Abstract
Plant photoreceptors mediate light suppression of the E3 ubiquitin ligase COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) to affect gene expression and photomorphogenesis. However, how photoreceptors mediate light regulation of COP1 activity remains unknown. We report here that Arabidopsis blue-light receptor cryptochrome 1 (CRY1) undergoes blue-light-dependent interaction with the COP1-interacting protein SPA1 (SUPPRESSOR OF PHYTOCHROME A). We further show that the CRY1–SPA1 interaction suppresses the SPA1–COP1 interaction and COP1-dependent degradation of the transcription factor HY5. These results are consistent with a hypothesis that photoexcited CRY1 interacts with SPA1 to modulate COP1 activity and plant development.
Keywords: blue-light receptor, cryptochrome, E3 ubiquitin ligase, protein degradation, photomorphogenesis
Cryptochromes are photolyase-like blue-light receptors first discovered in Arabidopsis but later found in all major evolutionary lineages (Cashmore 1997; Lin 2002; Sancar 2003). Arabidopsis cryptochrome 1 (CRY1) and cryptochrome 2 (CRY2) mediate primarily blue-light inhibition of hypocotyl elongation (Ahmad and Cashmore 1993) and photoperiodic control of floral initiation (Guo et al. 1998), respectively. It is known that cryptochromes regulate plant development via modulation of gene expression, but the initial photoreaction of the cryptochrome signal transduction is not fully understood. It has been proposed that cryptochromes undergo blue-light-dependent conformational changes to alter their physical interactions with signaling proteins (Partch et al. 2005; Yu et al. 2007). For example, Arabidopsis CRY2 undergoes blue-light-dependent interaction with the basic helix–loop–helix (bHLH) transcription factor CIB1 to regulate flowering time (Liu et al. 2008). However, no blue-light-dependent CRY1-interacting protein has been reported thus far.
Photoreceptors mediate light regulation of gene expression by several mechanisms, including suppression of the E3 ubiquitin ligase activity of COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) (Deng et al. 1991). COP1 is a RING finger E3 ubiquitin ligase that acts downstream from both phytochromes and cryptochromes (Ang and Deng 1994). COP1 catalyzes ubiquitination of various transcription regulators, such as the bZIP factor HY5, leading to their degradation in the dark (Osterlund et al. 2000; Yi and Deng 2005; Jiao et al. 2007). The photoreceptors mediate light suppression of COP1 activity, resulting in accumulation of the respective transcription factors in response to light and light-dependent gene expression changes (Sullivan et al. 2003; Jiao et al. 2007). COP1 acts in the context of a protein complex that contains multiple subunits, including the coiled-coil/WD repeat protein SPA1 (SUPPRESSOR OF PHYTOCHROME A) (Hoecker et al. 1999; Yi and Deng 2005). SPA1 interacts with COP1 to positively regulate COP1 activity (Hoecker and Quail 2001; Saijo et al. 2003; Seo et al. 2003; Laubinger et al. 2006), whereas light suppresses the COP1–SPA1 interaction and the E3 ubiquitin ligase activity of COP1 (Saijo et al. 2003). However, how photoreceptors mediate light suppression of the COP1–SPA1 interaction and COP1 activity remains unclear. It has been found that CRY1 interacts with COP1, but the interaction between CRY1 and COP1 is independent of light in both yeast and Arabidopsis (Wang et al. 2001; Yang et al. 2001). Therefore, the light-dependent mechanism underlying cryptochrome-mediated blue-light inhibition of COP1 activity was unclear.
