Abstract
Tumor cells gain a survival/growth advantage by adapting their metabolism to respond to environmental stress, a process known as metabolic transformation. The best-known aspect of metabolic transformation is the Warburg effect, whereby cancer cells up-regulate glycolysis under aerobic conditions. However, other mechanisms mediating metabolic transformation remain undefined. Here we report that carnitine palmitoyltransferase 1C (CPT1C), a brain-specific metabolic enzyme, may participate in metabolic transformation. CPT1C expression correlates inversely with mammalian target of rapamycin (mTOR) pathway activation, contributes to rapamycin resistance in murine primary tumors, and is frequently up-regulated in human lung tumors. Tumor cells constitutively expressing CPT1C show increased fatty acid (FA) oxidation, ATP production, and resistance to glucose deprivation or hypoxia. Conversely, cancer cells lacking CPT1C produce less ATP and are more sensitive to metabolic stress. CPT1C depletion via siRNA suppresses xenograft tumor growth and metformin responsiveness in vivo. CPT1C can be induced by hypoxia or glucose deprivation and is regulated by AMPKα. Cpt1c-deficient murine embryonic stem (ES) cells show sensitivity to hypoxia and glucose deprivation and altered FA homeostasis. Our results indicate that cells can use a novel mechanism involving CPT1C and FA metabolism to protect against metabolic stress. CPT1C may thus be a new therapeutic target for the treatment of hypoxic tumors.
Keywords: CPT1C, fatty acid homeostasis, metabolic stress, rapamycin resistance, xenograft tumors
The metabolism of human cancers shows adaptations that promote survival under conditions of metabolic stress (Brown and Wilson 2004; Gatenby and Gillies 2004; Pan and Mak 2007). Tumor cells typically undergo “metabolic transformation” that is modulated by AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) (Plas and Thompson 2005; Shaw 2006; Gwinn et al. 2008). AMPK is activated under poor nutrient conditions, whereas mTOR activation is triggered by high levels of glucose and amino acids. Upon induction of metabolic transformation, cancer cells limit their energy consumption and enhance energy production using a program characterized by increased glycolysis (Brown and Wilson 2004; Shaw 2006). However, several lines of evidence implicate fatty acid (FA) oxidation (FAO) as contributing to metabolic transformation (Yamashita et al. 2000; Liu 2006; Swinnen et al. 2006; Hirsch et al. 2010). Expression of genes involved in lipid metabolism is increased in tumors (Hirsch et al. 2010), and monoacylglycerol lipase (Maglione et al. 2001), an enzyme that liberates free FA from lipid stores, promotes cancer pathogenesis (Nomura et al. 2010). Interestingly, the cancer-promoting effect of MAGL does not seem to involve FAO mediated by the usual carnitine palmitoyltransferase 1 (CPT1) isozymes (Nomura et al. 2010). FAO is also increased in response to glucose deprivation by the p53-dependent induction of guanidinoacetate methyltransferase (GAMT), but the mechanism is unknown (Ide et al. 2009).
FAO is controlled at the step of FA import into the mitochondria, a process executed by tissue-specific isoforms of CPT1 (Kerner and Hoppel 2000). CPT1A functions in the liver and most other tissues, CPT1B functions predominantly in muscle, and CPT1C functions in the brain (Kerner and Hoppel 2000; Price et al. 2002; Ramsay and Zammit 2004). Like CPT1A and CPT1B, the brain-specific CPT1C isoform displays high-affinity binding to malonyl-CoA, but enzymatic activity has not been observed using conventional substrates (Price et al. 2002; Ramsay and Zammit 2004; Wolfgang et al. 2006; Sierra et al. 2008). Gene-targeting studies have demonstrated that Cpt1c is not essential in mice but that the animals exhibit reduced FAO (Wolfgang et al. 2006, 2008).
The precise roles of FAO and CPT1 enzymes in cancer cells experiencing metabolic stress remain unknown. In response to glucose deprivation, AMPK induces activation of a p53-dependent metabolic checkpoint that promotes cell survival (Jones et al. 2005). Similarly, treatment of human cancer and normal cells with the AMPK activators metformin or aminoimidazole carboxamide ribonucleotide (AICAR) increases FAO. Furthermore, both metformin and AICAR selectively inhibit the growth of p53-deficient tumors in vivo (Buzzai et al. 2007). These findings suggest that FAO induction downstream from AMPK activation may be a survival/growth strategy employed by cancer cells subjected to metabolic stress. However, many of the gene(s) and pathway(s) involved have yet to be elucidated.
In this study, we identify CPT1C as a gene that is frequently expressed in tumors and up-regulated in response to metabolic stress. Strikingly, CPT1C expression in tumor cells correlates inversely with both mTOR pathway activation and sensitivity to the mTOR inhibitor rapamycin. Enhanced CPT1C expression increases FAO and ATP production and protects cells from death induced by glucose deprivation or hypoxia. Furthermore, CPT1C expression is induced by metabolic stress in an AMPKα-dependent manner. Conversely, cells deficient in CPT1C show reduced ATP production, altered FA homeostasis, and heightened sensitivity to hypoxia or glucose deprivation. Our findings suggest a new approach for cancer therapies based on manipulating FA metabolism.
