Abstract
Vertebrate CpG islands (CGIs) are short interspersed DNA sequences that deviate significantly from the average genomic pattern by being GC-rich, CpG-rich, and predominantly nonmethylated. Most, perhaps all, CGIs are sites of transcription initiation, including thousands that are remote from currently annotated promoters. Shared DNA sequence features adapt CGIs for promoter function by destabilizing nucleosomes and attracting proteins that create a transcriptionally permissive chromatin state. Silencing of CGI promoters is achieved through dense CpG methylation or polycomb recruitment, again using their distinctive DNA sequence composition. CGIs are therefore generically equipped to influence local chromatin structure and simplify regulation of gene activity.
Keywords: CpG island, promoter, transcription, DNA methylation, polycomb
Vertebrate genomes are methylated predominantly at the dinucleotide CpG, and consequently are CpG-deficient owing to the mutagenic properties of methylcytosine (Coulondre et al. 1978; Bird 1980). The globally methylated, CpG-poor genomic landscape is punctuated, however, by CpG islands (CGIs), which are, on average, 1000 base pairs (bp) long and show an elevated G+C base composition, little CpG depletion, and frequent absence of DNA methylation. These shared properties have allowed CGIs to be isolated as a relatively homogeneous fraction of the genome, despite the heterogeneity of their individual nucleotide sequences (Bird et al. 1985; Cross et al. 1994; Illingworth et al. 2010). Approximately 70% of annotated gene promoters are associated with a CGI, making this the most common promoter type in the vertebrate genome (Saxonov et al. 2006). Included are virtually all housekeeping genes, as well as a proportion of tissue-specific genes and developmental regulator genes (Larsen et al. 1992; Zhu et al. 2008). Recent work has uncovered a large class of CGIs that are remote from annotated transcription start sites (TSSs), but nevertheless show evidence for promoter function (Illingworth et al. 2010; Maunakea et al. 2010). These findings emphasize the strong correlation between CGIs and transcription initiation.
In spite of their link with transcription, the functional significance of CGIs is only just beginning to emerge. CGI promoters turn out to have distinctive patterns of transcription initiation and chromatin configuration. Their regulation involves proteins (some of which specifically bind nonmethylated CpG) that influence the modification status of CGI chromatin. In addition, the CpG moieties themselves are sometimes subject to cytosine methylation, which correlates with stable shutdown of the associated promoter. Here we examine the properties shared by vertebrate CGIs and how transcription is regulated at these sites. Recent related reviews include Illingworth and Bird (2009), Mohn and Schubeler (2009), and Blackledge and Klose (2011).
Evolutionary conservation of CGIs
CGIs are distinct in vertebrates due to their lack of DNA methylation and absence of CpG deficiency, which sets them apart form bulk genomic DNA. Organisms such as the invertebrates Drosophila melanogaster and Caenorhabditis elegans and the fungus Saccharomyces cereviceae have little or no DNA methylation and, as a result, CpG occurs at the expected frequency throughout the genome. CGIs are not detectable in these genomes because, in a sense, the whole genome is CGI-like. On the other hand, many plant genomes are very highly methylated, and early research detected a nonmethylated CGI-like genomic fraction (Antequera and Bird 1988). Lack of DNA methylation in plants is evidently linked to genes, as isolation of nonmethylated DNA by “methylation filtration” greatly enriches for transcribed sequences (Palmer et al. 2003), despite the presence of gene body methylation in these organisms (Zhang et al. 2006; Zilberman et al. 2007). Extensive DNA methylome analysis has documented nonmethylated regions at both extremities of plant transcription units (Feng et al. 2010; Zemach et al. 2010). Whether these can be considered equivalent to or distinct from vertebrate CGIs is not presently known.
Unlike vertebrates, most invertebrate animals exhibit mosaically methylated genomes comprising alternating methylated and nonmethylated domains (Bird et al. 1979; Tweedie et al. 1997). The persistence over evolutionary time of discrete genomic regions with different DNA methylation states has partitioned DNA sequences into two fractions: (1) methylated and, consequently, CpG-deficient; and (2) nonmethylated, with the expected frequency of CpG. The origin of vertebrates appears to have coincided with a transition from mosaic to global DNA methylation, accompanied by concomitant CpG depletion throughout most of the genome (Tweedie et al. 1997). Well-studied among the mosaic genomes is the invertebrate Ciona intestinalis (sea squirt), which is evolutionarily close to the invertebrate–vertebrate boundary. C. intestinalis genes within methylated domains are sometimes associated with short nonmethylated CGI-like regions that colocalize with TSSs (Suzuki et al. 2007). CGIs may therefore predate the evolution of vertebrates.
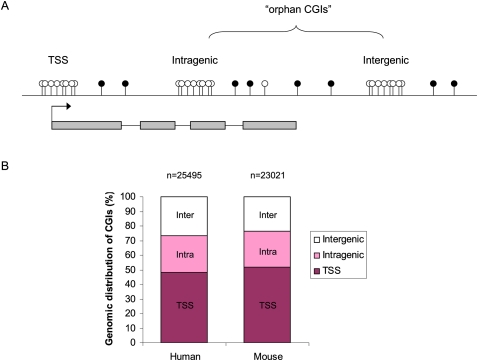
Until recently, it was not clear that CGIs were conserved in either number or genomic location between different vertebrates. Initially, far fewer CGIs were bioinformatically predicted in the mouse genome than in the human genome (Waterston et al. 2002), and this apparent lack of conservation called into question their regulatory importance. CGI prediction algorithms by necessity employ thresholds for detection, alteration of which dramatically changes the number predicted (Illingworth et al. 2008; Illingworth and Bird 2009). The algorithms are also unable to take into account the methylation status of CGIs. A biochemical approach has shed fresh light on the issue, using affinity purification with the CXXC protein domain to isolate clusters of unmethylated CpGs from genomic DNA (Illingworth et al. 2008). High-throughput DNA sequencing of this fraction identified a comprehensive CGI complement from both humans and mice (Illingworth et al. 2010) and revealed similar numbers of CGIs per haploid genome: 25,495 and 23,021, respectively. The reason for the initial discrepancy is that mouse CGIs show, on average, a slightly lower CpG content compared with human CGIs. Biochemical purification of clusters of nonmethylated CpG overcomes this weakness. Also conserved between humans and mice was the proportion of CGIs associating with annotated TSSs (∼50%). Moreover, the remaining half of CGIs were distributed equally between locations within gene bodies (intragenic) or between genes (intergenic) in both species (Fig. 1). The genomic position of many of these additional CGIs appears to have been maintained since the divergence of humans and mice ∼75 million years ago, implying functional importance.
