Abstract
Cryptochromes are a class of photosensory receptors that control important processes in animals and plants primarily by regulating gene expression. How photon absorption by cryptochromes leads to changes in gene expression has remained largely elusive. Three recent studies, including Lian and colleagues (pp. 1023–1028) and Liu and colleagues (pp. 1029–1034) in this issue of Genes & Development, demonstrate that the interaction of light-activated Arabidopsis cryptochromes with a class of regulatory components of E3 ubiquitin ligase complexes leads to environmentally controlled abundance of transcriptional regulators.
Keywords: photoreceptor, SPA1, COP1, cryptochrome 1 signaling, Arabidopsis, blue light
Living organisms have evolved mechanisms to sense, respond to, and adapt to changes in their environment. Alterations in the light environment provide important cues regarding the time of the day, the season, and the surrounding habitat (e.g., water depth and shade from the vegetation) (Kami et al. 2010; Zoltowski and Gardner 2011). Organisms in all kingdoms of life possess a variety of photoreceptors that trigger these light responses. Among these is the cryptochrome family of blue-light sensors, which are present in both animals and plants (Cashmore 2003; Liu et al. 2010). The founding member of this family was uncovered in Arabidopsis almost 20 years ago (Ahmad and Cashmore 1993).
In plants, cryptochromes control important developmental transitions. These include de-etiolation, a transition during which seedlings switch from using energy within seed reserves to photoautotrophic growth, and photoperiodic induction of floral initiation (Fig. 1A). In addition, cryptochromes are a component of the light input pathway that resets the circadian oscillator. In mammals and insects, cryptochromes are well known for their role in the circadian clock, where they act as either light-regulated or light-independent components of the oscillator (Cashmore 2003). Interestingly, cryptochromes have also been implicated as magnetoreceptors, and have been shown to be essential for the time-compensated sun compass of the monarch butterfly (Reppert et al. 2010).
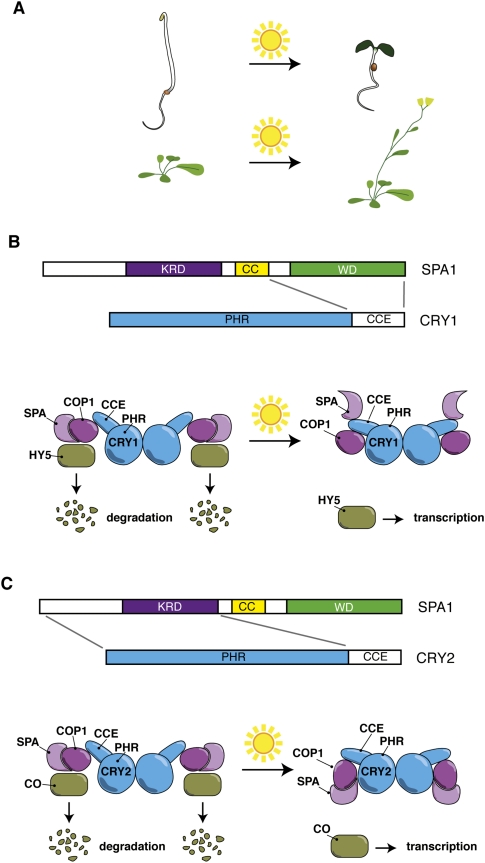
Figure 1.
