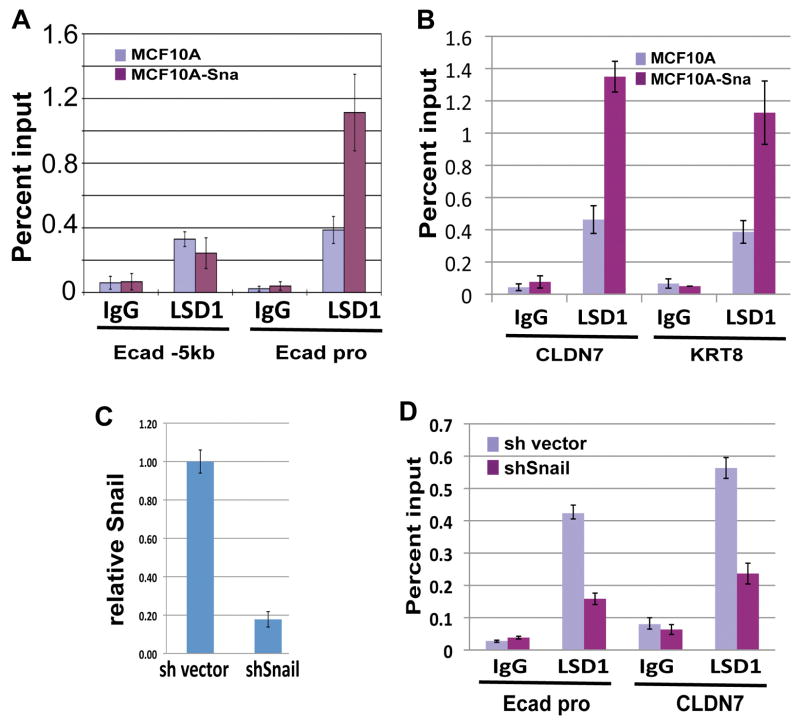
Snai1 recruits LSD1 to its target genes
Snai1 binds to the epithelial gene promoters and causes a decrease of H3K4m2. Given the direct association between Snai1 and LSD1, LSD1 may be recruited to these promoters by Snai1 where it demethylates H3K4m2. To validate this hypothesis, we examined LSD1 occupancy at the E-cadherin promoter region by ChIP analysis. LSD1 is enriched at the E-cadherin promoter specifically in the Snai1-expressing cells but not in the control MCF10A cells (Fig. 4A). Consistent with this observation, LSD1 is also enriched in the promoters of CLDN7 and KRT8 in Snai1-expressing cells (Fig. 4B). The levels of LSD1 binding inversely correlate with H3K4m2 (compare to Fig. 2).
Figure 4. LSD1 is recruited to epithelial gene promoters by Snai1.
(A) Occupancy of LSD1 at the E-cadherin promoter is increased in Snai1-expressing cells compared to control MCF10A cells as shown by ChIP analysis with the LSD1 antibody.
(B) The binding of LSD1 to the CLDN7 and KRT8 promoters is increased in MCF10A cells expressing Snai1.
(C) Quantitative RT-PCR analysis shows efficient depletion of Snai1 in MDA-MB-231 cells by a retroviral shRNA.
(D) The presence of LSD1 at the E-cadherin and CLDN7 promoters is reduced in MDA-MB-231 cells depleted of Snai1.
The invasive MDA-MB-231 breast cancer cells express high levels of Snail-like factors and are negative for epithelial markers (Hajra et al, 2002). We asked whether the binding of LSD1 to epithelial gene promoters might be dependent on Snai1 in these cells. We first used short hairpin RNA (shRNA) to deplete Snai1 (Fig. 4C), and then compared LSD1 occupancy at epithelial gene promoters in control and Snai1-knockdown cells. As expected, the binding of LSD1 to the E-cadherin and CLDN7 promoters decreased in the Snai1-depleted cells (Fig. 4D). The remaining binding might be due to the presence of Snai2 in these cells (Hajra et al, 2002). Taken together, these data suggest that LSD1 is brought to the E-cadherin and other epithelial gene promoters through interaction with Snai1.