Figure 1.
The genomic distribution of CGIs. (A) CGIs can be located at annotated TSSs, within gene bodies (Intragenic), or between annotated genes (Intergenic). Intragenic and intergenic CGIs of unknown function are classed as “orphan” CGIs. (Empty circles) Unmethylated CpG residues. (Filled circles) Methylated CpG residues. (B) The genomic distribution of CGIs in the human and mouse genome as determined by Illingworth and colleagues (2010). The total number of CGIs is given at the top of each graph.
CGIs are sites of transcriptional initiation
About half of all CGIs self-evidently contain TSSs, as they coincide with promoters of annotated genes. The other half are either within or between characterized transcription units and have been termed “orphan” CGIs to reflect uncertainty over their significance (Illingworth et al. 2010). Do orphan CGIs weaken the correlation between CGIs and transcriptional activity, or do they also mark hitherto unsuspected promoters? The available evidence shows that many orphan CGIs are also sites of transcriptional initiation. Specific examples of intragenic CGI promoters have been known for many years. For example, CGIs at the 3′ end of the Pomc gene and exon 2 of the MHC class II I-Aβ gene both initiate transcripts of unknown function whose coding potential is minimal (Gardiner-Garden and Frommer 1994; Macleod et al. 1998). More is known functionally about the role played by the Air transcript in imprinting of the Igf2r gene. Air is a noncoding RNA (ncRNA) that initiates at a CGI within intron 2 of Igf2r and is essential for silencing of the paternal allele (Sleutels et al. 2002). Similarly, analysis of a CGI in intron 10 of the imprinted Kcnq1 gene identified it as the origin of a noncoding transcript (Kcnq1ot1) that is required for imprinting of several genes within this domain (Mancini-DiNardo et al. 2003, 2006). In these and other cases, the presence of a CGI in an unexpected location stimulated a successful search for an associated transcript.
Genome-wide analyses have confirmed that many orphan CGIs represent novel promoters (Illingworth et al. 2010; Maunakea et al. 2010). Criteria for transcript annotation include colocalization of a CGI with bound RNA Polymerase II (RNAPII) as detected by chromatin immunoprecipitation (ChIP) or, more tellingly, RNA sequence data showing that transcripts originate within orphan CGIs (Fig. 2; Illingworth et al. 2010; Maunakea et al. 2010). Examples of the latter data include cap analysis of gene expression (CAGE), which uses the 5′ cap to isolate and sequence full-length transcripts (Shiraki et al. 2003), and global run-on sequencing (GRO-seq) (Core et al. 2008), which detects in vitro elongation of engaged RNAPII. Altogether, evidence for transcriptional initiation has been found at ∼40% of orphan CGIs (∼5000) (Illingworth et al. 2010). Less directly, CGIs are frequently marked by trimethylation of histone H3 (H3K4me3), which is a signature of active promoters (see Fig. 2 and below). A subset of intergenic H3K4me3 peaks (∼1600), many of which are likely to correspond to orphan CGIs, were found to represent TSSs for long ncRNAs (Guttman et al. 2009).
Figure 2.
Orphan CGIs are sites of transcriptional initiation. High-throughput sequencing data showing colocalization of orphan CGIs with sites of transcriptional initiation taken from Illingworth et al. (2010). CXXC affinity purification identifies the locations of CGIs that overlap with H3K4me3, RNAPII, GRO-seq (Core et al. 2008), and CAGE tags (Faulkner et al. 2009). Genes (RefSeq) are annotated below the sequencing profiles, with those mapped to the positive and negative strand displayed above and below the chromosome line, respectively. Orphan CGIs are denoted by asterisks.
Many orphan CGI promoters are active in a tissue-specific manner (Illingworth et al. 2010), suggesting that they are tightly regulated. Because only a few tissues have been investigated so far, it is likely that most, if not all, orphan CGIs will be associated with a novel transcript. What is the functional significance of this transcription? Some orphan CGIs probably represent alternative promoters of nearby annotated genes (Maunakea et al. 2010). Others may initiate ncRNAs that regulate gene expression. For example, the study of imprinted genes has already uncovered key examples of the regulatory relevance of CGI-derived ncRNAs (see above). In addition to Air and Kcnq1ot1, the Xist and Tsix ncRNAs function in X-chromosome inactivation (Herzing et al. 1997; Lee et al. 1999), and the ncRNA HOTAIR has been reported to regulate Hox gene expression (Rinn et al. 2007). Given the hitherto unsuspected abundance of CGI promoters in the genome, it seems likely that involvement of ncRNAs in gene regulation may be widespread. Recent studies have shown that enhancers are often associated with transcription of ncRNAs, although these have not been linked so far to CGIs (Kim et al. 2010). In a different vein, CGI promoters within MHC class II genes coincide precisely with the hypervariable exon 2, whose polymorphism is caused by gene conversion between members of the class II gene family. It has been suggested that the “open” CGI chromatin structure in germ cells enhances gene conversion in this exon, thereby benefiting the immune system (Macleod et al. 1998). Potential roles as boundaries or insulators of transcriptional units remain as yet unexplored. In summary, the biological roles played by CGI transcripts may be diverse, but the stage is now set for rigorous testing of hypotheses concerning their function.
It is clear from the above discussion that CGIs act as promoters in mammalian genomes. This leaves two possibilities regarding their significance: (1) CGIs are evolutionary footprints of molecular events that occur at many eukaryotic promoters, but are only visible in organisms that have extensive genomic DNA methylation; or (2) CGIs are important regulatory structures that have evolved under selection in genomes where DNA methylation plays a regulatory role. The weight of evidence now appears to support the second of these possibilities. In what follows, we summarize this emerging data and discuss ways in which CGIs are adapted for promoter function.
Characteristics of CGI promoters
DNA sequence motifs
CGIs colocalize with the majority of promoters in both the human and mouse genomes. Early studies suggested that CGI promoters may often lack TATA boxes and display heterogeneous TSSs (Reynolds et al. 1984). CAGE analysis has shown, on a genome-wide scale, the broad correlation between this distributive pattern of transcription initiation, typically over a region of 50–100 bp, and the presence of CGIs (Carninci et al. 2006). These observations are compatible with the idea that CGI promoters adopt a transcriptionally permissive state within which initiation can occur at a number of locations. In general, TATA boxes, along with other core promoter elements (such as the BRE, DPE, and DCE), tend to be associated with focused transcriptional initiation, whereas CGIs tend to lack these elements and display dispersed initiation patterns (for review, see Juven-Gershon et al. 2008). There are, however, exceptions to this generalization. The human genes for α-globin, MyoD1, and erythropoietin, for example, have CGI promoters, yet possess TATA boxes.