A light-induced conformational change in the CCE domain of plant cryptochromes inhibits COP1-mediated degradation of transcription factors. (A) In plants, the cryptochromes (CRY1 and CRY2 in Arabidopsis) control two major developmental transitions, which are controlled by the light-regulated abundance of transcription factors. In the dark, seedlings grow from their seed reserves. Upon transfer into the light, several photoreceptors, including the cryptochromes, promote photoautotrophic growth that is accompanied by major developmental adaptations, such as development of chloroplasts, expansion of the leaves, and inhibition of stem growth. Flowering in many plant species is controlled by day length. The cryptochromes, particularly CRY2 in Arabidopsis, are major contributors of photoperiodic induction of reproductive development. (B) A model depicting how plant CRY1 regulates the activity of a multimeric E3 ubiquitin ligase composed of COP1, SPA proteins, RBX1, DDB1, and cullin 4 (the latter three are not shown on the figure). Schematic depiction of the domain structure of SPA1 and CRY1. (KRD) Kinase-related domain; (CC) coiled-coil domain; (WD) WD40 repeat domain. Parts of the proteins involved in the interaction are indicated. Cryptochromes act as dimers and are composed of two domains, the PHR and the CCE, the latter of which changes conformation upon light activation. In the dark, the CCE domain comes into direct contact with COP1 but not SPA proteins. However, COP1 and SPA interact with each other, promoting E3 ubiquitin ligase activity of the complex, leading to degradation of important transcription factors such as HY5. In the light, SPA interacts directly with the CRY1 CCE domain, preventing further interaction with COP1. This inhibits E3 ligase activity, leading to the accumulation of HY5 during de-etiolation. (C) A model depicting the mechanism of CRY2-controlled CO accumulation. Schematic depiction of the domain structure of SPA1 and CRY2. (KRD) Kinase-related domain; (CC) coiled-coil domain; (WD) WD40 repeat domain. Parts of the proteins involved in the interaction are indicated. In short day conditions, the SPA/COP multimeric E3 ligase degrades the transcription factor CO. Increased day length leads to longer CRY2 activation, and thus interaction between SPA and the PHR domain of CRY2. This leads to a tighter interaction between COP1 and CRY2 and, subsequently, inhibition of ubiquitin E3 ligase activity, resulting in stabilization of CO.
Mechanism of cryptochrome light activation
Cryptochromes are related to a class of DNA repair enzymes known as DNA photolyases. A key difference between DNA photolyases and cryptochromes is that, in the former, light energy is used to initiate DNA repair, while, in the latter, light is used as an informational cue (Liu et al. 2010). Proteins of the photolyase/cryptochrome superfamily share the N-terminal photolyase homologous region (PHR) that harbors the light-sensing chromophores. Photolyases possess an antenna pigment (e.g., methenyltetrahydrofolate) and a flavin cofactor (FADH−) that, upon light perception, transfers an electron to reverse cross-linking between bases for DNA repair. Cryptochromes also possess a primary FAD chromophore, but the presence and identity of a second antenna pigment is not always clearly established (Liu et al. 2010). Moreover, the photocycle of cryptochromes is not yet fully understood, and this topic remains a matter of debate (Liu et al. 2010). Importantly, the redox state of the flavin chromophore is at the heart of light-induced events triggered upon cryptochrome excitation. Such a redox-based signaling system could also be influenced by other factors, such as oxygen concentration, the cellular midpoint potential, and temperature. Therefore, it is possible that cryptochrome signaling is also affected by additional factors other than light (Yang et al. 2008). Another distinguishing feature of cryptochromes is the presence of a C-terminal CCE (cryptochrome C-terminal extension) domain of varying length (Cashmore 2003; Liu et al. 2010). In Arabidopsis cryptochrome 1 (CRY1) and CRY2, the CCE appears to be largely unstructured, yet is known to act as an effector domain (Yang et al. 2000; Partch et al. 2005; Kottke et al. 2006; Yu et al. 2007b; Liu et al. 2010; Zoltowski and Gardner 2011).
Light-controlled cryptochrome signaling in plants
Cryptochromes are widespread in plants, where they control important developmental transitions throughout the plant life cycle (Cashmore 2003; Liu et al. 2010). Higher plants typically possess two cryptochromes with clear functional diversification. This has been studied extensively in the model plant Arabidopsis thaliana, where light-stable CRY1 mediates mostly high light responses and light-labile CRY2 specializes in responses to low light (Lin and Shalitin 2003). CRY1 and CRY2 are both nuclear proteins, but CRY1 is also present in the cytosol. Consistent with their role in blue-light-regulated gene expression, cryptochromes act primarily in the nucleus (Wu and Spalding 2007; Yu et al. 2007a). Both Arabidopsis cryptochromes are phosphorylated upon blue-light perception (Shalitin et al. 2002, 2003). For CRY2, this post-translational modification triggers proteasome-mediated degradation (Shalitin et al. 2002; Yu et al. 2007a). Phosphorylation has also been related to the activation of cryptochromes, but the link between phosphorylation and cryptochrome activity remains poorly understood (Shalitin et al. 2003; Yu et al. 2007a,b). Despite the absence of a canonical kinase domain, CRY1 binds to ATP and autophosphorylates in vitro (Liu et al. 2010). However, light-induced cryptochrome phosphorylation does not necessarily depend on the kinase activity of cryptochrome itself, and the functional significance of this activity remains to be determined (Liu et al. 2010).