Results and Discussion
CRY1 interacts with SPA1 in response to blue light
In a previous study to search for proteins that interact with Arabidopsis CRY2 in response to blue light (Liu et al. 2008), we found several blue-light-specific CRY2-interacting clones corresponding to the SPA1 gene (Zuo et al. 2011). Given the structural and functional conservation of CRY1 and CRY2, and the important role of CRY1 and SPA1 in the blue-light-dependent de-etiolation responses (Lin 2002; Laubinger et al. 2004; Yang et al. 2005; Fittinghoff et al. 2006; Yang and Wang 2006), we investigated the relationship between SPA1 and CRY1 in more detail. We first examined and confirmed that SPA1 interacts with CRY1 in a blue-light-dependent manner in yeast cells using both the auxotrophy marker (LEU2) (Supplemental Fig. S1A) and the colorimetric marker (LacZ) (Fig. 1A,B; Supplemental Fig. S1B) in yeast two-hybrid assays. As shown in Figure 1A, SPA1 interacts with CRY1 in yeast cells illuminated with blue light (Fig. 1A, B40). In contrast, little CRY1–SPA1 interaction was detected in yeast cells kept in the dark (Fig. 1A,D) or illuminated with red light (Fig. 1A, R40). These results show the blue-light specificity of the CRY1–SPA1 interaction. The intensity of the CRY1–SPA1 interaction increases as the fluence rates of blue light increased from 5 μmol m−2 sec−1 to 50 μmol m−2 sec−1 (Fig. 1B), demonstrating that the SPA1–CRY1 interaction in yeast cell is dependent on not only the wavelength, but also the photon density of light. SPA1 is one of the four “SPA quartet” genes (SPA1, SPA2, SPA3, and SPA4) that play partially redundant functions in photomorphogenesis of Arabidopsis (Laubinger et al. 2004; Zhu et al. 2008). We found that, among the SPA quartet gene products, only SPA1 and SPA4 interacted with CRY1 strongly in response to blue light in yeast cells (Supplemental Fig. S2). We focused on the analysis of the CRY1–SPA1 interaction for the rest of this study.
Figure 1.
CRY1 undergoes blue-light-dependent interactions with SPA1. (A) β-Galactosidase assays showing the CRY1–SPA1 interaction in yeast cells treated with blue light (B40, 40 μmol m−2 sec−1), red light (R40, 40 μmol m−2 sec−1), or darkness (D). (B) β-Galactosidase assays showing the CRY1–-SPA1 interaction in response to 5 μmol m−2 sec−1 (B5), 25 μmol m−2 sec−1 (B25), or 50 μmol m−2 sec−1 (B50) of blue light for the durations indicated. (C) Colocalization of CRY1 and SPA1 in the nucleus of Arabidopsis cells. Nuclei were isolated from transgenic plants expressing MycSPA1. Samples were probed with anti-CRY1 (rabbit polyclonal IgG), anti-Myc (mouse monoclonal IgG), or preimmune serum (Preim), followed by Rhodaine red-X-conjugated goat-against-rabbit IgG (red) and Diaminotriazinylaminofluorescein-conjugated goat-against-mouse IgG (green). The images of the same cell from separate color channels were merged by the merge program of Photoshop and are shown (Merge). Bar, 5 μm.
We next examined whether SPA1 and CRY1 may colocalize in the nuclei of Arabidopsis cells by coimmunolocalization assay using transgenic plants expressing the MycSPA1 fusion protein under control of the constitutive 35S promoter (35S::MycSPA1). As reported previously, SPA1 and CRY1 accumulate in the nucleus (Yang et al. 2001; Seo et al. 2003; Laubinger et al. 2006). At least a fraction of the CRY1 and SPA1 proteins colocalized in the nuclear bodies. Blue light appears to induce a modest increase in colocalization of the two proteins (Fig. 1C, yellow dots). We further investigated whether CRY1 and SPA1 colocalize in the same protein complex and whether blue light stimulates formation of the CRY1–SPA1 complex using coimmunoprecipitation (co-IP) assays. It is known that CRY1 is constitutively expressed but SPA1 expression is light-inducible in response to prolonged light treatment of etiolated seedlings (Lin et al. 1998; Hoecker et al. 1999; Fittinghoff et al. 2006; Zhu et al. 2008). However, the expression of the 35S::MycSPA1 transgene showed no change in etiolated seedlings or dark-adapted and red-light-adapted adult plants exposed to blue light for up to 180 min (Fig. 2A; Supplemental Figs. S3, S4). We treated 5-d-old etiolated transgenic seedlings with blue light (20 μmol m−2 sec−1) for 15 or 30 min (Fig. 2A, B15 and B30) and collected samples for the co-IP assays. After blue-light treatment for 30 min, we also transferred a portion of the plants to the dark or red light for up to 60 min and collected more samples for additional analysis of the light effect (Fig. 2A, D30, D60, and R60). As shown in Figure 2, similar levels of MycSPA1 and CRY1 were detected in the immunoblot from etiolated seedlings exposed to blue light (Fig. 2A, Input). Comparable amounts of CRY1 were also immunoprecipitated from those samples (Fig. 2A, CRY1-IP). Little MycSPA1 was coimmunoprecipitated with CRY1 by the anti-CRY1 antibody in etiolated seedlings (Fig. 2A,D). In contrast, MycSPA1 was coimmunoprecipitated with CRY1 in plants treated with blue light. The amount of MycSPA1 coprecipitated by CRY1 increased when plants were exposed to blue light for a longer time (Fig. 2A, B15 and B30), and decreased when plants were transferred to darkness or red light (Fig. 2A, D30 and R60). The CRY1–SPA1 complex was no longer detected after the blue-light-treated plants were transferred to darkness or red light for 60 min (Fig. 2A, D60 and R60). Similarly, the blue-light-specific CRY1–SPA1 complex was also detected by the co-IP assay in adult plants (Supplemental Fig. S4A). Taking into account the results from the yeast two-hybrid, coimmunostaining, and co-IP experiments, we conclude that CRY1 undergoes blue-light-dependent interaction with SPA1.