The idea that many CGI promoters are transcriptionally permissive is supported by genome-wide ChIP and transcriptome analysis. RNAPII is bound at the CGI promoters of many inactive genes in embryonic stem (ES) cells (Guenther et al. 2007) and at silent lipopolysaccharide (LPS)-inducible genes in unstimulated macrophages (Hargreaves et al. 2009). Also, global nuclear run-on analysis examining the products of transcriptionally engaged RNAPII molecules detected bidirectional transcription of short nonproductive RNAs as well as full-length transcripts at many CGI promoters (Core et al. 2008; Seila et al. 2008). Much emphasis had been placed previously on the recruitment of RNAPII as the rate-limiting step in transcription, but these results suggest that regulation of many CGI promoters takes place downstream from polymerase binding. One way of achieving this is by regulating transcriptional elongation and mRNA processing, as in the case of CGI-associated inducible genes in macrophages (Hargreaves et al. 2009). In the latter study, RNAPII recruitment was dependent on the transcription factor Sp1, but the initiating form of RNAPII (phosphorylated at Ser 5 of the C-terminal domain [CTD]) was promoter-bound even when the genes were inactive and low levels of full-length transcripts were produced (Hargreaves et al. 2009). A switch from nonproductive to productive transcription was triggered by inducible transcription factor-dependent recruitment of P-TEFb and subsequent Ser 2 phosphorylation of the RNAPII CTD. This resulted in the production of mature, processed transcripts. Similarly, gene regulation at the level of transcriptional elongation via RNAPII pausing (controlled by the pause factors DSIF and NELF) and release of RNAPII mediated by P-TEFb (for review, see Peterlin and Price 2006) is reported to be widespread in ES cells (Rahl et al. 2010). CGI promoters, it seems, attract RNAPII and, unless actively restrained, will engage in transcription of some kind.
Transcription factor binding at CGIs
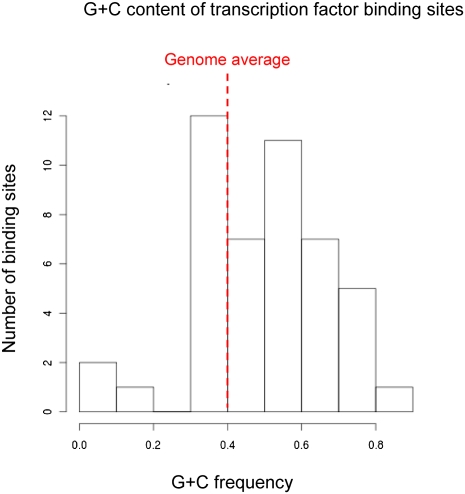
CGIs share little long-range sequence conservation, apart from an elevated CpG density and G+C content, and often lack core promoter elements such as the TATA box (Carninci et al. 2006; Juven-Gershon et al. 2008). What features adapt them to promoter function? A simplistic possibility is that GC richness increases the probability that ubiquitous transcription factors will bind. In general, mammalian transcription factor-binding sites are more GC-rich than the bulk genome (see Fig. 3) and many contain CpG in their recognition sequence. These include the general transcription factor Sp1, which has been shown to recruit TATA-binding protein (TBP) to promoters lacking a TATA box (Butler and Kadonaga 2002). Transient reporter gene assays examining the activity of 4575 human promoters found that ubiquitously active CGI promoters tended to be enriched for Sp1, Nrf-1, E2F, and ETS transcription factor-binding motifs, each of which contains a CpG (Landolin et al. 2010). Consistent with this, potential ETS and E2F family binding sites, as characterized in vitro, are overrepresented in mouse CGIs (Jaeger et al. 2010). Similarly, CpG-containing ETS, NRF-1, BoxA, SP1, CRE, and E-box motifs are enriched in the CGI promoters of housekeeping genes (Rozenberg et al. 2008), and Sp1 binding on human chromosomes 21 and 22 is focused predominantly at CGIs (Cawley et al. 2004). In the case of the α-globin gene, DNA footprinting studies detected little difference in transcription factor-binding patterns whether the gene was active or inactive (Cuadrado et al. 2001), suggesting that, even in the case of a highly tissue-specific gene, transcription factor binding can be constitutive.
Figure 3.
Transcription factor-binding sites are, on average, GC-rich. The G+C content of binding sites for 46 mouse transcription factors was calculated using fasta sequences of actual binding sites obtained from the JASPAR database (Bryne et al. 2008). G+C frequency is given on the X-axis, while the Y-axis shows the number of each of these 46 binding sites with a given G+C frequency.
The chromatin signature of CGIs
Unstable nucleosomes
There is evidence that nonmethylated CGIs are organized in a characteristic chromatin structure that predisposes them toward promoter activity. A study of chromatin at LPS-inducible genes in macrophages found that CGIs are relatively nucleosome-deficient (Ramirez-Carrozzi et al. 2009). Inducible “primary response genes” fall into two classes: those that require SWI/SNF chromatin remodeling complexes for their activation and those that do not. It was noticed that these groups corresponded with non-CGI and CGI promoters, respectively, suggesting that DNA in CGI chromatin is intrinsically accessible without the need for ATP-dependent nucleosome displacement (Ramirez-Carrozzi et al. 2009). In macrophages, the CGIs showed a reduced density of histone H3 even in the uninduced state. Accordingly, in vitro nucleosome assembly indicated that a set of these CGIs is significantly more reluctant to assemble into nucleosomes than other genomic DNA (Ramirez-Carrozzi et al. 2009). An attractive interpretation of the in vitro instability of CGI chromatin is that weakening of this barrier allows greater accessibility of the underlying DNA to transcriptional regulators in vivo. Other evidence has shown that nucleosome deficiency is a feature of CGI promoters in general. Early analysis of CGI chromatin detected abundant nonnucleosomal DNA that was absent in preparations of bulk chromatin (Tazi and Bird 1990). The same conclusion emerged from a re-examination of genome-wide nucleosome mapping data (Schones et al. 2008; Choi 2010). In addition to chromatin instability, nucleosome deficiency in vivo may also arise because CGI promoters, in common with all eukaryotic promoters, typically possess a nucleosome-free region surrounding the TSS (Schones et al. 2008). In other words, active promoters by definition may be nucleosome-deficient whether or not they are CGIs. It is not yet certain whether nucleosome deficiency at CGIs is due primarily to intrinsic chromatin instability or nucleosome exclusion due to the presence of the transcription initiation complex. It could, of course, be a mixture of both, and may even vary between individual CGIs.