The CCE domain is central to the activity of plant cryptochromes. This was first demonstrated using plants expressing the CCE domain fused to a dimerization domain, but lacking the PHR domain (Yang et al. 2000). These plants showed a phenotype referred to as constitutively photomorphogenic, where seedlings develop as they would normally under light conditions, despite being grown in darkness. This result suggested that the CCE domain of plant cryptochromes is active in the absence of light when it is separated from the regulatory PHR domain. This hypothesis was further supported by reports demonstrating that the CCE domain undergoes light-induced conformational changes, suggesting that CCE domain activity is controlled by light (Partch et al. 2005; Kottke et al. 2006; Yu et al. 2007b; Liu et al. 2010; Zoltowski and Gardner 2011). The ensuing hunt for proteins interacting with the CCE domain rapidly identified the E3 ubiquitin ligase component CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) (Wang et al. 2001; Yang et al. 2001). COP1 exists in both animals and plants, where it controls accumulation of important transcription factors (Yi and Deng 2005). In contrast to plant cryptochromes, there is currently no experimental evidence suggesting an interaction between animal cryptochromes and COP1. Surprisingly, however, plant cryptochromes interact with COP1 independently of light (Wang et al. 2001; Yang et al. 2001). Thus, although it was proposed that cryptochromes inhibit COP1 activity specifically upon light perception, the primary light-mediated mechanism through which COP1 activity is controlled has remained elusive. This perspective highlights three important studies (Lian et al. 2011; Liu et al. 2011; Zuo et al. 2011) that further our knowledge in this regard.
Cryptochromes interact with SPA family proteins in a blue-light-dependent manner to regulate COP1-mediated protein degradation
COP1 is part of a multimeric E3 ubiquitin ligase complex that includes CULLIN 4 (CUL4), DAMAGED DNA-BINDING PROTEIN 1 (DDB1), RING-BOX 1 (RBX1), and SUPPRESSOR OF PHYA (SPA) proteins (SPA1–SPA4 in Arabidopsis) (Chen et al. 2010). Consistent with the notion that SPAs and COP1 are part of one multimeric E3 ubiquitin ligase, the quadruple spa mutant displays a phenotype similar to cop1 alleles. Members of this complex repress light responses in both seedlings and mature plants (Laubinger et al. 2004, 2006). COP1 is comprised of an N-terminal RING finger domain, followed by a coiled-coil domain and C-terminal WD40 repeats. SPA proteins also have C-terminal WD40 repeats and a central coiled-coil domain, but possess an N terminus that resembles a protein kinase domain (Fig. 1B,C). COP1 interacts with substrates via its WD40 repeats, whereas the interaction between SPA and COP1 involves the coiled-coil domains of the two proteins (Hoecker and Quail 2001). Importantly, structure–function analyses have identified the coiled-coil domain and WD40 repeats of SPA1 as essential for its activity (Fittinghoff et al. 2006; Yang and Wang 2006).