Results
Identification of CPT1C as a potential contributor to tumor cell metabolic transformation
Our isolation of CPT1C as a gene potentially involved in tumor cell metabolic transformation occurred as the result of the unexpected confluence of two independent screening methods designed to detect transcripts of interest in cancer biology. The first method, intended to identify novel p53-activated transcripts, employed a cDNA microarray screen based on the differential activation of a temperature-sensitive form of p53 in transformed mouse erythroleukemia cells (DP16.1/p53ts) that lack endogenous p53. The second method, intended to identify genes conferring rapamycin resistance in mouse mammary tumor lines, was based on genetically engineered murine primary tumors driven by mammary-targeted overexpression of a human ERBB2 cDNA in the context of Ink4a deletion. In this second screen, rapamycin sensitivity correlated positively with high expression of a set of coregulated genes (Pik3ca, Frap1, Pik3r1, Erbb3, and Pik3r3) that drive the mTOR pathway; the sum of the mRNA expression levels of these genes was termed the mTOR index (Supplemental Fig. S1A, top).
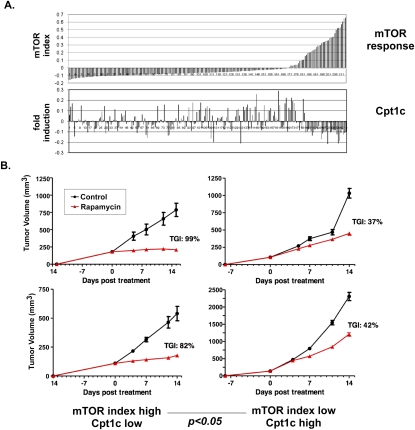
The results of the first screening method revealed that a partial cDNA encoding Cpt1c was up-regulated fourfold upon p53 activation (Supplemental Fig. S1B), and that Cpt1c was the only Cpt gene to show putative responsiveness to p53 (Supplemental Fig. S1C). Serendipitously, the results of the second screening method also identified Cpt1c as a gene of interest (Supplemental Fig. S1A, bottom). Most significantly, in the 168 tumors of the 214 analyzed for which the mTOR index fell below the mean, 108 of them exhibited Cpt1c expression greater than the overall mean for this gene (Fig. 1A). The emergence of Cpt1c in both screens pinpointed this molecule as being of potential importance to the bioenergetics of cancer cells.
Figure 1.
Cpt1c expression correlates inversely with mTOR activation and protects cancer cells against rapamycin. (A) Correlation of Cpt1c expression with mTOR index. Gene expression microarray profiling was performed for 214 murine primary tumors engineered to express human ERBB2 cDNA. The mTOR index is the average of the mean centered expression (MCE) values of the mTOR pathway genes Pik3ca, Frap1, Pik3r3, Pik3r1, and Erbb3. Cpt1c results are the mean MCE value obtained for each tumor using two separate probes. All expression data were determined using Agilent two-color microarray and are reported as the log2 ratio of the signal intensity of cy3-labeled versus cy5-labeled hybridizations. (B) Correlation of Cpt1c expression with rapamycin sensitivity in vivo. Cells from selected tumors in A with a high mTOR index and low Cpt1c or cells from tumors in A with a low mTOR index and high Cpt1c were injected into nude mice and the animals were treated with vehicle (control) or rapamycin for 14 d. New growths were measured at the indicated times; two examples for each group are shown. (TGI) Tumor growth inhibition index.
Validation of Cpt1c as a rapamycin resistance factor
To validate the correlation between low Cpt1c expression and rapamycin sensitivity, we examined the rapamycin sensitivity of several of our 214 primary tumors in mouse xenograft models. Cells from tumors representative of either low mTOR index/high Cpt1c expression (Cpt1c high) or high mTOR index/low Cpt1c expression (Cpt1c low) were implanted into nude mice. These mice were treated with rapamycin for 14 d and new tumor formation was monitored. Malignancies derived from Cpt1c-high tumors were more resistant to rapamycin (tumor growth inhibition [TGI] index of ∼40% vs. vehicle-treated controls) than those derived from Cp1c-low tumors (TGI of >80%) (Fig. 1B). Thus, in general, the Cpt1c mRNA level in a given tumor correlates inversely with its mTOR index and rapamycin sensitivity.
CPT1C is up-regulated in human lung cancers
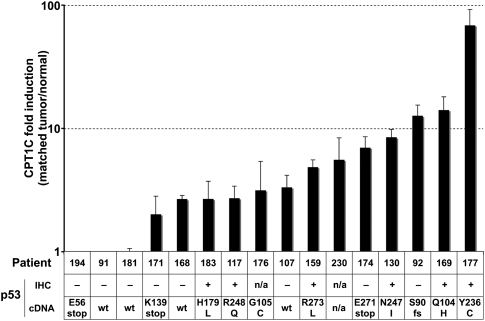
Since Cpt1c mRNA expression appeared to give tumors a growth advantage in mice, we next asked whether human CPT1C might be up-regulated in patient cancer samples. We used real-time RT–PCR to determine CPT1C mRNA levels in paired normal and tumor tissues from 19 patients with non-small-cell lung carcinoma (NSCLC). Where both the normal and tumor tissues of a patient showed detectable levels of CPT1C, CPT1C mRNA was up-regulated in tumor tissue compared with matched normal lung tissue in 81% (13 out of 16) of cases (Fig. 2, top). Because our early studies suggested that CPT1C might be a p53-related transcript, we examined whether CPT1C up-regulation might be a bystander effect of p53 activation. We examined the status of p53 in our NSCLCs by immunohistochemistry (IHC) and cDNA sequencing and found no correlation between p53 expression or mutation and CPT1C mRNA expression (Fig. 2, bottom). Thus, CPT1C up-regulation in human NSCLCs is not a bystander effect of p53 activation. These data imply that CPT1C provides a p53-independent growth advantage to a tumor, and that CPT1C expression must be controlled by additional factors that are as yet unknown.