Figure 2.
The biochemical and genetic interaction between CRY1 and SPA1. (A) Co-IP analyses showing blue-light-dependent formation of the CRY1–SPA1 complex in Arabidopsis. The wild-type (WT) and transgenic seedlings expressing the 35S::MycSPA1 transgene were grown in the dark for 5 d, or exposed to blue light (20 μmol m−2 sec−1) for 15 min (B15) or 30 min (B30). Aliquots of plant samples were then transferred to darkness for 30 min (D30) or 60 min (D60), or transferred to red light (20 μmol m−2 sec−1) for 60 min (R60). Total protein extractions (Input) and immunoprecipitation products prepared by the anti-CRY1 antibody (CRY1-IP) or preimmune serum (Preim) were fractionated in a SDS-PAGE gel, blotted to membranes, probed with the anti-Myc antibody (MycSPA1), stripped, and reprobed with the anti-CRY1 antibody (CRY1). (B,C) A phenotypic analysis of the cry1, spa1spa4, and spa1spa4cry1 mutants. Seedlings of the indicated genotypes were grown in the dark or continuous blue light (25 μmol m−2 sec−1) for 5 d. Images of the representative seedlings are shown in B, and the hypocotyl lengths of the indicated genotypes grown in different conditions were measured and are shown in C. Standard deviations (n ≥ 20) are indicated. (D) Immunoblot showing blue-light-dependent stabilization of the HY5 protein in wild-type plants and spa1spa4cry1 mutant plants but not in cry1 mutant plants. Five-day-old etiolated transgenic seedlings expressing 35::MycHY5 in the Col, cry1, and spa1spa4cry1 backgrounds were transferred to blue light (50 μmol m−2 sec−1) for 5 h. Immunoblots were probed with anti-Myc antibody (MycHY5), stripped, and reprobed with the anti-CRY1 antibody (CRY1) to verify the cry1 genotype. The blot was stripped again and reprobed with the anti-HSP90 antibody (HSP90) to access relative loading of samples. The numbers shown at the top (B/D) indicate relative abundance of MycHY5 in light-treated samples, which was calculated by the formula (MycHY5/HSP90 [blue light])/(MycHY5/HSP90 [dark]). The relative abundance of MycHY5 in etiolated seedlings was set to 1 for each genotype. (E) Quantitative PCR (qPCR) results showing mRNA expression of the chalcone synthase (CHS) gene in the wild type (Col4), and in cry1, spa1sap4, and spa1spa4cry1 mutants. Three-week-old light-grown plants were treated in the dark for 3 d and transferred to blue light (25 μmol m−2 sec−1) for 4 h. The relative level of CHS mRNA was normalized by that of the wild-type seedlings kept in the dark, which was set to 1. The standard deviations were derived from three independent experiments.