COP1 acts downstream from multiple plant photoreceptors—such as cryptochromes, phytochromes, and the UV-B receptor—and controls the light-regulated abundance of numerous transcription factors (Yi and Deng 2005; Favory et al. 2009). In most cases, target proteins are degraded in a COP1-dependent manner in the dark. Upon light exposure, these proteins then rapidly accumulate due to photoreceptor-mediated inhibition of COP1 (Yi and Deng 2005). The bZIP transcription factor ELONGATED HYPOCOTYL5 (HY5), which is required for normal light-regulated seedling development under all light conditions, was the first known target of COP1-mediated degradation (Yi and Deng 2005). The mechanism by which phytochromes rapidly inhibit COP1 in response to red or far-red light is still largely unknown. However, in response to UV-B, COP1 interacts in a light-dependent manner with UVR8, a protein that selectively controls plant responses to UV-B exposure. The interaction with UVR8 presumably inhibits COP1-mediated degradation of target proteins and leads to the accumulation of HY5 in the presence of UV-B (Favory et al. 2009). The recently described process by which COP1 activity is controlled by cryptochromes bears some resemblance to the control mechanism identified for COP1 in response to UV-B radiation. However, in blue-light signaling, it is not COP1 but SPA1, another component of the multimeric E3 ligase complex, that exhibits light-mediated interaction with cryptochromes (Lian et al. 2011; Liu et al. 2011; Zuo et al. 2011).
The blue-light-dependent CRY1–SPA1 interaction inhibits COP1–SPA1 binding
Two recent studies (Lian et al. 2011; Liu et al. 2011) have shown that the light-regulated interaction between Arabidopsis CRY1 and SPA1 is mediated by the CCE domain of CRY1 that interacts with the WD40 domain of SPA1 (Fig. 1B). This regulated protein–protein interaction presumably depends on light-induced conformational changes that take place in the CRY1 CCE domain (Partch et al. 2005; Kottke et al. 2006; Zoltowski and Gardner 2011). Consistent with this notion, the light-regulated CRY1–SPA1 interaction was reconstituted in a heterologous system (Lian et al. 2011; Liu et al. 2011). This raised the question of how the CRY1–SPA1 interaction is able to modulate ubiquitin ligase activity of the multimeric COP1 E3 ligase. It was shown previously that SPA1 modulated the ubiquitin ligase activity of COP1 in vitro (Saijo et al. 2003; Seo et al. 2003), and there is genetic evidence that indicates that SPA proteins are required for COP1 function in vivo (Laubinger et al. 2004). Moreover, there exists a strong interaction between SPA1 and COP1 in the dark, as this interaction is inhibited by light (Saijo et al. 2003). Recent studies in the Lin (Liu et al. 2011) and Yang (Lian et al. 2011) laboratories in this issue of Genes & Development show that the light-induced interaction between CRY1 and SPA1 inhibits the interaction between SPA1 and COP1. This network of light-regulated interactions can be reconstituted in yeast using a three-hybrid assay, which indicates that no other plant-specific factors are required for these interactions to take place (Lian et al. 2011; Liu et al. 2011). Furthermore, in cry1 mutants, the accumulation of downstream transcription factors is no longer regulated by blue light (Liu et al. 2011; Zuo et al. 2011). Taken together, these two studies suggest that CRY1-mediated disruption of the SPA1–COP1 interaction inhibits the activity of this multimeric E3 ligase (Fig. 1B).
Blue-light-dependent protein–protein interactions underlie CRY2-mediated flowering
In another study conducted in parallel in the Lin laboratory (Zuo et al. 2011), a variation on the same theme is described for CRY2-regulated gene expression. CRY2, COP1, and SPA1 play a very important role in a later stage of the plant life cycle; namely, the initiation of flowering. The photoperiodic control of plant reproduction requires an integration of information from both the circadian clock and day length (Turck et al. 2008). Central to this process is the B-box-type zinc finger transcription factor CONSTANS (CO). Only when day length is appropriate does CO protein accumulate sufficiently to trigger flowering. Both COP1 and SPA1 are required for degradation of CO under short days, which do not promote flowering in Arabidopsis (Laubinger et al. 2006; Jang et al. 2008; LJ Liu et al. 2008). Under long days, a developmental switch takes place whereby proteolytic activity of the COP1/SPA1 E3 ligase is inhibited by CRY2 (Turck et al. 2008). Until recently, the mechanism through which CRY2 controls E3 ligase activity of the COP1/SPA1 complex had remained unknown. However, two studies now show that, similar to CRY1, CRY2 also interacts with SPA1 in a blue-light-dependent manner (Lian et al. 2011; Zuo et al. 2011). Interestingly, and in contrast to the CRY1/SPA1 mode of action, the light-regulated CRY2–SPA1 interaction does not affect the interaction of COP1 with SPA1, but rather strengthens the CRY2/COP1 complex (Fig. 1C). This enhanced interaction of COP1 with the photoreceptor is thought to suppress proteolytic activity of the COP1/SPA1 complex, thereby allowing accumulation of CO and initiation of flowering under long days (Zuo et al. 2011). In contrast to the light-regulated CRY1–SPA1 interaction described above, formation of the CRY2/SPA1 complex requires both the PHR domain of the photoreceptor and the N terminus of SPA1 (Fig. 1B,C; Zuo et al. 2011). Regardless of the notable differences between CRY1 and CRY2 modes of action, the light-regulated activity of both proteins results in accumulation of a key transcriptional regulator.