Figure 2.
CPT1c is overexpressed in human lung tumors. (Top) Levels of CPT1C mRNA were assayed in human NSCLC tumors and matched normal lung tissues using real-time RT–PCR. Results are the fold change in CPT1C mRNA levels in tumor tissue compared with the matched normal tissue from the same patient. (Bottom) p53 status and mutations are shown for the tumors in the top panel. (IHC) Immunohistochemical staining to detect p53; (cDNA) sequence of p53 exons 4–7; (wt) only wild-type p53 detected; (fs) frameshift mutation.
CPT1C up-regulation induces resistance to nutrient deficiency
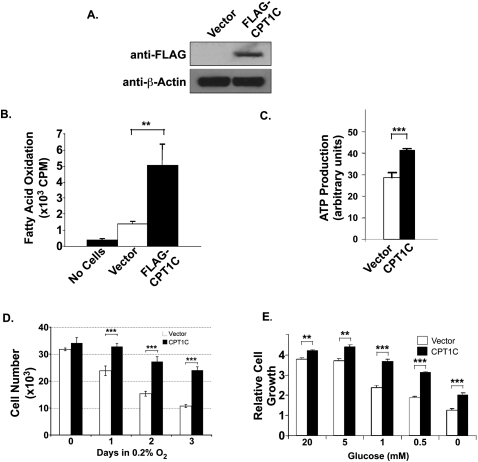
In order to directly examine the functional consequences of CPT1C expression, we generated a line of MCF-7 cancer cells stably overexpressing Flag-tagged CPT1C (Fig. 3A). We assessed whether CPT1C overexpression had a direct effect on mTOR signaling by examining the phosphorylation of S6K and 4E-BP1, two key effectors of the mTOR pathway. However, no differences were detected between CPT1C-overexpressing and control cells (Supplemental Fig. S2). Thus, CPT1C does not directly modulate mTOR signaling.
Figure 3.
CPT1C overexpression alters FAO, ATP production, and responses to metabolic stress. (A) Validation. MCF-7 cells were stably transfected with control vector or vector expressing Flag-tagged CPT1C protein. Flag-CPT1C expression was confirmed by immunoblotting. (B) Increased FAO. FAO was determined in MCF-7 cells overexpressing CPT1C or control vector as described in the Materials and Methods. Results are mean counts per minute (cpm) ± SD of triplicates. (*) P < 0.05; (**) P < 0.01; (***) P < 0.005 for all figures. (C) Increased ATP production. MCF-7 cells overexpressing CPT1C or control vector were evaluated for ATP production as described in the Materials and Methods. Results are the mean ATP production ± SD of triplicates. (D) Increased resistance to hypoxia. MCF-7 cells overexpressing CPT1C or control vector were cultured for the indicated number of days in 0.2% O2, and cell growth was measured by sulforhodamine B (SRB) staining (see the Materials and Methods). Results shown are the mean growth ± SD of triplicates relative to untreated controls. (E) Increased resistance to glucose deprivation. MCF-7 cells overexpressing CPT1C or control vector were cultured in the indicated concentrations of glucose for 5 d. Cell growth was measured by SRB staining as for D.
Because CPT1 proteins are involved in the catabolism of FA to produce ATP, we tested whether our CPT1C-overexpressing MCF-7 cells demonstrated increased FAO and ATP production. When FAO was measured using 14C-palmitic acid as a substrate (see the Materials and Methods), we found that FAO was significantly increased in CPT1C-overexpressing cells compared with vector-transfected controls (Fig. 3B). CPT1C-overexpressing cells also produced significantly more ATP than control cells (Fig. 3C). In addition, CPT1C-overexpressing cells grew significantly better than controls under hypoxia (Fig. 3D) and in limited glucose (Fig. 3E). Taken together, our data suggest a model in which a high level of CPT1C expression facilitates energy production from FA and provides advantages that enhance cell growth under conditions of poor nutrient supply.
CPT1C depletion confers sensitivity to rapamycin and metabolic stress
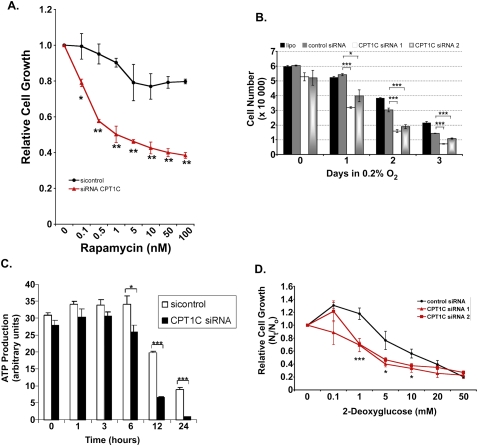
We next asked whether CPT1C loss of function might confer sensitivity to factors that exert metabolic stress. We designed siRNAs to deplete CPT1C in cancer cells, and used quantitative RT–PCR to confirm an ∼70%–80% reduction in CPT1C mRNA in siRNA-treated MCF-7 cells compared with controls (Supplemental Fig. S3A). In order to determine whether CPT1C depletion might alter rapamycin sensitivity, we treated the HCT116 human colon cancer cell line with our CPT1C siRNAs. HCT116 cells are partially resistant to rapamycin, with a GI50 > 10 nM (Buck et al. 2006). However, upon CPT1C depletion, these cells showed a significant increase in rapamycin sensitivity as compared with HCT116 cells treated with control siRNA (Fig. 4A).