LSD1 is essential for Snai1-initiated transcriptional repression
Because LSD1 is capable of removing the active H3K4m2 mark, it is conceivable that LSD1 may contribute to Snai1-mediated transcriptional repression. Several histone-modifying enzymes, namely HDAC, EZH2, and the arginine methyltransferase PRMT5 (Peinado et al, 2004; Herranz et al, 2008; Hou et al, 2008), have been identified as corepressors of Snai1, it was unclear whether LSD1 might play a significant role in Snai1-dependent transcriptional repression.
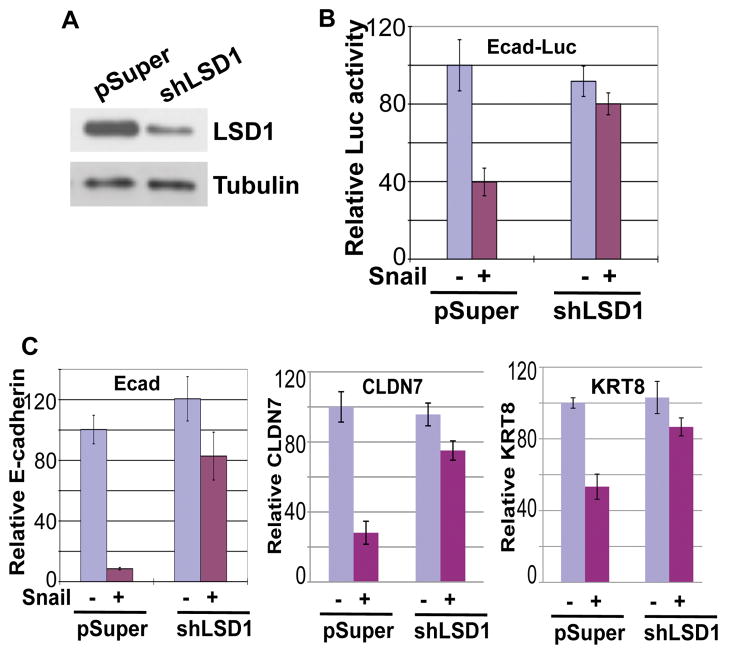
Snai1 has been reported to function as a potent transcriptional repressor. We speculated that the ability of exogenous Snai1 to repress transcription could be attributed to endogenous LSD1 present in cells. To check whether endogenous LSD1 might contribute to Snai1’s repressive activity, we applied shRNA to substantially deplete endogenous LSD1 in the MCF7 breast cancer epithelial cells (Fig. 5A). Using a luciferase reporter driven by the E-cadherin promoter carrying three E-boxes recognized by Snai1, we found that Snai1 could repress this reporter when transfected into the control MCF7 epithelial cells. However, in MCF7 cells with much reduced LSD1 levels, the Snai1-mediated inhibitory effect on the E-cadherin reporter was essentially eliminated (Fig. 5B).
Figure 5. LSD1 is essential for Snai1-mediated repression.
(A) Verification of LSD1 knockdown by Western blotting. MCF7 cells were infected with retroviral empty vector pSuper or with shRNA targeting LSD1, followed by Western blotting assays.
(B) Depletion of LSD1 impairs the repressive activity of Snai1 in reporter-based assays. Snai1 fails to repress a luciferase reporter driven by the E-cadherin promoter in LSD1-depleted cells. Error bars indicate S.D. from three independent experiments.
(C) LSD1 is essential for Snai1-mediated repression of endogenous epithelial genes. Expression of E-cadherin, CLDN7, and KRT8 was determined by quantitative RT-PCR.
We further studied whether inhibition of endogenous epithelial genes by Snai1 depended on LSD1. Control or LSD1-depleted MCF7 cells were transduced with lentiviral Snai1-IRES-GFP, which simultaneously expresses both Snai1 and GFP. GFP-positive cells were sorted and purified by flow cytometry, and RNA expression of epithelial genes in these cells was analyzed. Ectopic Snai1 expression potently reduced RNA levels of endogenous E-cadherin, CLDN7, and KRT8 in MCF7 cells (Fig. 5C). By contrast, in cells depleted of LSD1, Snai1’s suppressive effect on these genes was significantly diminished (Fig. 5C). Together, these observations suggest that Snai1-initiated inhibition of epithelial genes is dependent on LSD1. Therefore, LSD1 is an essential effector of Snai1-mediated transcriptional repression.