Characteristic histone modifications
Early biochemical studies of isolated CGI chromatin showed high levels of histone H3 and H4 acetylation, which are characteristic of transcriptionally active chromatin. Histone H1, on the other hand, which is regarded as antagonistic to transcription, was depleted in this fraction (Tazi and Bird 1990). Genome-wide studies have confirmed this association at high resolution (Birney et al. 2007; Wang et al. 2008) and have revealed that H3K4me3 is a signature histone mark of CGI promoters, often persisting even when the associated gene is inactive (Guenther et al. 2007; Mikkelsen et al. 2007). Recent work has established a biochemical connection between the abundance of CpG in CGIs and H3K4me3, mediated by a CXXC domain protein that binds specifically to nonmethylated CpG (Voo et al. 2000). Cfp1 (CXXC finger protein 1; also known as CGBP) is an integral component of the Setd1 H3K4 methyltransferase complex (Lee and Skalnik 2005) and localizes to the vast majority of CGIs in the mouse genome, suggesting dependence of this histone modification on the DNA sequence (Thomson et al. 2010). In keeping with this model, depletion of Cfp1 reduces H3K4me3 at many CGIs. Importantly, insertion of an artificial CGI-like DNA sequence into the genome results in recruitment of Cfp1 and creates a novel peak of H3K4me3 in the absence of RNAPII (Thomson et al. 2010). Further support for a mechanistic link between unmethylated CpG residues, Cfp1, and H3K4me3 comes from the finding that CpG density in CGIs correlates positively with H3K4me3 levels (Illingworth et al. 2010). The ability of CpG density alone to directly influence the chromatin modification state (Thomson et al. 2010) is likely to be a key function of CGIs.
The presence of H3K4me3 appears to facilitate transcription in a number of ways. The H3K4me3 tail has been shown to interact with the NuRF chromatin remodeling complex (Li et al. 2006; Wysocka et al. 2006; Ruthenburg et al. 2007) as well as ING4-containing histone acetyltransferase complexes (Saksouk et al. 2009). Also, H3K4me3 interacts with the transcriptional machinery directly, as the core transcription factor TFIID has an affinity for the H3K4me3 mark (Vermeulen et al. 2007; van Ingen et al. 2008). Core transcriptional machinery can recruit H3K4 methyltransferases to chromatin (for review, see Ruthenburg et al. 2007), so it is likely that transcription also contributes to H3K4me3 at CGIs. The relative contributions of CpG-mediated and transcription-mediated H3K4me3 to CGI chromatin modification have yet to be determined.
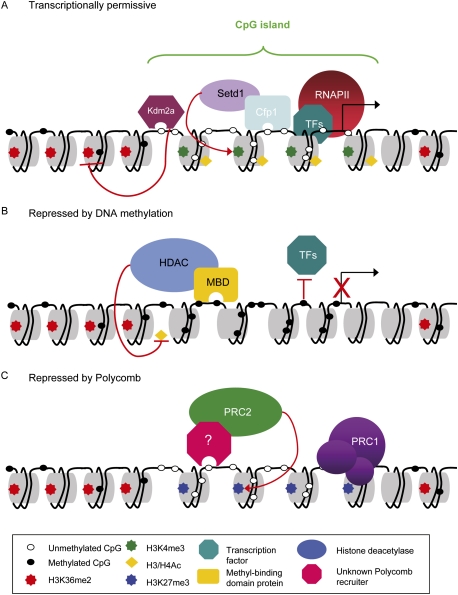
Another distinctive feature of CGI chromatin is depletion of histone H3K36 dimethylation (H3K36me2) compared with non-CGI promoters and gene bodies (Blackledge et al. 2010). The H3K36me2 histone demethylase Kdm2a (Tsukada et al. 2006) is a CXXC domain protein that, like Cfp1, binds specifically to nonmethylated CpG. Accordingly, Kdm2a is bound in vivo at ∼90% of CGIs in mouse ES cells and mediates demethylation of H3K36me2 at these regions (Blackledge et al. 2010). Why H3K36me2 should be depleted at CGIs is uncertain, but this modification has been reported to inhibit transcriptional initiation through histone deacetylase (HDAC) recruitment in yeast (Strahl et al. 2002; Youdell et al. 2008; Li et al. 2009). H3K36me2 depletion may therefore contribute to a transcriptionally permissive state at CGIs (Fig. 4A). Both Cfp1 and Kdm2a have the characteristics of CGI-specific proteins that use CpG density to influence chromatin modification. It is likely that more factors of this kind are yet to be identified, for example, by a comprehensive characterization of the CGI proteome.
Figure 4.
The chromatin state at CGIs. (A) CGIs usually exist in an unmethylated transcriptionally permissive state. They are marked by histone acetylation (H3/H4Ac) and H3K4me3, which is directed by Cfp1, and show Kdm2a-dependent H3K36me2 depletion. Nucleosome deficiency and constitutive binding of RNAPII may also contribute to this transcriptionally permissive state. (B) DNA methylation is associated with stable long-term silencing of CGI promoters. This can be mediated by MBD proteins, which recruit corepressor complexes associated with HDAC activity, or may be due to directed inhibition of transcription factor binding by DNA methylation. (C) CGIs can also be silenced by PcG proteins and may be key elements involved in polycomb recruitment. An unknown CGI-binding factor could be responsible for recruiting PRC2 to CGIs that then trimethylates H3K27. This H3K27me3 is recognized by PRC1 complexes that act to impede transcriptional elongation, thereby silencing genes. Note that the transcriptionally permissive and polycomb-repressed states can coexist at bivalent CGIs, predominantly in totipotent embryonic cells.
CGI promoter silencing by DNA methylation
CGIs are typically in a nonmethylated state in an otherwise heavily methylated genome, even when the corresponding gene is transcriptionally inactive. There are, however, well-known examples of CGIs that become methylated during normal development, leading to stable silencing of the associated promoter (Stein et al. 1982; Mohn et al. 2008; Payer and Lee 2008). Silencing is thought to be either due to direct inhibition of transcription factor binding by DNA methylation or mediated by methyl-binding domain (MBD) proteins that recruit chromatin-modifying activities to methylated DNA (see Fig. 4B; for reviews, see Klose and Bird 2006; Bogdanovic and Veenstra 2009). It seems that CGI methylation is not the initiating event in gene silencing, but acts to lock in the silent state. For example, during X-chromosome inactivation in female eutherian mammals, X-linked CGIs do not become methylated until after gene silencing and the acquisition of several silencing chromatin modifications, such as H3K27me3 (for reviews, see Payer and Lee 2008; Okamoto and Heard 2009). CGI methylation is, however, essential for maintenance of leak-proof X-chromosome inactivation, as inhibition of DNA methylation leads to gene reactivation in a fraction of cells (Sado et al. 2000; Csankovszki et al. 2001). As discussed above, CGI methylation also has well-characterized roles in genomic imprinting where parent-of-origin monoallelic expression is controlled by CGI methylation marks (for review, see Edwards and Ferguson-Smith 2007). In several instances, the CGIs concerned act as promoters for ncRNAs whose expression is silenced by DNA methylation. Expression of these ncRNAs—including Air and Kcnq1ot1—is responsible for the silencing of neighboring genes (Sleutels et al. 2002; Mancini-DiNardo et al. 2003, 2006).