Figure 4.
CPT1C depletion confers sensitivity to rapamycin and metabolic stress. (A) Rapamycin sensitivity. HCT116 cells were transfected with siRNA against CPT1C or control siRNA (sicontrol). Transfected cells were treated with the indicated concentrations of rapamycin for 5 d and cell growth was measured using SRB staining as for Figure 3D. (B) Sensitivity to hypoxia. MCF-7 cells were transfected with no siRNA (lipo), luciferase siRNA (control), or CPT1C siRNA1 or siRNA2. Transfected cells were exposed to hypoxia for the indicated number of days and cell growth was measured by SRB staining as for Figure 3D. (C) Reduced ATP production. PC3 cells were transfected with CPT1C or sicontrol siRNA and cultured in glucose-free medium for the indicated times. ATP production was evaluated as for Figure 3C. (D) Sensitivity to glycolytic inhibition. MCF-7 cells were transfected with control siRNA or CPT1C siRNA1 or siRNA2 and the indicated concentrations of 2-DG were added at 24 h post-transfection. After 5 d culture, cell growth was measured by SRB staining as for Figure 3D.
To test whether loss of CPT1C also hindered growth in a hypoxic environment, MCF-7 cells transfected with control or CPT1C-specific siRNA were grown for up to 3 d in 0.2% oxygen. Fewer cells were detected in cultures of CPT1C siRNA-expressing cells than in cultures of control siRNA-expressing cells after 1 d in hypoxia, and the magnitude of this difference increased as the time in hypoxia was extended (Fig. 4B). Similar results were obtained when CPT1C was depleted in Hs578T cells (Supplemental Fig. S3B). CPT1C depletion did not affect the growth of either MCF-7 or Hs578T cells under normoxia (Supplemental Fig. S3C,D). Notably, under conditions of glucose deprivation, ATP production was markedly impaired in MCF-7 cells transfected with CPT1C siRNA (Fig. 4C).
Our earlier results implied that CPT1C likely stimulates ATP production by effects on FAO. We therefore investigated whether CPT1C depletion sensitized cells to a glycolysis inhibitor. We cultured MCF-7 cells transfected with one of two different CPT1C siRNAs (or control siRNA) for 5 d in the presence of increasing doses of 2-deoxyglucose (2-DG). At intermediate doses of 2-DG, the CPT1C-depleted cells showed significantly reduced growth compared with controls (Fig. 4D). Similar results were obtained with CPT1C-depleted A549 human lung cancer cells (Supplemental Fig. S3E). These data support our hypothesis that CPT1C expression can facilitate ATP production from FAO and enhance the growth of cancer cells subjected to various forms of metabolic stress.
CPT1C depletion reduces tumor growth in xenograft models
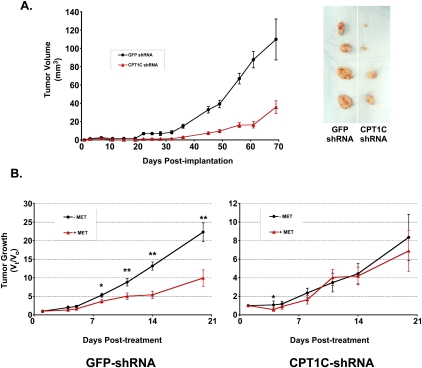
Because many solid tumors show the ability to overcome metabolic stress, we examined the impact of CPT1C depletion on tumor growth in mouse xenograft models. We generated CPT1C shRNA-expressing retroviruses based on our CPT1C siRNA1 sequence and transduced them—or retroviruses expressing control GFP-shRNA—into the MDA-MB-468 human breast cancer cell line. These cells were then implanted into nude mice and tumor growth was monitored for ∼10 wk. Tumors arising from cells expressing CPT1C shRNA grew much more slowly than did control tumors (Fig. 5A), indicating that CPT1C contributes significantly to solid tumor growth.
Figure 5.
Sustained depletion of CPT1C reduces tumor growth in xenografts. (A) CPT1C depletion inhibits human breast cancer cell growth in vivo. MDA-MB-468 cells were infected with retroviruses expressing pRS-Cpt1c shRNA or pRS-GFP shRNA (control) and injected s.c. into nude mice (n = 5 per group). Tumors were measured twice per week for ∼10 wk. (Left) Results are the mean tumor volume ± SD of all tumors in a group on the indicated day. (Right) Representative images of tumors after excision on day 70 post-implantation. (B) CPT1C-depleted tumors are not responsive to metformin treatment in vivo. HCT116 cells were infected with retroviruses expressing pRS-GFP shRNA or pRS-CPT1C shRNA, and the infected cells were injected s.c. into nude mice (n = 5 per group). The animals were treated once daily with PBS or metformin (250 mg/kg) for 20 d. Results are the mean relative tumor growth of all tumors in a group on the indicated day after treatment.