It should also be mentioned that CRY2 controls initiation of flowering by an additional mechanism. Light-activated CRY2 specifically interacts with the basic helix–loop–helix (bHLH) class transcription factor CIB1 (CRYPTOCHROME-INTERACTING bHLH) (H Liu et al. 2008). This mechanism seems analogous to the light-dependent interaction between phytochromes and PIFs (PHYTOCHROME-INTERACTING PROTEINS), which are bHLH transcription factors related to CIB1 (H Liu et al. 2008). Interestingly, light-regulated interaction of CRY2 with either SPA1 or CIB1 requires the PHR domain (Kennedy et al. 2010; Zuo et al. 2011), whereas the light-regulated interaction of CRY1 with SPA1 occurs via the CCE domain (Fig. 1B,C; Liu et al. 2011; Zuo et al. 2011). CIB1 and related bHLH transcription factors then induce flowering in a CRY2-dependent, but CO-independent, mechanism (H Liu et al. 2008). However, similar to CO, CIB1 also interacts with the promoter of FLOWERING LOCUS T (FT), inducing gene expression and thus flowering (H Liu et al. 2008; Turck et al. 2008). How the interaction between the CRY2 PHR domain and CIB1 controls CIB1 activity remains unknown at present.
Additional roles of the SPA1–COP1 E3 ligase in light signaling
To better understand how blue light controls plant growth and development, it is necessary to characterize the transcription factors controlled by cryptochromes via suppression of COP1 activity. In addition to HY5, as described above, several other transcription factors, including the bHLH factor HFR1, are required for de-etiolation to take place. Some of these transcription factors are also degraded in a COP1-dependent mechanism in the dark (Henriques et al. 2009). Interestingly, SPA1 also interacts with HFR1, and it has been shown that HFR1 is important for cryptochrome-mediated de-etiolation, suggesting that a similar mode of regulation exists for HFR1 as for HY5 (Henriques et al. 2009). The regulated abundance of multiple transcription factors during de-etiolation may thus contribute to the severe phenotype displayed by cop1 mutants during young seedling development. In addition, given that COP1 controls accumulation of HFR1, which inhibits the activity of the photomorphogenesis-regulating bHLH proteins from the PIF family, COP1-mediated degradation indirectly controls the activity of other transcription factors (Leivar et al. 2008; Hornitschek et al. 2009).
Interestingly, the COP1-associated degradation machinery has also been implicated in feedback mechanisms controlling the level of activated light sensors. During phytochrome signaling, COP1 contributes to the degradation of the light-activated photoreceptor, indicating the presence of a feedback loop (Seo et al. 2004; Jang et al. 2010). In UV-B signaling, a feedback loop has also been described, but operates via a different mechanism. To begin with, two WD40 proteins related to COP1 are transcriptionally induced by UV-B. Once accumulated, these WD40 proteins then interact with UVR8 to negatively regulate its function (Gruber et al. 2010). It is currently unknown whether a related feedback loop mechanism exists in cryptochrome-regulated gene expression. Also, the exact role of COP1 in cryptochrome stability has not been elucidated.