Until recently, methylation of CGIs during imprinting and X-chromosome inactivation were suspected to be special cases, most CGIs remaining nonmethylated regardless of gene expression. However, genome-wide studies focusing on CGIs at annotated TSSs have uncovered numerous instances of CGI methylation in normal somatic cells. CGIs in the germline are almost invariably nonmethylated, but a small proportion acquire methylation in somatic tissues (Schilling and Rehli 2007; Shen et al. 2007; Weber et al. 2007). Similarly, a small number of CGI promoters acquire methylation during differentiation of ES cells into neurons, with most of the changes taking place during the early stages of differentiation (Mohn et al. 2008). The majority of CGIs that gain methylation during differentiation are already silent in ES cells (Mohn et al. 2008), providing further evidence that silencing precedes DNA methylation. The genes affected are often expressed only in the germline, such as the MAGE family of testis-specific antigens (De Smet et al. 1999). Differences in gene-associated CGI methylation between one somatic tissue and another have also been reported, although these are relatively rare compared with differences between germline and somatic CGIs (Meissner et al. 2008).
In contrast to the rarity of methylated CGIs at the promoters of annotated genes, orphan CGIs are methylated much more frequently. About 17% of orphan CGIs have been found so far in a methylated state, compared with ∼3% of CGIs at annotated gene promoters (Illingworth et al. 2008, 2010; Rauch et al. 2009; Maunakea et al. 2010). By further separating orphan CGIs into intragenic and intergenic categories, it becomes apparent that intragenic CGIs are especially prone to methylation (∼20%–34%) (Illingworth et al. 2010; Maunakea et al. 2010). Accordingly, CGIs located within gene bodies show the greatest number of DNA methylation differences between different somatic cells and tissues (Maunakea et al. 2010; AM Deaton, S Webb, ARW Kerr, RS Illington, J Guy, R Andrews, and A Bird, in prep.). Functionally, it can be speculated that some of the transcripts initiating from gene body CGIs are regulatory ncRNAs whose presence or absence affects expression of the associated protein-coding gene or a nearby gene (Dinger et al. 2008; Mercer et al. 2009; Orom et al. 2010). Another possibility is that these sites of unusual chromatin and transcription affect alternative splicing of the gene in which they are located in a manner that differs with methylation status (Kornblihtt 2006). It is also possible that a methylated CGI within a gene body down-regulates transcriptional elongation, as reported in a transgenic cell model (Lorincz et al. 2004). Further studies are required to elucidate the consequences of methylation at these sites.
In addition to its occurrence during normal development, CGI methylation has been well documented in cancer. The CGIs of several tumor suppressor genes acquire cancer-specific methylation, and many genes involved in familial forms of cancer undergo DNA methylation-associated silencing in sporadic cancers (for review, see Jones and Baylin 2002, 2007). These changes are thought to contribute to uncontrolled proliferation and thus tumor development. However, whether DNA methylation is the initiating event in gene silencing or is acquired at already silenced genes remains unclear. Genome-wide studies have demonstrated that many of the CGIs that acquire aberrant methylation in cancer are not associated with tumor suppressor genes (Weber et al. 2005; Keshet et al. 2006; Illingworth et al. 2010; Ruike et al. 2010). A key issue regarding cancer-specific CGI methylation is whether it mirrors normal developmental methylation. Profiling DNA methylation in the flanks of CGIs suggested that cancer-specific methylation patterns resemble those occurring in normal tissues (Irizarry et al. 2009). A recent study, however, suggested that cancer-specific CGI methylation can be distinguished from that in normal tissues. Unlike normal tissues, where orphan CGIs are preferentially methylated, in colorectal cancers, the proportion of TSSs, intragenic CGIs, and intergenic CGIs acquiring tumor-specific methylation was approximately equal (Illingworth et al. 2010).
CGIs and polycomb-mediated silencing
In addition to DNA methylation, CGI promoters can be silenced by polycomb group proteins (PcG). There are two PcG complexes in mammals: polycomb-repressive complex 1 (PRC1) and PRC2. PRC2 mediates H3K27me3, and this mark is recognized by PRC1, which is thought to inhibit transcriptional elongation by a mechanism involving H2A ubiquitylation (Stock et al. 2007; Zhou et al. 2008) and/or chromatin compaction (Eskeland et al. 2010). Differentiation of ES cells into neurons is accompanied by losses and gains of H3K27me3 at many promoters at various stages of differentiation. In contrast, DNA methylation is gained at a relatively small number of promoters only during the early stages of differentiation, remaining relatively stable thereafter (Mohn et al. 2008). This indicates that polycomb is a more dynamic repression system than DNA methylation, at least during later developmental stages. In ES cells, CGIs silenced by polycomb possess the “active” mark H3K4me3 as well as H3K27me3 (Azuara et al. 2006; Bernstein et al. 2006; Mikkelsen et al. 2007). These “bivalent” CGI promoters are poised between two alternative states: either active transcription or stable repression. Upon differentiation, they can lose H3K27me3 and become active or be subject to more stable transcriptional repression. Bivalent CGI promoters account for approximately one-fifth of CGI promoters in ES cells (Ku et al. 2008) but are also found in other cell types, although to a lesser extent (Roh et al. 2006; Mikkelsen et al. 2007).
Acquisition of CGI methylation in cancer has been found to occur preferentially at genes marked by H3K27me3 in ES cells (Ohm et al. 2007; Widschwendter et al. 2007; Illingworth et al. 2010). By mirroring the pattern of polycomb-dependent gene silencing established in ES cells, DNA methylation may hypothetically facilitate maintenance of a “pseudo-pluripotent state,” thereby favoring unrestrained proliferation of cancer cells (Keshet et al. 2006; Ohm et al. 2007; Schlesinger et al. 2007). It has also been found that promoters that are positive for H3K27me3 in ES cells are more than four times more likely to acquire DNA methylation, supporting the idea that there is a relationship of some sort between the two systems (Mohn et al. 2008). One report claimed that the PcG protein Ezh2, which catalyzes H3K27me3, can recruit DNA methyltransferases directly (Vire et al. 2006), but this mechanistic link remains unproven. In many respects, DNA methylation and polycomb behave as alternative silencing mechanisms in ES cells (Fouse et al. 2008; Mohn et al. 2008).