SPA1 is required for the CRY1-mediated blue-light suppression of HY5 degradation and hypotocotyl elongation
To understand the role of the blue-light-dependent CRY1–SPA1 interaction in the function of CRY1, we investigated the genetic interaction between the CRY1 and SPA1 genes. Because both SPA1 and SPA4 interact with CRY1 in a blue-light-dependent manner (Fig. 1; Supplemental Fig. S2), they may function redundantly to mediate the action of CRY1. Therefore, we tested the genetic interaction of the three recessive mutations (cry1, spa1, and spa4) for blue-light inhibition of hypocotyl growth response (Fig. 2B,C; Supplemental Fig. S5). As shown in Figure 2, the cry1 mutant seedlings grew markedly taller than the wild type in continuous blue light, and the spa1spa4 double mutant was slightly shorter than the wild type (Fig. 2B,C). Importantly, the spa1spa4cry1 triple mutant exhibited a hypocotyl phenotype indistinguishable from that of the spa1spa4 double mutant (Fig. 2B,C), suggesting that spa1spa4 is epistatic to cry1, and that CRY1 mediates blue-light inhibition of hypocotyl elongation at least partially through SPA1 (and SPA4).
It is known that the SPA1-interacting protein COP1 catalyzes ubiquitination and degradation of the transcription regulator HY5, whereas CRY1 mediates blue-light suppression of COP1 to promote accumulation of HY5, leading to altered transcription of blue-light-regulated genes such as CHS (chalcone synthase) (Ang and Deng 1994; Osterlund et al. 2000). To examine whether SPA1 is involved in the CRY1-dependent and blue-light-dependent accumulation of the HY5 protein, we compared blue-light regulation of the abundance of Myc-tagged HY5 (MycHY5) transgenically expressed in the wild type, the cry1 mutant, and the spa1spa4cry1 mutant (Fig. 2D). Figure 2D shows that blue light induces an approximately threefold increase in the abundance of the MycHY5 protein (3.1×), but this blue-light stimulation of MycHY5 protein accumulation was not found in the cry1 mutant (1.2×). Importantly, the defect of the cry1 mutation in blue-light-induced HY5 accumulation was rescued in the spa1spa4cry1 triple mutant. In the spa1spa4cry1 mutant plants exposed to blue light, the MycHY5 protein accumulated to a level similar to that of the wild type (2.6×) (Fig. 2D; Supplemental Fig. S4B). Consistent with this result, the impairment of the blue-light-induced mRNA expression of the HY5 target gene CHS was similarly impaired in the cry1 monogenic mutant but rescued in the spa1spa4cry1 triple mutant (Fig. 2E). Taken together, we conclude that SPA1 is required for CRY1-dependent blue-light regulation of COP1 activity.
The CRY1-intercting domain of SPA1 suppresses blue-light sensitivity of transgenic seedlings
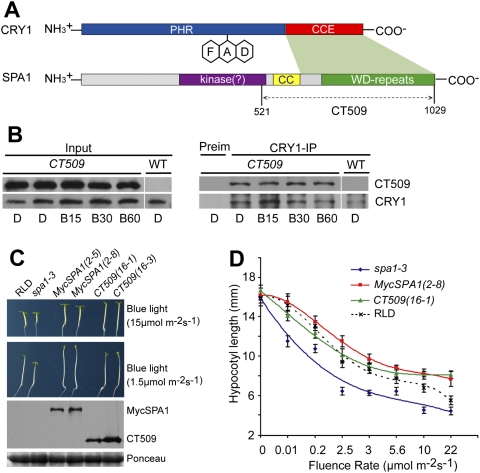
CRY1 has two domains: the N-terminal PHR (photolyase homologous region) domain and the C-terminal CCE (cryptochrome C-terminal extension) domain (Fig. 3A; Yu et al. 2010). The PHR is the evolutionarily conserved chromophore-binding domain; the CCE domain interacts with COP1 and is critical for the function of CRY1 in de-etiolation responses (Yang et al. 2000, 2001; Wang et al. 2001; Lin and Shalitin 2003; Brautigam et al. 2004). SPA1 is composed of three domains (Fig. 3A): the N-terminal kinase-like domain, the central coiled-coil domain, and the C-terminal WD repeat domain (Fig. 3A). The coiled-coil domain and the WD repeat domain interact with COP1 and its substrate, HY5, respectively (Hoecker and Quail 2001; Saijo et al. 2003). A yeast two-hybrid analysis indicates that the CCE domain of CRY1 interacts with the WD repeat domain of SPA1 (Fig. 3A, Supplemental Fig. S1B) or SPA4 (Supplemental Fig. S2C). It has been reported previously that a SPA1 fragment that contains only the coiled-coil and WD repeat domains—referred to as CT509 (Fig. 3A) or SCT1 (Lian et al. 2011)—is necessary and sufficient for interacting with COP1 and suppressing photomorphogenesis (Yang and Wang 2006). We found that the CT509 fragment of SPA1 (and the SPA4 equivalent) constitutively interacts with CRY1 in yeast and plant cells (Fig. 3A,B; Supplemental Figs. S1B, S2C). Transgenic seedlings expressing either MycSPA1 or CT509 grew taller in blue light than wild type or the spa1 mutant parent (Fig. 3C). Interestingly, the CT509-expressing seedlings grew modestly shorter than the MycSPA1-expressing seedlings under blue light with the relatively low fluence rates (<3 μmol m−2 sec−1, P < 0.0025) (Fig. 3C,D), but the difference between the two genotypes diminished under blue light of higher fluence rates (>3 μmol m−2 sec−1) (Fig. 3D) and disappeared at the highest fluence rate of blue light tested (22 μmol m−2 sec−1) (Fig. 3D). Because MycSPA1 and CT509 have a similar physiological activity promoting hypocotyl elongation, and the level of CT509 expression is not lower than that of MycSPA1 (Fig. 3C), the blue-light-dependent CRY1–SPA1 interaction and constitutive CRY1–CT509 interaction appear to best explain their different blue-light sensitivities observed in Figure 3D. According to this interpretation, in low light, CRY1 interacts relatively stronger with CT509 than with full-length SPA1 (MycSPA1), causing more severe suppression of CT509. In high light, CRY1 interacts similarly with CT509 and full-length SPA1 (MycSPA1), resulting in a similar extent of suppression of the activity of CT509 and MycSPA1 and a similar hypocotyl growth of the respective transgenic lines.
Figure 3.
The C-terminal domain of SPA1 interacts with CRY1 to affect the blue-light sensitivity of Arabidopsis seedlings. (A) A diagram depicting the domain organization of the SPA1 and CRY1 proteins. The different domains and regions involved in the CRY1–SPA1 interaction (green shade) are indicated. (B) A co-IP assay showing that CT509, which contains C-terminal 509 residues of SPA1, constitutively interacts with CRY1 in transgenic plants. The wild-type and MycCT509-expressing plants were grown in white light under a long-day photoperiod (16 h light /8 h dark) for 2 wk; plants were transferred to darkness for ∼18 h and exposed to blue light (20 μmol m−2 sec−1) for 15 min (B15), 30 min (B30), or 60 min (B60). Total extracts (Input), CRY1 (CRY1-IP), and control (Preim) immunoprecipitation products were fractionated by a SDS-PAGE gel, transferred to membranes, probed with the anti-Myc antibody (MycCT509), stripped, and reprobed with the anti-CRY1 antibody. (C) Five-day-old transgenic seedlings expressing MycSPA1 or MycCT509 (in the spa1-3 mutant background) and the controls were grown in continuous blue lights. Two independent lines of each genotype expressing the respective protein were tested, for which the respective levels of MycSPA1 or MycCT509 are shown in an immunoblot. (D) Hypocotyl lengths of the indicated genotypes grown in the dark or continuous blue light with different fluence rates for 5 d were measured and are shown. Standard deviations (n ≥ 20) are indicated.