We next examined whether depletion of CPT1C in a xenograft model could alter the effects of the AMPK agonist metformin on tumors. Metformin is thought to exert its anti-cancer effects by reducing free glucose within the tumor microenvironment; however, this agent concurrently enhances cellular responses to metabolic stress. We introduced CPT1C or control shRNA into HCT116 cells, implanted them in nude mice, and treated the animals with PBS or metformin for 21 d. We found that the CPT1C-depleted tumors grew much more slowly than controls and were not responsive to metformin treatment (Fig. 5B). In order to confirm that this metformin was indeed active within xenografted mice, we isolated tumors from metformin-treated mice implanted with untransduced HCT116 cells and immunoblotted protein lysates to examine the status of AMPK. An increase in active phospho-AMPK was detected, as well as enhanced phosphorylation of the AMPK substrate ACC (Supplemental Fig. S4). Thus, metformin successfully activated AMPK in cancer cells, as expected.
Taken together, these data indicate that depletion of CPT1C reduces the growth of tumors in vivo, and imply that CPT1C may function in a pathway downstream from metformin in cancer cell metabolic transformation.
CPT1C is up-regulated in response to hypoxia in vivo
Our data indicated that CPT1C expression provided a growth advantage to tumor cells under conditions of metabolic stress. We therefore hypothesized that such stress might alter the regulation of the CPT1C gene. To test this theory, we first cultured HCT116 cells in 0.2% oxygen for 72 h and monitored levels of CPT1C protein. Immunoblotting was performed using an antiserum raised against a CPT1C peptide and validated in mutant mouse tissues (see Fig. 7B, below). We found that CPT1C protein was clearly induced after 24 h in hypoxia and sustained over 72 h (Fig. 6A). CPT1C protein induction was not apparent for cells in normoxia at any time point. We then investigated whether hypoxia applied in vivo could also up-regulate CPT1C. We subjected tumor-bearing PyMT transgenic mice (Guy et al. 1992) to chronic hypoxia or normoxia (see the Materials and Methods) and analyzed tumors and normal tissues from these animals for EF5 (hypoxia marker) and Cpt1c mRNA. As expected, tumors from hypoxic PyMT mice showed an ∼300% increase in EF5 staining compared with normoxic controls (Supplemental Fig. S5A). Strikingly, in situ hybridization revealed that Cpt1c expression was increased only in EF5+ hypoxic tumors (Fig. 6B). RT–PCR analysis confirmed that three out of four tumors from hypoxic PyMT mice showed higher Cpt1c mRNA expression than did tumors from normoxic controls (Supplemental Fig. S5B). Cpt1c expression was not induced in any normal tissue under hypoxia (Supplemental Fig. S5C,D). Thus, Cpt1c mRNA is induced by hypoxia in vivo in tumor tissues but not in normal tissues, suggesting that low oxygen tension combined with the unique circumstances of the tumor microenvironment can trigger CPT1C up-regulation.
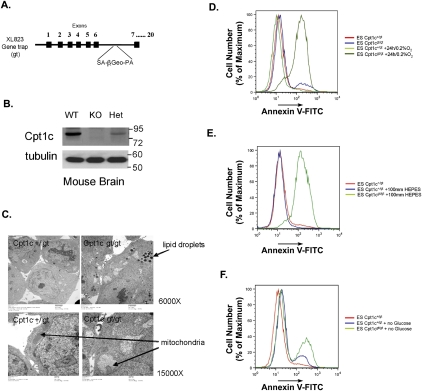
Figure 7.
Characterization of Cpt1c-deficient ES cells. (A) Diagram of the gene trap Cpt1c allele in ES cell clone XL823 (BayGenomics) showing the splice acceptor (SA) site, β-Geo, and polyadenylation site (PA) integrated into intron 6. (B) Immunoblotting of CPT1C in extracts of brain tissues from Cpt1c+/+ (WT), Cpt1cgt/gt (KO), and Cpt1c+/gt (Het) mice. (Tubulin) Loading control. (C) Altered morphology. The morphology of Cpt1c+/gt and Cpt1cgt/gt ES cells was examined by electron microscopy. Cpt1cgt/gt cells show cytoplasmic lipid droplets (top) and swollen mitochondria lacking internal structure (bottom). Results are representative of two trials. (D) Increased death under hypoxia. Cpt1c+/gt and Cpt1cgt/gt cells were cultured for 24 h under normoxia or hypoxia (0.2% O2) and cell death was detected using Annexin V staining and flow cytometry. (E) Hypoxia-induced acidosis does not cause the death of Cpt1cgt/gt cells. Cpt1c+/gt and Cpt1cgt/gt ES cells were treated as in C with the addition of 100 mM HEPES. Cell death was measured as for C. (F) Increased death upon glucose withdrawal. Cpt1c+/gt and Cpt1cgt/gt ES cells were cultured for 48 h in DMEM or DMEM with no glucose. Cell death was measured as for C. For D–F, results are representative of two trials.