LSD1 is required for maintenance of the silenced state of Snai1 target genes
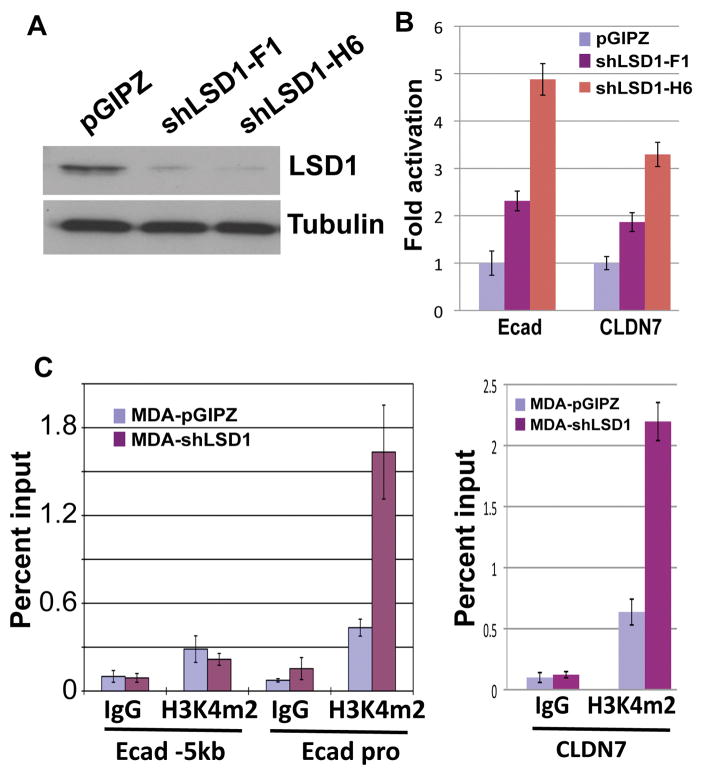
In highly invasive cancer cells such as MDA-MB-231, E-cadherin and other epithelial genes are already silenced. To investigate whether LSD1 is required for maintenance of the silenced state of epithelial genes in these cells, we knocked down endogenous LSD1 with two lentiviral shRNAs (Fig. 6A). Consequently, upregulation of E-cadherin and CLDN7 mRNA levels was detected in LSD1-deprived cells (Fig. 6B), suggesting that loss of LSD1 leads to de-repression of these genes. However, very much like depletion of the EZH2-containing PRC2 complex (Herranz et al, 2008), depletion of LSD1 only resulted in partial activation of E-cadherin. E-cadherin protein could not be detected by Western blotting and there was no mesenchymal to epithelial morphological change in LSD1-depleted cells (not shown). It is likely that in addition to LSD1, other repressive mechanisms keep these genes from full activation.
Figure 6. LSD1 is required to maintain the silenced status of Snai1 target genes in invasive cancer cells.
(A) Validation of LSD1 depletion in MDA-MB-231 cells with two lentiviral shRNAs by Western blotting with denoted antibodies. Tubulin serves as a loading control.
(B) Expression of E-cadherin and CLDN7 is upregulated in MDA-MB-231 cells depleted of LSD1. The RNA levels of the two genes were normalized against GAPDH by quantitative RT-PCR analysis.
(C) LSD1 depletion in MDA-MB-231 cells increases H3K4m2 levels specifically at the E-cadherin and CLDN7 promoters, as determined by ChIP assays.
As LSD1 is involved in demethylation of H3K4m2, we examined the H3K4m2 levels at the promoters of E-cadherin and CLDN7. A marked increase of H3K4m2 specifically at the E-cadherin and CLDN7 promoters was observed in the LSD1-depleted cells (Fig. 6C), suggesting that LSD1 constantly erases the H3K4m2 mark to prevent its accumulation at the epithelial gene promoters in these cells. These results suggest that an active demethylation process by LSD1 is necessary to reinforce silencing of Snai1 target genes.