The CRY1–SPA1 interaction suppresses the COP1–SPA1 interaction in response to blue light
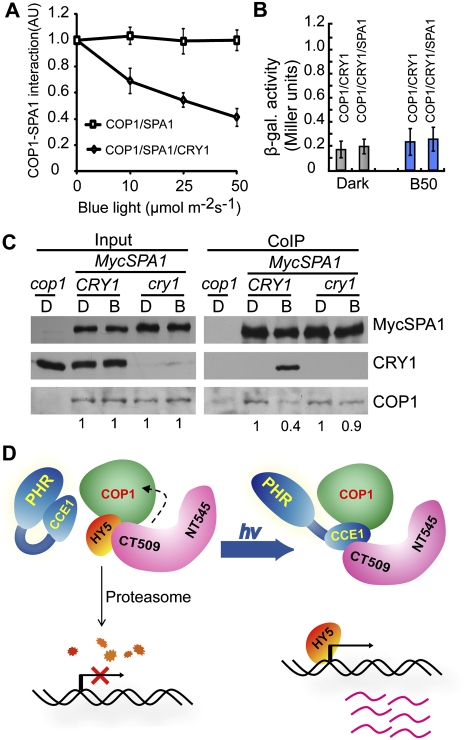
We then asked the question of how blue-light-dependent CRY1–SPA1 interaction conveys CRY1-mediated blue-light response in plants. We reasoned that, because SPA1 interacts with COP1 to positively regulate COP1 activity (Hoecker and Quail 2001; Saijo et al. 2003; Seo et al. 2003; Laubinger et al. 2006), light suppresses SPA1–COP1 interaction (Saijo et al. 2003), and that, since SPA1 interacts with CRY1 and COP1 via adjacent domains, the CRY1–SPA1 interaction might interfere with the COP1–SPA1 interaction to suppress the COP1 activity in response to blue light. We tested this possibility using a yeast three-hybrid assay (Fig. 4; Supplemental Fig. S6; Tirode et al. 1997). In this experiment, the protein–protein interaction between the bait (BD-COP1) and prey (AD-SPA1) was tested in the absence or presence of the third protein bait mate (CRY1) in response to blue light. Expression of CRY1 was controlled by the methionine-suppressible Met25 promoter in yeast cells (Supplemental Fig. S6A; Tirode et al. 1997). As shown in Figure 4A, the COP1–SPA1 interaction was not affected by blue light in the absence of CRY1 expression (Fig. 4A, COP1/SPA1). Importantly, the COP1–SPA1 interaction was also not affected by CRY1 in the absence of blue light (0 μmol m−2 sec−1) (Fig. 4A, COP1/SPA1/CRY1). However, the SPA1–COP1 interaction was suppressed in the presence of CRY1 when yeast cells were exposed to blue light (Fig. 4A; Supplemental Fig. S6B). The CRY1-dependent suppression of the SPA1–COP1 interaction is dependent on the fluence rate of blue light: The level of SPA1–CRY1 interaction reached the lowest level in yeast cells illuminated with the highest fluence rate of blue light tested (50 μmol m−2 sec−1), which is ∼40% that of the yeast cells grown in the dark or in the absence of CRY1 (Fig. 4A). A control experiment showed that the CRY1–COP1 interaction was not affected by SPA1 or blue light (Fig. 4B), which is consistent with a previous report that CRY1 interacts with COP1 constitutively (Wang et al. 2001; Yang et al. 2001).
Figure 4.
The CRY1–SPA1 interaction suppresses the COP1–SPA1 interaction in response to blue light. (A) Yeast three-hybrid assays showing the fluence rate-dependent CRY1 inhibition of the SPA1–COP1 interaction in yeast cells exposed to blue light. Relative β-galactosidase activity of yeast cells expressing the GAL4 DB-COP1 (bait) and GAL4 AD-SPA1 (prey) in the absence (square) or presence (diamond) of the CRY1 bait mate, grown in the dark or blue light of different fluence rates, were analyzed, calculated, and plotted. The relative extents of the SPA1–COP1 interaction were calculated and are presented as arbitrary units (AU) {AU = (Miller units [light])/(Miller unit [dark])}, with the AU of dark-treated samples set to 1. Standard deviations are derived from three independent experiments (n = 3). (B) A yeast three-hybrid assay with COP1 as bait, CRY1 as prey, and SPA1 as bait mate shows that the CRY1–COP1 interaction was not affected by SPA1 or blue light in yeast cells. (C) A co-IP experiment showing a reduction of the SPA1–COP1 interaction in the wild-type but not the cry1 mutant seedlings in response to blue light. Transgenic seedlings expressing MycSPA1 in the wild type (CRY1) or cry1 mutant (cry1) and the cop1 control were grown in continuous white light for 4 d, transferred to dark for 2 d, and exposed to blue light (50 μmol m−2 sec−1) for 60 min. Total protein extractions (Input) and immunoprecipitation product (Myc-IP) prepared by the anti-CRY1 antibody (CRY1-IP) or preimmune serum (Preim) were fractionated in a SDS-PAGE gel, blotted to membranes, probed with the anti-Myc antibody (MycSPA1), stripped, and reprobed with the anti-CRY1 antibody (CRY1), followed by anti-COP1 antibody (COP1). The relative levels of the COP1 co-IP product of dark-treated samples (set to 1) or blue-light-treated samples were calculated by the formula (COP1/MycSPA1 of blue light-treated sample)/(COP1/MycSPA1 of dark-treated sample) and are indicated at the bottom. (D) A working model depicting how CRY1 mediates blue-light suppression of COP1 activity. In the dark, SPA1 interacts with COP1 to activate COP1, and COP1 facilitates degradation of its substrates, such as HY5. In response to blue light, photoexcited CRY1 adopts an open conformation to interact with SPA1 via the CCE domain of CRY1 and the WD repeat domain of SPA1 encompassed in the C-terminal 509 residues of SPA1 (CT509). The CRY1–SPA1 interaction suppresses the SPA1–COP1 interaction and consequentially reduces COP1 activity, resulting in the accumulation of COP1 substrates and light-induced gene expression changes.