Light controls activity of the SPA/COP1 E3 ligase via regulated protein–protein interactions (Fig. 1B,C). Additional layers of regulation result from the light-controlled abundance of SPA family members and the distinct functions among members of this family, which can all interact with COP1 (Chen et al. 2010). For example, the SPA1 transcript increases rapidly and transiently during de-etiolation (Fittinghoff et al. 2006). In contrast, SPA2, which is required in dark-grown seedlings but is dispensable in the light, is constitutively expressed (Fittinghoff et al. 2006). Moreover, both SPA1 and SPA2 proteins are degraded in a proteasome-dependent manner in response to light, although SPA2 is degraded more rapidly than SPA1 (Balcerowicz et al. 2011). Finally, SPA1 and SPA2 proteins have distinct roles, with SPA2 being inherently incapable of repressing photomorphogenesis in the light (Balcerowicz et al. 2011). This difference is apparently not due to an inability of SPA2 to interact with CRY1, given that all members of the SPA family can interact with the CCE domain of CRY1 (Lian et al. 2011). However, the regulated abundance of SPA family members is predicted to lead to different COP1/SPA complexes that may have distinct functions (Chen et al. 2010).
Animal cryptochromes control degradation of transcriptional regulators
The ability of light acting via the cryptochromes to reset the circadian clock is best understood in Drosophila. As in numerous species, the circadian clock in Drosophila relies on a transcriptional negative feedback loop. CLOCK and CYCLE act as heterodimeric transcription factors to activate expression of the negative elements PERIOD and TIMELESS (Young and Kay 2001). The Drosophila type I cryptochrome dCRY synchronizes the circadian clock through light-regulated protein degradation events that have been elucidated only recently. This regulation mechanism shares astonishing similarities with how plant cryptochromes control transcription factor abundance. dCRY controls light-dependent degradation of TIMELESS in a manner requiring the E3 ubiquitin ligase JETLAG, which will also ultimately lead to dCRY degradation (Koh et al. 2006; Peschel et al. 2009; Ozturk et al. 2011). Both TIMELESS and JETLAG interact with dCRY in a light-dependent manner. The interaction between JETLAG and dCRY is enabled by a light-induced conformational change of the short dCRY CCE domain (Ozturk et al. 2011). Interestingly, a dCRY mutant lacking the CCE domain is constitutively active and interacts with JETLAG independently of light, highlighting the importance of this distinguishing feature of cryptochromes (Busza et al. 2004; Ozturk et al. 2011). Thus, in Drosophila, a light stimulus causes a conformational change in the CCE domain of dCRY. This then leads to an interaction between dCRY and an E3 ubiquitin ligase, which controls the abundance of the transcriptional regulator TIMELESS. The analogy between recently described plant mechanisms and the mode of action for dCRY is noteworthy. In both cases, light induces a conformational change in the CCE domain, which is central for cryptochrome activity, and, also in both cases, this conformational change directly affects downstream degradation of transcription factors. However, in plants, constitutive signaling is conferred by the isolated CCE domain, whereas, in Drosophila, the isolated PHR domain is constitutively active. Furthermore, in Drosophila, dCRY activation leads to degradation of TIMELESS, while, in Arabidopsis, CRY activation leads to stabilization of HY5 and CO.
Concluding remarks
Recent studies showing how plant cryptochromes inactivate the COP1 complex in response to blue light give rise to a number of further questions that need to be addressed: Can a similar molecular mechanism be projected to phytochrome and UV-B receptor pathways? How, exactly, do SPA proteins regulate E3 ubiquitin ligase activity of the COP1 complex? How can a light-induced interaction between SPA1 and the CRY2 PHR be reconciled with the constitutively active form of the CRY2 CCE domain? Finally, although the genetic and molecular experiments presented in the aforementioned studies show that the cryptochromes suppress COP1/SPA-mediated degradation of HY5 and CO, it is surprising to observe that both proteins still accumulate in a blue-light-dependent manner in spa cry mutants (Liu et al. 2011; Zuo et al. 2011). It will be interesting to uncover the mechanism behind this additional layer of light regulation.