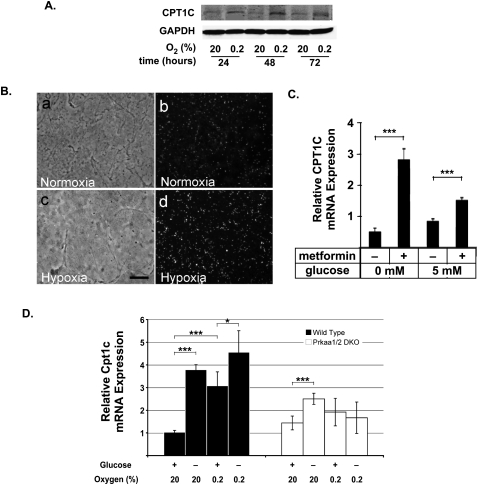
Figure 6.
CPT1C is induced by metabolic stress and regulated by AMPK. (A) Induction of CPT1C protein in a cancer cell line subjected to hypoxia. At 24 h post-seeding, HCT116 cells were cultured in 20% or 0.2% O2 for 24 h, 48 h, or 72 h, and CPT1C protein was detected by immunoblotting. (GAPDH) Loading control. (B) Up-regulation of Cpt1c expression in a hypoxia model in vivo. Tumor-bearing PyMT mice were injected with the hypoxia marker EF5 and subjected to hypoxia or normoxia (see the Materials and Methods). Tumors from the normoxic (panels a,b) and hypoxic (panels c,d) animals were examined by bright-field (panels a,c) and dark-field (panels b,d) microscopy. Cpt1c expression was detected by in situ hybridization. Bar, 100 μm. (C) Metformin treatment induces CPT1C expression. MCF-7 cells were cultured for 48 h in medium with or without 10 mM metformin and/or 5 mM glucose, as indicated. Results are mean relative mRNA levels ± SD normalized to Hprt1 expression. (D) Involvement of AMPK in Cpt1c induction upon glucose withdrawal or hypoxia. SV40-transformed wild-type or AMPK-deficient (Prkα1/α2 double knockout; DKO) MEFs were cultured for 24 h in DMEM with (+) or without (−) 25 mM glucose and in 20% or 0.2% O2, as indicated. Cpt1c mRNA levels were assayed by quantitative PCR. Results are mean relative mRNA levels ± SD normalized to Hprt1 expression.
CPT1C mRNA can be regulated by AMPK
To investigate how CPT1C might be regulated in cells exposed to hypoxia or other metabolic stress, we examined Cpt1c mRNA induction in cultured mouse embryonic fibroblasts (MEFs). Since AMPK is a central regulator of metabolic stress, we first determined whether metformin treatment could induce CPT1C mRNA expression in MCF-7 cells. Indeed, metformin increased CPT1C mRNA levels in these cells in both the presence and absence of 5 mM glucose (Fig. 6C). We then determined Cpt1c mRNA levels in MEFs carrying a compound mutation of Prkaa1 and Prkaa2, the genes encoding the α subunits of mouse AMPK. Whereas wild-type MEFs showed an approximately threefold to fivefold induction of Cpt1c mRNA over controls under conditions of glucose deprivation and/or hypoxia, Cpt1c induction was impaired in hypoxic AMPK-deficient MEFs (Fig. 6D). A modest level of Cpt1c induction occurred in AMPK-deficient MEFs deprived of glucose, implying that Cpt1c transcription is also regulated by an AMPK-independent mechanism. These data show that the induction of CPT1C expression in response to metabolic stress is at least partly dependent on AMPK.
Loss of Cpt1c function leads to alterations in apoptosis, FAO, and ATP production
To understand how CPT1C might protect cells against metabolic stress, we analyzed a murine embryonic stem (ES) cell line (clone XL823; BayGenomics) heterozygous for a single gene trap vector insertion into intron 6 of the Cpt1c gene (Fig. 7A) that prematurely terminates Cpt1c transcription. We generated ES cells homozygous for the gt mutation (Supplemental Fig. S6A) and verified their loss of Cpt1c mRNA using real-time RT–PCR and primers specific for sequences within exons 3, 7, or 9 of the Cpt1c gene. Cpt1cgt/gt ES cells were hypomorphic but retained <1% of normal expression levels of the full-length Cpt1c mRNA (Supplemental Fig. S6B). Immunoblotting of brain tissue of Cpt1cgt/gt mice generated from these ES cells confirmed the absence of CPT1C protein (Fig. 7B).
Despite the normal health and life span of Cpt1cgt/gt mice, long-term cultures of Cpt1cgt/gt ES cells showed decreased total cell numbers (Supplemental Fig. S6C) and reduced ATP production (Supplemental Fig. S6D) compared with Cpt1c+/gt cells. Enhanced apoptosis, increased caspase-3 and caspase-9 activation, and impaired mitochondrial membrane potential were also observed (Supplemental Table S1). Most striking was a dramatic difference in lipid composition between Cpt1cgt/gt and Cpt1c+/gt ES cells, in that Cpt1c deficiency led to increased levels of linoleic, arachidonic, and docosotetraenoic acids, and decreased levels of oleic acid (Supplemental Table S2). Electron microscopy of Cpt1cgt/gt cells revealed the presence of swollen mitochondria exhibiting abnormal internal membrane structure and loss of internal cristae density (Fig. 7C). The mutant mitochondria also contained small vesicles not found in the mitochondria of Cpt1c+/gt cells. Intriguingly, the cytoplasm of Cpt1cgt/gt cells showed an accumulation of lipid droplets that was not present in either Cpt1c+/gt cells (Fig. 7C) or wild-type ES cells (data not shown). These results are consistent with our siRNA experiments and demonstrate that Cpt1c deficiency impairs FA metabolism, reduces ATP production, and increases apoptosis.