To further investigate the hypothesis that CRY1 mediates blue-light suppression of COP1 activity by inhibiting the SPA1–COP1 interaction, we tested whether CRY1 affects formation of the COP1–SPA1 complex in Arabidopsis cells using a co-IP assay. In this experiment, transgenic seedlings expressing the 35S::MycSPA1 transgene in the wild-type (CRY1) or cry1 mutant background were examined. The dark-adapted seedlings were illuminated with blue light for 1 h, and the SPA1–COP1 interaction was compared in the presence (CRY1) or absence (cry1) of CRY1 in response to blue light. Similar levels of MycSPA1 and COP1 proteins were detected regardless of blue-light treatment in both the CRY1 and cry1 backgrounds (Fig. 4C). As expected, CRY1 was coprecipitated by MycSPA1 from the blue-light-treated plants but not from the dark-adapted control (Fig. 4C, CRY1). Importantly, a relatively lower amount of COP1 was coprecipitated by MycSPA1 in the wild-type (CRY1) seedlings exposed to blue light (Fig. 4C, COP1). We calculated the relative amount of COP1 coprecipitated with MycSPA1 in response to blue light in the presence or absence of CRY1 (Fig. 4C). According to this calculation, the COP1–SPA1 interaction in blue-light-treated wild-type seedlings is ∼40% that of the dark control, which is consistent with the results of the yeast three-hybrid experiment (Fig. 4A). The blue-light suppression of the SPA1–COP1 interaction was not observed in the cry1 mutant seedlings (Fig. 4C), demonstrating that CRY1 mediates blue-light suppression of the SPA1–COP1 interaction. Because blue light stimulates the CRY1–SPA1 interaction, it is conceivable that photoexcited CRY1 acts as a competitive inhibitor of the SPA1–COP1 interaction to suppress COP1 activity in response to blue light.
Taking into account the results that CRY1 undergoes a blue-light-dependent interaction with SPA1 (Figs. 1, 2), that SPA1 is required for the CRY1 function and CRY1-mediated blue-light suppression of the COP1-dependent HY5 degradation (Figs. 2, 3), and that the CRY1–SPA1 interaction suppresses the SPA1–COP1 interaction in response to blue light (Fig. 4), we propose a working hypothesis to explain how CRY1 mediates blue-light suppression of COP1 activity (Fig. 4D). According to this model, SPA1 interacts with COP1 via the coiled-coil domain of SPA1 to activate COP1-dependent ubiquitination and degradation of transcription factors such as HY5, resulting in suppression of light-dependent transcription in etiolated seedlings. In response to blue light, the photoexcited CRY1 interacts with SPA1 via the CCE domain of CRY1 and the WD repeat domain of SPA1, resulting in suppression of the SPA1–COP1 interaction. A weaker SPA1–COP1 interaction reduces COP1 activity, leading to the accumulation of transcription factors such as HY5, light-dependent transcription, and de-etiolation in response to blue light. In addition to this mode of action, other mechanism(s) may also be involved in the action of CRY1 in the de-etiolation response. For example, given the light-independent CRY1–COP1 interaction reported previously (Wang et al. 2001; Yang et al. 2001), photoexcited CRY1 may also directly suppress COP1 activity. Additional studies are needed to further elucidate the biochemical mechanism of CRY1.