Interestingly, polycomb tends to be targeted to CGI-containing regions of the genome (Mikkelsen et al. 2007; Ku et al. 2008), whereas non-CGI promoters do not often associate with H3K27me3 (Mikkelsen et al. 2007; Mohn et al. 2008). The observation that some CGIs are specifically targeted by polycomb led to the hypothesis that these DNA sequences are prone to polycomb recruitment (Fig. 4C). Although H3K27me3 often tends to mark large genomic domains, binding of Ezh2 (the catalytic component of PRC2) in human ES cells strongly correlates with CGIs and, in particular, those lacking motifs for activating transcription factors (such as Ets1 and 2, NFY, YY1, and c-Myc) (Ku et al. 2008). Intriguingly, a polycomb recruitment element identified in human cells is a nucleosome-deficient sequence that includes a CGI (Woo et al. 2010). A recent study provided functional evidence of a role for CGIs in polycomb recruitment. The CGI of the bivalent Zfpm2 locus was found to be sufficient to recruit H3K27me3, H3K4me3, and PRC2 to a human gene desert region inserted into mouse ES cells (Mendenhall et al. 2010). Interestingly, GC-rich DNA from a bacterial source also had the ability to recruit PRC2, suggesting that base composition plays an important role.
As the majority of CGIs do not normally recruit polycomb, base composition per se is very unlikely to be the only factor involved in attracting or excluding polycomb. To explain this selectivity, it was proposed that the presence of transcription factor-binding motifs within a CGI might be sufficient to protect it from PRC2 recruitment (Ku et al. 2008). Consistent with this hypothesis, deletion of activating motifs at a CGI promoter that normally lacks H3K27me3 in ES cells resulted in PRC2 recruitment and modification of H3K27 (Mendenhall et al. 2010). This suggests that transcription factor binding or productive transcription may be sufficient to protect against silencing by polycomb. A corollary of this hypothesis is that polycomb-mediated silencing, like DNA methylation, is secondary to gene silencing by other mechanisms. This fits with early work on Drosophila showing that polycomb group proteins do not affect the establishment of developmental gene expression patterns, but are essential to maintain those patterns through time. Indeed, recent experiments in Drosophila indicate that polycomb is preferentially targeted to stalled promoters of coding and noncoding transcripts (Beisel and Paro 2011; Enderle et al. 2011) or to forms of RNA polymerase that are not competent to couple RNA synthesis with cotranscriptional modification (Brookes and Pombo 2009). Transcriptionally silent CGIs may not always be sufficient to invite polycomb, however, as PRC2-recruitment is proposed to depend on transcription of ncRNAs from CGIs (Rinn et al. 2007; Zhao et al. 2008). Short transcripts produced from bivalent CGIs in ES and T cells are reported to form stem–loop structures that may be involved in recruiting polycomb to these CGIs (Kanhere et al. 2010). In addition, the JmjC domain-containing protein Jarid2 has been identified as a novel component of PRC2 and has been implicated in polycomb targeting in mammals (for review, see Herz and Shilatifard 2010). We have clues about the mechanisms behind polycomb recruitment to specific sites in the mammalian genome and the role played by CGIs in this process, but much remains to be learned.
What protects CGIs from DNA methylation?
CGIs are generally unmethylated CpG-enriched domains that occur against a backdrop of genome-wide DNA methylation and consequent CpG depletion. How are they protected from the waves of de novo DNA methylation that occur during development? A simple explanation would be that CGIs are intrinsically refractory to the action of DNA methyltransferases, but this is very unlikely. Some CGIs do become methylated during normal development; for example, hundreds of CGIs are heavily methylated on one female X chromosome, but are nonmethylated on the other. An alternative possibility is that methylation at CGIs is actively removed by a DNA demethylase (Wu and Zhang 2010), but uncertainty concerning the identity of the DNA demethylating activities in animals has made this hypothesis difficult to test. The discovery that 5-methylcytosine can be converted to 5-hydroxymethylcytosine (hmC) poses the tantalizing possibility that hmC is an intermediate in the demethylation process (Ito et al. 2010; Wu and Zhang 2010). The demethylating enzyme Tet1 possesses a domain related to the CpG-binding CXXC domain found in Cfp1 (Tahiliani et al. 2009), and could in theory be targeted to CGIs. Compatible with this hypothesis, recent studies locate Tet1 preferentially at CGIs in mouse ES cells (Ficz et al. 2011; Wu et al. 2011a,b), and depletion of Tet1 results in increased CpG methylation at CGIs (Wu et al. 2011a). An attractive scenario is that CGIs are subject to sporadic de novo methylation, but are continually swept clean by a mechanism involving oxidation of 5-methylcytosine. Defects in such a system may predispose to de novo CGI methylation, as seen in many cancers. Indeed, mutations in the TET2 gene are frequent in leukemias and compromise the hydroxylation reaction (Ko et al. 2010).
Regardless of the detailed molecular mechanism, there is evidence that the methylation-free state of CGIs is causally related to their function as promoters. Deletion or mutation of Sp1 transcription factor-binding sites in the mouse Aprt promoter results in a failure to maintain the unmethylated state of the Aprt CGI in transgenic experiments (Brandeis et al. 1994; Macleod et al. 1994). This suggests that the binding of transcription factors, or the act of transcription itself, during early development is required for establishment of the DNA methylation-free state. A prediction of this hypothesis is that all CGI promoters should be active during the waves of de novo methylation that occur at the blastocyst stage and in developing germ cells of the embryo (Morgan et al. 2005). Indeed, studies of a small number of CGI promoters for highly tissue-specific CGI-associated genes showed that several are expressed in early embryonic cells (Daniels et al. 1997). Large-scale analyses have strengthened this relationship, showing that 90% of genes with CGI promoters are expressed in the early embryo or testis (Sequeira-Mendes et al. 2009). The idea that transcription is antagonistic to CGI methylation also fits well with the observation that the presence of RNAPII at CGIs, irrespective of gene activity, is associated with resistance to DNA methylation in cancer (Takeshima et al. 2009). DNA sequence-specific transcription factors also have been implicated in preventing DNA methylation, as cooperative binding of the transcription factors Sp1, Nrf-1, and YY1 in normal monocytes correlates with protection from CGI methylation in leukemia cells (Gebhard et al. 2010). In this case, CGI promoters bound by these factors also had the highest expression levels. A survey of human CGIs reinforced the correlation with transcription factor binding (Straussman et al. 2009). Subsets of CGIs that become methylated or remain nonmethylated during development were screened for DNA sequence motifs that might influence this decision. An algorithm based on these motifs, several of which were transcription factor-binding sites, was able to predict which CGIs would be immune from DNA methylation, again suggesting that the switch is dependent on cis-acting factors.