CPT1C protects ES cells from hypoxia-induced apoptosis
We next examined the responses of Cpt1c+/gt and Cpt1cgt/gt ES cells to hypoxia and glucose deprivation. When cultured for 24 h under hypoxia, 79% of Cpt1cgt/gt cells underwent apoptosis, whereas only 11% of Cpt1c+/gt cells did so (Fig. 7D). Growth under hypoxia enhances the acidosis of cultured cells, but the addition of HEPES buffer to the medium did not prevent the excessive death of hypoxic Cpt1cgt/gt cells (Fig. 7E). Similarly, withdrawal of glucose resulted in the death of 34% of Cpt1cgt/gt cells and 17.8% of Cpt1c+/gt cells but only 5% of control wild-type ES cells (Fig. 7F; data not shown). Thus, loss of CPT1C function renders ES cells prone to apoptosis under conditions of metabolic stress.
Discussion
Solid tumors frequently contain regions of poor oxygenation and experience metabolic stress (Brown and Wilson 2004; Gatenby and Gillies 2004; Pan and Mak 2007). Cancer cells in these regions are driven to adapt to survive and grow under these conditions, and this metabolic transformation usually takes the form of increased glycolysis (Pelicano et al. 2006; Shaw 2006). However, other cellular components can serve as energy sources for cancers when nutrient depletion or other metabolic stress triggers the activation of alternative pathway(s) to maintain bioenergetic supply (Pan and Mak 2007). We sought to target genes in these alternative metabolic pathways, and demonstrate here that CPT1C is a novel gene that protects cells from metabolic stress.
Our work shows that CPT1C is a potential alternative energy supply gene that is induced in tumors during the process of metabolic transformation. Constitutive expression of CPT1C increases FAO and ATP generation, whereas depletion of CPT1C decreases ATP production. Knockdown of CPT1C in human cancer cell lines reduces growth in vitro under conditions of hypoxia or limited glucose, as well as new tumor formation in vivo. Moreover, Cpt1c mRNA is induced by hypoxia and glucose deprivation in murine cells as well as in tumors, and this regulation depends on a mechanism that involves activated AMPK. Finally, murine ES cells lacking Cpt1c exhibit mitochondrial membrane abnormalities, altered lipid homeostasis, decreased growth, and increased caspase-mediated cell death. Taken together, our results suggest that CPT1C may be a regulator of FA homeostasis and involved in the modulation of bioenergetics that occurs in tumors experiencing metabolic stress.
Our results expand our knowledge of the function of this unusual CPT family member. CPT1C is expressed predominantly in normal mammalian brains, particularly in neurons (Price et al. 2002; Wolfgang et al. 2006, 2008; Sierra et al. 2008). It has been suggested previously that CPT1C plays an important role in maintaining the energy homeostasis of the whole mouse (Wolfgang et al. 2006, 2008). Our finding that CPT1C may mediate the adaptation of nonneuronal tumor cells exposed to metabolic stress suggests that CPT1C has functions at both the organismal level (controlling nutrient intake and energy homeostasis) and the cellular level (protection against acute metabolic stress). It is not yet clear why this particular CPT1 gene is induced in malignancies, but we speculate that its gene product may act on uncharacterized FA substrates that may be very beneficial for cell survival during metabolic stress. Consistent with this hypothesis, the FA composition of cells is altered in cells lacking CPT1C.
Three lines of evidence arising from our study suggest that one link between CPT1C and tumorigenic adaptation may be mediated through AMPK. First, mutant MEFs lacking AMPK showed impaired Cpt1c mRNA induction in response to hypoxia or glucose deprivation. Second, metformin, an AMPK agonist, was able to induce Cpt1c expression. Third, Cpt1c originally came to our attention in a screen designed to detect p53-related transcripts. Since AMPK and p53 reportedly activate each other (Jones et al. 2005; Bungard et al. 2010), we propose that a p53–AMPK–CPT1C axis exists for sensing and responding to metabolic stress. This p53–AMPK–CPT1C axis might permit the adaptive utilization of FA as a fuel source to support cell growth. In tumors where p53 is lost, AMPK may work with additional factors to induce CPT1C and maintain cell survival. Consistent with this hypothesis, we showed that CPT1C depletion sensitized cancer cells expressing either wild-type or mutant p53 to glycolytic inhibition, glucose deprivation, or hypoxia. Conversely, ectopic expression of CPT1C in cells expressing wild-type p53 conferred resistance to glucose deprivation or hypoxia. However, the induction of Cpt1c mRNA by glucose withdrawal was not completely eliminated in cells lacking AMPK, suggesting that the p53–AMPK–CPT1C axis likely involves additional, still unidentified players.