Acknowledgments
We apologize to colleagues whose work could not be cited in this brief perspective. We are grateful to Séverine Lorrain, Emilie Demarsy, and Kimberley Tilbrook for their comments on this manuscript. Work in the Fankhauser laboratory is supported by the University of Lausanne, the Swiss National Science Foundation (grant no. 31003A-124747), the NCCR “Plant Survival in Natural and Agricultural Ecosystems,” and the SystemsX.ch RTD “Plant growth in a changing environment.” Work in the Ulm laboratory is supported by the University of Geneva, the Emmy Noether Programme, and the Swiss National Science Foundation (grant no. 31003A-132902).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2053911.
References
- Ahmad M, Cashmore AR 1993. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Balcerowicz M, Fittinghoff K, Wirthmueller L, Maier A, Fackendahl P, Fiene G, Koncz C, Hoecker U 2011. Light exposure of Arabidopsis seedlings causes rapid de-stabilization as well as selective post-translational inactivation of the repressor of photomorphogenesis SPA2. Plant J. 65: 712–723 [DOI] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P 2004. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science 304: 1503–1506 [DOI] [PubMed] [Google Scholar]
- Cashmore AR 2003. Cryptochromes: enabling plants and animals to determine circadian time. Cell 114: 537–543 [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al. 2010. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. 2009. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittinghoff K, Laubinger S, Nixdorf M, Fackendahl P, Baumgardt RL, Batschauer A, Hoecker U 2006. Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J 47: 577–590 [DOI] [PubMed] [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R 2010. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci 107: 20132–20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Jang IC, Chua NH 2009. Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Quail PH 2001. The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 276: 38173–38178 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C 2009. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G 2008. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH 2010. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C 2010. Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL 2010. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 7: 973–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Zheng X, Sehgal A 2006. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312: 1809–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottke T, Batschauer A, Ahmad M, Heberle J 2006. Blue-light-induced changes in Arabidopsis cryptochrome 1 probed by FTIR difference spectroscopy. Biochemistry 45: 2472–2479 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U 2004. The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U 2006. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH 2008. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ 2011. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev (this issue). doi:10.1101/gad.2025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Shalitin D 2003. Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ 2008. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Liu H, Zhong D, Lin C 2010. Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol 13: 578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C 2011. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev (this issue). doi:10.1101/gad.2025011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A 2011. Reaction mechanism of Drosophila cryptochrome. Proc Natl Acad Sci 108: 516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Clarkson MW, Ozgur S, Lee AL, Sancar A 2005. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 44: 3795–3805 [DOI] [PubMed] [Google Scholar]
- Peschel N, Chen KF, Szabo G, Stanewsky R 2009. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol 19: 241–247 [DOI] [PubMed] [Google Scholar]
- Reppert SM, Gegear RJ, Merlin C 2010. Navigational mechanisms of migrating monarch butterflies. Trends Neurosci 33: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW 2003. The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH 2003. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH 2004. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C 2002. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767 [DOI] [PubMed] [Google Scholar]
- Shalitin D, Yu X, Maymon M, Mockler T, Lin C 2003. Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15: 2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G 2008. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW 2001. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wu G, Spalding EP 2007. Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc Natl Acad Sci 104: 18813–18818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang H 2006. The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J 47: 564–576 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR 2000. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR 2001. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Zuo ZC, Zhao XY, Li X, Klejnot J, Li Y, Chen P, Liang SP, Yu XH, Liu XM, et al. 2008. Blue-light-independent activity of Arabidopsis cryptochromes in the regulation of steady-state levels of protein and mRNA expression. Mol Plant 1: 167–177 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW 2005. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA 2001. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715 [DOI] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C 2007a. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19: 3146–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C 2007b. Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci 104: 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski BD, Gardner KH 2011. Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein–protein interactions. Biochemistry 50: 4–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C 2011. Arabidopsis cryptochrome 2 undergoes blue light-dependent interaction with the SPA1–COP1 complex to regulate floral initiation in plants. Curr Biol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]