There is evidently a close relationship between transcription in the early embryo and lack of CGI methylation, but mechanisms that relate the two are unknown. It has been proposed that origins of DNA replication (ORIs) are the missing link. Based on evidence that CGIs often colocalize with ORIs (Delgado et al. 1998), it was speculated that intermediates in the process of replication initiation lead to local exclusion of DNA methylation and, over time, an altered base composition (Antequera and Bird 1999). A large fraction of CGIs (83%) colocalizes with ORIs in ES cells (Sequeira-Mendes et al. 2009), which may reflect the situation in the early embryo. A causal relationship between ORI function and CGIs has yet to be established, however, and therefore other kinds of DNA-based metabolism might be responsible for excluding DNA methylation from these regions. For example, CGI promoters are typically loaded with polymerases that create short abortive transcripts (Core et al. 2008) even when the associated gene is inactive (Kanhere et al. 2010). This “futile” transcription cycle may somehow protect CGIs from the action of DNA methyltransferases, allowing these “silent” promoters to exclude DNA methylation.
Another possible explanation for the immunity of most CGIs to DNA methylation is that their signature chromatin mark, H3K4me3, interferes with DNA methyltransferase activity. Chromatin binding of Dnmt3L, a partner protein of the de novo DNA methyltransferases Dnmt3a and Dnmt3b, is inhibited by H3K4me3 (Ooi et al. 2007). Importantly, the ADD domains of both Dnmt3a and Dnmt3b also fail to interact with H3K4me3 and are catalytically less active in vitro on chromatin containing this modification compared with unmodified or H3K9me3-modified chromatin (Zhang et al. 2010). Multiple potential mechanisms for preventing CGI methylation, including those discussed above, are not mutually exclusive, but may act in concert. It is noteworthy that a promoterless CpG-rich sequence marked by Cfp1 binding and H3K4me3 was only partially immune to DNA methylation, suggesting that this chromatin modification by itself is insufficient (Thomson et al. 2010). It may be that a combination of factors—including, perhaps, initiation of transcription—is required to exclude DNA methylation from CGIs.
Concluding remarks regarding CGI function
CGIs represent a dispersed but coherent DNA sequence class whose members function as genomic platforms for regulating transcription at their associated promoters. These properties depend on the shared features of their DNA sequence; notably, CpG richness and a higher than average G+C content. Paradoxically, CpG richness by itself attracts protein complexes that promote H3K4me3 (Thomson et al. 2010), but G+C-rich DNA also recruits H3K27me3 (Mendenhall et al. 2010). An equivocal activity state due to the coexistence of these contradictory tendencies prevails at bivalent CGIs, particularly in ES cells, where transcription decisions at many promoters are pending (Azuara et al. 2006; Bernstein et al. 2006). Of course, CpG is also the substrate for DNA methyltransferases, providing another opportunity for regulation. Why should CGI DNA be poised to set up both active and inactive chromatin structures? A possible reason is to create tension between these opposing states and thus facilitate decisive switching. A nonbiological analogy is the spring in an electrical switch, which ensures that a light can be flicked easily between on and off states that are subsequently stable. A key part of such a “spring” model is that features of the silent or active condition should be self-reinforcing once a decision has been made. Biochemically, this can be achieved by positive feedback on pathways that emphasize the chosen state, or by inhibition of opposing pathways. For example, H3K4me3 can attract the ING4 zinc finger protein, which indirectly augments acetylation of histone H3 tails (Hung et al. 2009), thereby emphasizing the active chromatin configuration. At the same time, this mark inhibits the action of Dnmts, which might otherwise impose transcriptional silence. It is likely that a cascade of related positive and negative feedback loops ensures spring-like stability. Indeed, the more we learn about the marking of chromatin, the more loops of this kind are uncovered, suggesting that consolidation of transcriptional states could be a major role for these epigenetic systems. How is the decision between activity or silence made? The evidence suggests that DNA sequence-specific transcriptional regulators are often ultimately responsible, and that their influence, and that of the proteins they recruit, is dominant over the tendencies imposed by CGI DNA. It is notable that the ability of GC-rich DNA to recruit H3K4me3 and H3K27me3 was detected using promoterless exogenous DNA domains (Mendenhall et al. 2010; Thomson et al. 2010). Thus, factors that repress or activate transcription appear to exaggerate the chromatin changes to which CGIs are already prone. The construction of abrupt switches based on biochemical processes that are often continuously variable is a challenge in many biological processes. CGIs may provide one solution at the level of transcriptional regulation.
Acknowledgments
We thank Shaun Webb for analyzing the base composition of transcription factor-binding sites. We are also grateful to Thomas Clouaire, Jim Selfridge, and Elisabeth Wachter for helpful comments on the manuscript. The Bird laboratory is funded by The Wellcome Trust, the Medical Research Council, and Cancer Research UK.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2037511.