Discussion
In the present report, we have investigated the epigenetic mechanism underlying Snai1-mediated transcriptional repression of epithelial genes. We show that Snai1 directly interacts with the LSD1 histone demethylase and recruits the LSD1 complex to the E-cadherin and other epithelial gene promoters, resulting in downregulation of the active H3K4m2 mark and promoter activity. As depletion of LSD1 substantially impairs Snai1’s ability to repress its targets, LSD1 serves as an essential effector of Snai1-dependent transcriptional repression of epithelial genes. Mouse embryos deficient for LSD1 die at an early embryonic stage (Wang et al, 2007). As Snai1 and Snai2 play critical roles in EMT events during mesoderm and neural crest development, it is interesting to examine whether LSD1 mutant embryos have the corresponding defects. Recent studies show that LSD1 is highly expressed in clinically advanced breast tumors and in poorly differentiated neuroblastomas (Schulte et al, 2009; Lim et al, 2010). Together with the prominent role of Snail family members in initiation of EMT and tumor invasion (Peinado et al, 2007), targeting the enzymatic components of the LSD1 complex with pharmacological agents may offer a new avenue to thwart tumor progression and dissemination.
While this work highlights the significance of an epigenetic silencing mechanism in Snai1’s function, the exact mechanism underlying Snai1-dependent transcriptional repression is complex. In addition to the LSD1 complex, Snai1 has been reported to directly or indirectly associate with repressor complexes containing HDAC, PRC2, and Ajuba-PRMT5, through its SNAG domain (Peinado et al, 2004; Herranz et al, 2008; Hou et al, 2008). All these complexes are somewhat essential for Snai1’s action. Although we show that the Snai1-LSD1 interaction is probably direct, it remains to be determined whether all these components are in the same complex, or they interact with Snai1 independently. As a consequence, Snai1 leaves multiple repressive histone modifications at its target genes, including histone hypoacetylation, decreased H3K4m2, and increased H3K27m3. Because the H3K4m3 mark is not a substrate of LSD1 and remains intact in Snai1-expressing cells (Fig. 2C), Snai1 apparently induces a “bivalent” histone modification pattern with co-existence of both H3K4m3 and H3K27m3. Promoters with bivalent histone modifications are inactive but are poised for activation (Bernstein et al, 2006). This implies that Snai1-repressed epithelial genes are primed for re-activation, which is consistent with the reversibility of EMT (Thiery and Sleeman JP, 2006).
Materials and Methods
Plasmids and Antibodies
Full-length human Snai1 cDNA was PCR amplified and cloned into pcDNA3 (Invitrogen) with a Flag epitope tag at its carboxyl end. The Snai1-P2A mutant was generated using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene). Lentiviral expression vector for Snai1-IRES-GFP was made in pCSCGW2. The expression plasmids for SNAG-GST and GST-ZF were based on pGEX-KG. The human E-cadherin promoter was PCR amplified and cloned into the pGL3 vector for luciferase reporter assays. shRNA constructs specifically targeting human LSD1 were in the retroviral pSuper and lentiviral pGIPZ (OpenBiosystems) vectors.
The following antibodies were used in this study: anti-H3K4m2, anti-H3K4m3, anti-Snai1, anti-LSD1, and anti-E-cadherin (Cell Signaling); anti-LSD1, anti-RNA polymerase II (Millipore); anti-Snai1 (Santa Cruz); anti-Flag, anti-Occludin, and anti-Tubulin (Sigma).
Cell culture, Transfection, Infection, Luciferase assay, and RT-PCR
The human cell lines HEK293, MCF7 and MDA-MB-231 were grown in regular DMEM medium (Cellgro) supplemented with 10% bovine calf serum (BCS). MCF10A cells were cultured in DMEM/F12 medium (Cellgro) supplemented with 5% horse serum (Sigma), 20ng/mL epidermal growth factor (EGF, Sigma), 10μg/ML insulin (Sigma), 0.5μg/mL hydrocortisone (Sigma).