Materials and methods
Plant materials, seedling photoresponse assays, yeast two-hybrid assays, immunostaining, immunoblots, and co-IP are as described previously (Liu et al. 2008; Yu et al. 2009) and in the Supplemental Material.
Yeast three-hybrid assay
The pBridge vector (Clontech, catalog no. 630404) expressing both the bait-BD fusion protein and the third protein (bait mate), and the PGADT7 vector (Clontech, catalog no. K1612-1) expressing the prey-AD fusion protein were used. The yeast strain MAV203 was transformed with one pair (PB-COP1 and PG-SPA1) or the other pair (PB-COP1–CRY1 and PG-SPA1) of plasmids. Colonies were selected and cultured at 180 rpm at 28°C in the dark until they reached OD600 ∼0.1 in a 100-mL flask containing 40 mL of SD medium (−Leu/−Met/−Trp/+Asp). The yeast culture was split and cultured at 160 rpm at 21°C under different light conditions until OD600 ∼0.5∼0.8. The SPA1–COP1 interaction was presented as an arbitrarily unit (AU), which is defined by the formula AU = (Miller units [light])/(Miller unit [dark]), with the AU of dark-treated samples set to 1.
Acknowledgments
We thank Dr., Xingwang Deng for kindly providing anti-COP1 antibody, and Drs. Haiyang Wang and Hongquan Yang for other experimental materials used in this study. Work in our laboratories is supported in part by the National Institutes of Health (GM56265 to C.L.), UCLA faculty research grants, Sol Leshin BGU-UCLA Academic Cooperation programs, and the National Transgenic Crop Initiative (to ICS-CAAS).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2025011.
Supplemental material is available for this article.
References
- Ahmad M, Cashmore AR 1993. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Ang LH, Deng XW 1994. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6: 613–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J 2004. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci 101: 12142–12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR 1997. A cryptochrome family of photoreceptors. Plant Cell Environ 20: 764–767 [Google Scholar]
- Deng XW, Caspar T, Quail PH 1991. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev 5: 1172–1182 [DOI] [PubMed] [Google Scholar]
- Fittinghoff K, Laubinger S, Nixdorf M, Fackendahl P, Baumgardt RL, Batschauer A, Hoecker U 2006. Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J 47: 577–590 [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C 1998. Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Quail PH 2001. The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 276: 38173–38178 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH 1999. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496–499 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW 2007. Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U 2004. The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U 2006. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222 [DOI] [PubMed] [Google Scholar]
- Lian HL, He SH, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ 2011. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev (this issue). doi: 10.1101/gad.2025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C 2002. Blue light receptors and signal transduction. Plant Cell 2002S: S207–S225 doi: 10.1105/tpc.000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Shalitin D 2003. Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR 1998. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW 2000. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Partch CL, Clarkson MW, Ozgur S, Lee AL, Sancar A 2005. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 44: 3795–3805 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW 2003. The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 103: 2203–2237 [DOI] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH 2003. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Sullivan JA, Shirasu K, Deng XW 2003. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat Rev Genet 4: 948–958 [DOI] [PubMed] [Google Scholar]
- Tirode F, Malaguti C, Romero F, Attar R, Camonis J, Egly JM 1997. A conditionally expressed third partner stabilizes or prevents the formation of a transcriptional activator in a three-hybrid system. J Biol Chem 272: 22995–22999 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW 2001. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Yang J, Wang H 2006. The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J 47: 564–576 [DOI] [PubMed] [Google Scholar]
- Yang H-Q, Wu Y-J, Tang R-H, Liu D, Liu Y, Cashmore AR 2000. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR 2001. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Hoecker U, Liu B, Xu L, Wang H 2005. Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J 43: 131–141 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW 2005. COP1—from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C 2007. Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci 104: 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C 2009. Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C 2010. The cryptochrome blue-light receptors. The Arabidopsis Book 8: e0135 doi: 10.1199/tab.0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW 2008. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C 2011. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]