References
- Antequera F, Bird AP 1988. Unmethylated CpG islands associated with genes in higher plant DNA. EMBO J 7: 2295–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F, Bird A 1999. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr Biol 9: R661–R667 doi: 10.1016/S0960-9822(99)80418-7 [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Beisel C, Paro R 2011. Silencing chromatin: comparing modes and mechanisms. Natl Rev 12: 123–135 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bird AP 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8: 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP, Taggart MH, Smith BA 1979. Methylated and unmethylated DNA compartments in the sea urchin genome. Cell 17: 889–901 [DOI] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D 1985. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell 40: 91–99 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Klose RJ 2011. CpG island chromatin: a platform for gene regulation. Epigenetics 6: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ 2010. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Veenstra GJ 2009. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 118: 549–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371: 435–438 [DOI] [PubMed] [Google Scholar]
- Brookes E, Pombo A 2009. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep 10: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A 2008. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res 36: D102–D106 doi: 10.1093/nar/gkm955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev 16: 2583–2592 [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635 [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 [DOI] [PubMed] [Google Scholar]
- Choi JK 2010. Contrasting chromatin organization of CpG islands and exons in the human genome. Genome Biol 11: R70 doi: 10.1186/gb-2010-11-7-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274: 775–780 [DOI] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP 1994. Purification of CpG islands using a methylated DNA binding column. Nat Genet 6: 236–244 [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R 2001. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol 153: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado M, Sacristan M, Antequera F 2001. Species-specific organization of CpG island promoters at mammalian homologous genes. EMBO Rep 2: 586–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, Lowell S, Bolton V, Monk M 1997. Transcription of tissue-specific genes in human preimplantation embryos. Hum Reprod 12: 2251–2256 [DOI] [PubMed] [Google Scholar]
- Delgado S, Gomez M, Bird A, Antequera F 1998. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J 17: 2426–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet C, Lurquin C, Lethe B, Martelange V, Boon T 1999. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol 19: 7327–7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC 2007. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol 19: 281–289 [DOI] [PubMed] [Google Scholar]
- Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R 2011. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res 21: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. 2010. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38: 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571 [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W 2011. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature doi: 10.1038/nature10008 [DOI] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G 2008. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2: 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M 1994. Transcripts and CpG islands associated with the pro-opiomelanocortin gene and other neurally expressed genes. J Mol Endocrinol 12: 365–382 [DOI] [PubMed] [Google Scholar]
- Gebhard C, Benner C, Ehrich M, Schwarzfischer L, Schilling E, Klug M, Dietmaier W, Thiede C, Holler E, Andreesen R, et al. 2010. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res 70: 1398–1407 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138: 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Shilatifard A 2010. The JARID2–PRC2 duality. Genes Dev 24: 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzing LB, Romer JT, Horn JM, Ashworth A 1997. Xist has properties of the X-chromosome inactivation centre. Nature 386: 272–275 [DOI] [PubMed] [Google Scholar]
- Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O 2009. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell 33: 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP 2009. CpG islands—‘a rough guide.’ FEBS Lett 583: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. 2008. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6: e22 doi: 10.1371/journal.pbio.0060022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr ARW, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP 2010. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet 6: e1001134 doi: 10.1371/journal.pgen.1001134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger SA, Chan ET, Berger MF, Stottmann R, Hughes TR, Bulyk ML 2010. Conservation and regulatory associations of a wide affinity range of mouse transcription factor binding sites. Genomics 95: 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2002. The fundamental role of epigenetic events in cancer. Natl Rev 3: 415–428 [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2007. The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT 2008. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol 20: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, et al. 2006. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 38: 149–153 [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR 2006. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol 13: 5–7 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolin JM, Johnson DS, Trinklein ND, Aldred SF, Medina C, Shulha H, Weng Z, Myers RM 2010. Sequence features that drive human promoter function and tissue specificity. Genome Res 20: 890–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen F, Gundersen G, Lopez R, Prydz H 1992. CpG islands as gene markers in the human genome. Genomics 13: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG 2005. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem 280: 41725–41731 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442: 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A 2009. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J Biol Chem 284: 7970–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MC, Dickerson DR, Schmitt M, Groudine M 2004. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol 11: 1068–1075 [DOI] [PubMed] [Google Scholar]
- Macleod D, Charlton J, Mullins J, Bird AP 1994. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes & Dev 8: 2282–2292 [DOI] [PubMed] [Google Scholar]
- Macleod D, Ali RR, Bird A 1998. An alternative promoter in the mouse major histocompatibility complex class II I-Aβ gene: implications for the origin of CpG islands. Mol Cell Biol 18: 4433–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini-DiNardo D, Steele SJ, Ingram RS, Tilghman SM 2003. A differentially methylated region within the gene Kcnq1 functions as an imprinted promoter and silencer. Hum Mol Genet 12: 283–294 [DOI] [PubMed] [Google Scholar]
- Mancini-DiNardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 20: 1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244 doi: 10.1371/journal.pegn.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: insights into functions. Natl Rev 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Schubeler D 2009. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet 25: 129–136 [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W 2005. Epigenetic reprogramming in mammals. Hum Mol Genet 14: R47–R58 doi: 10.1093/hmg/ddi114 [DOI] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. 2007. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39: 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Heard E 2009. Lessons from comparative analysis of X-chromosome inactivation in mammals. Chromosome Res 17: 659–669 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LE, Rabinowicz PD, O'Shaughnessy AL, Balija VS, Nascimento LU, Dike S, de la Bastide M, Martienssen RA, McCombie WR 2003. Maize genome sequencing by methylation filtration. Science 302: 2115–2117 [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT 2008. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP 2009. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci 106: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds GA, Basu SK, Osborne TF, Chin DJ, Gil G, Brown MS, Goldstein JL, Luskey KL 1984. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5′ untranslated regions. Cell 38: 275–285 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K 2006. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci 103: 15782–15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenberg JM, Shlyakhtenko A, Glass K, Rishi V, Myakishev MV, FitzGerald PC, Vinson C 2008. All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genomics 9: 67 doi: 10.1186/1471-2164-9-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G 2010. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics 11: 137 doi: 10.1186/1471-2164-11-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E 2000. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol 225: 294–303 [DOI] [PubMed] [Google Scholar]
- Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Cote V, et al. 2009. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell 33: 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci 103: 1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling E, Rehli M 2007. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics 90: 314–323 [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236 [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446 doi: 10.1371/journal.pgen.1000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP 2007. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 3: 2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, et al. 2003. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci 100: 15776–15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813 [DOI] [PubMed] [Google Scholar]
- Stein R, Razin A, Cedar H 1982. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci 79: 3418–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H 2009. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol 16: 564–571 [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Kerr AR, De Sousa D, Bird A 2007. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res 17: 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Yamashita S, Shimazu T, Niwa T, Ushijima T 2009. The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res 19: 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J, Bird A 1990. Alternative chromatin structure at CpG islands. Cell 60: 909–920 [DOI] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Tweedie S, Charlton J, Clark V, Bird A 1997. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol Cell Biol 17: 1469–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ingen H, van Schaik FM, Wienk H, Ballering J, Rehmann H, Dechesne AC, Kruijzer JA, Liskamp RM, Timmers HT, Boelens R 2008. Structural insight into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure 16: 1245–1256 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439: 871–874 [DOI] [PubMed] [Google Scholar]
- Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG 2000. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol Cell Biol 20: 2108–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. 2007. Epigenetic stem cell signature in cancer. Nat Genet 39: 157–158 [DOI] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE 2010. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y 2010. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y 2011a. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature doi: 10.1038/nature09934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y 2011b. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25: 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J 2008. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol Cell Biol 28: 4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D 2010. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci 107: 18729–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, et al. 2010. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res 38: 4246–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG 2008. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, He F, Hu S, Yu J 2008. On the nature of human housekeeping genes. Trends Genet 24: 481–484 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]