To establish a Snai1-expressing stable cell line, MCF10A cells were transfected with the Snai1-Flag expression plasmid and stable transfectants were picked and verified for Snai1 expression by Western blotting. All the transfections were done using Turbofect transfection reagent (Fermentas) according to the manufacturer’s instructions.
For the reporter assay, MCF7 cells were transfected with the E-cadherin promoter Luciferase reporter and SV40-driven Renilla reporter plasmids, and harvested for Luciferase assays 48 hours later. Luciferase activities were determined using the dual Luciferase Reporter Assay System (Promega). Renilla luciferase was used as a reference to normalize transfection efficiencies in all experiments. All values are means ± S.D. from three independent experiments.
For RNA isolation and RT-PCR assays, Snai1-IRES-GFP expression lentivirus was used to infect control or LSD1-depleted MCF7 cells. The transduced cells were sorted by flow cytometry based on the GFP signal. Total RNA from cells was extracted with TRIzol reagent (Invitrogen) following the manufacturer’s instruction, and cDNA was synthesized using M-MuLV reverse transcriptase with random primers. The expression levels of selected genes were determined by quantitative PCR.
GST pull-down, co-immunoprecipitation, and Chromatin immunoprecipitation (ChIP)
Both GST pull down and co-immunoprecipitation assays were performed as previously described (Ao et al, 2008).
For ChIP assay, cells were cross-linked with 1% formaldehyde for 10 min. The reaction was stopped by the addition of glycine. Cross-linked cells were washed in 1x PBS and collected. Cell pellets were washed in washing buffer (0.25% TritonX-100, 10mM EDTA, 0.5mM EGTA, 10mM Tris pH8.0), resuspended in sonication buffer (1mM EDTA, 0.5mM EGTA, 10mM Tris pH8.0), mixed with glass beads, and subjected to sonication. The sonicated samples were diluted in ChIP buffer (0.01% SDS, 1.0% TX-100, 1.0mM EDTA, 20mM Tris pH 8.1, 150 mM NaCl) and incubated with specific antibodies and protein A slurry (Invitrogen). The immunoprecipitates were subjected to a series of washing steps to remove non-specific binding. After reverse-crosslinking, the DNA samples were purified and then analyzed by real-time quantitative PCR. Final results represent percentage of input chromatin and error bars indicate standard deviations (S.D.) from triplicate experiments.
Acknowledgments
We appreciate technical assistance from Linbao Ai and Heiman Wang. We are grateful to Mien-Chie Hung (MD Anderson Cancer Center, Texas), M. Angela Nieto (Instituto de Neurociencias CSIC-UMH, Spain), Suming Huang, Yi Qiu, and Lizi Wu for kindly providing reagents. We thank Mike Kilberg and Jorg Bungert for critical reading of the manuscript. This work was supported by grants to J.L. from Stop!Children’s Cancer, Florida Bankhead-Coley Cancer Research Program (09BN-12-23092), and the National Cancer Institute (R01CA137021), and to A.P. from the University Scholars Program at University of Florida.
Footnotes
Conflict of Interest:
The authors declare no conflict of interest.
References
- Ao A, Wang H, Kamarajugadda S, Lu J. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc Natl Acad Sci USA. 2008;105:7821–7826. doi: 10.1073/pnas.0711677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- De Craene B, Gilbert B, Stove C, Bruyneel E, van Roy F, Berx G. The transcription factor snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res. 2005;65:6237–6244. doi: 10.1158/0008-5472.CAN-04-3545. [DOI] [PubMed] [Google Scholar]
- Francí C, Takkunen M, Dave N, Alameda F, Gómez S, Rodríguez R, et al. Expression of Snail protein in tumor-stroma interface. Oncogene. 2006;25:5134–5144. doi: 10.1038/sj.onc.1209519. [DOI] [PubMed] [Google Scholar]
- Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol. 1996;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- Hay E. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Díaz VM, Francí C, Gutierrez A, Dave N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumor progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial–mesenchymal transitions in tumor progression. Nature Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]