Our results are consistent with several reports emphasizing the emerging importance of FA metabolism in tumorigenesis (Menendez and Lupu 2007; Hirsch et al. 2010; Nomura et al. 2010). MAGL up-regulation has been documented in aggressive cancer cell lines and correlates with increased free FAs and FA metabolites. Moreover, pathological MAGL expression can be replaced by the provision of high fat levels both in vitro and in vivo, indicating that free FAs are themselves pathogenic (Nomura et al. 2010). We showed that cancer cells under metabolic stress express CPT1C, and that this CPT1C expression can alter FA homeostasis. It seems likely that FAs freed by CPT1C-driven events might be used as a fuel source to drive metabolic transformation and tumor growth. Under normal nutrient conditions, we speculate that CPT1C-mediated FAO induction in a tumor might actually impair its growth because the availability of the FAs needed to serve as membrane precursors might be reduced. Thus, as tumors progress, CPT1C induction may be restricted to areas where nutrient conditions are poor and the bioenergetics of the cell demand fuel from the alternative source represented by FAs.
Our data indicate that CPT1C may be an attractive target for therapeutic intervention where tumors are hypoxic and deprived of nutrient sources. We showed in human lung tumors that CPT1C mRNA is up-regulated independently of p53, and in mouse tumors that Cpt1c expression correlates inversely with mTOR pathway activation and tumor sensitivity to rapamycin. Our xenograft experiments demonstrated that CPT1C depletion greatly retards the growth of both breast cancer- and colon cancer-derived tumors. These results suggest that a CPT1C inhibitor, used either as a monotherapy or in combination with other anti-cancer agents, may be a promising new avenue of cancer treatment.
Materials and methods
Cell lines
Cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). Human cancer cell lines MCF-7, Hs578T, A549, PC3, MDA-MB-468, and HCT116 (Bunz et al. 1998) and mouse leukemia cell line DP16.1 were obtained from various sources. MCF-7, A549, PC3, and HCT116 cells are known to carry wild-type p53; MDA-MB-468 and Hs578T cells express mutant p53 R273H and V157F, respectively. DP16.1 lacks both alleles of p53. AMPKα-deficient MEFs and matched controls were transformed with SV40 transforming region (Laderoute et al. 2006). XL823, a gene trap ES cell line targeting Cpt1c (BayGenomics), was maintained as described previously (Okada et al. 2002).
Murine primary mammary tumors
Primary mammary tumors of 129SV origin Ink4a/ARF−/−; Tet-O-Her2V66E; MMTV-rtTA ES cells were induced in adult chimeric mice by tetracycline administration. Tumors were surgically resected and minced. Tumor cells were isolated from individual tumors and passaged in Matrigel (BD BioSciences) prior to in vivo propagation for two rounds of s.c xenografting into NCR-nude mice. Tumors were then cryopreserved for later analysis.
For microarray analysis (see legend for Supplemental Fig. S1), individual primary tumor sections were thawed and dissociated, and ∼1 × 105 cells were injected s.c. into BalbC-nude mice (approximately five mice per tumor line). When these tumors reached a volume of ∼500–800 mm3, mice were sacrificed and tumors were surgically removed.
mTOR index
The mTOR index was determined based on the method of Perou et al. (2000). A simple Pearson correlation algorithm was used to identify components of the mTOR pathway exhibiting correlated expression, as described in the legend for Supplemental Figure S1.
Human lung tumor and normal lung tissue samples
Matched tumor and normal lung tissue samples were harvested from 19 NSCLC patients treated at the UHN by surgical resection without adjuvant chemotherapy. Tumor samples were snap-frozen and stored in liquid nitrogen prior to RNA extraction. p53 IHC was performed on tumor tissues that were formalin-fixed and paraffin-embedded.
RNA isolations
Unless otherwise stated, RNA was extracted from cultured cells using the standard protocols of the RNeasy Kit (Qiagen). RNA was extracted from NSCLC samples using phenol-chloroform and was purified using the RNeasy kit (Qiagen). For mouse tumors, tissue was homogenized in Trizol reagent (Invitrogen) and chloroform-extracted, and RNA was prepared using the RNeasy kit.
Blotting
Southern and Northern blotting were performed according to standard protocols (Sambrook et al. 1989). For immunoblotting, standard protocols (Sambrook et al. 1989) were performed using affinity-purified rabbit polyclonal antiserum directed against a 15-amino-acid C-terminal peptide of CPT1C, anti-GADPH antibody (MAB374, Millipore), anti-tubulin antibody (AA2, Millipore), or anti-Flag antibody (F1804, Sigma).
Real-time PCR
Unless otherwise stated, RNA was reverse-transcribed using the SuperScript II kit (Invitrogen). Real-time PCR was performed using an SDS 7900 (Becton Dickinson), an ABI 7900HT, or an ABI PRISM 7900-HT (Applied Biosystems) plus SYBR Green reagent (Applied Biosystems). Raw values were normalized against external control housekeeping genes (Gapdh, Hprt, or β-actin) for the same cDNA sample. Specific primer sequences are available on request.
Sulforhodamine B (SRB) cell proliferation assay
Cells were fixed in situ using cold 10% trichloroacetic acid (TCA). Plates were washed, 50 μL of SRB solution (0.4% w/v) dissolved in 1% (v/v) acetic acid was added to each well, and plates were incubated for 30 min at room temperature. SRB stains were solubilized in 10 mM Tris-HCl (pH 10.5) and absorbance was read at 570 nm. Cell numbers were calculated based on standard curves of known cell dilutions.
siRNA transfection
For CPT1C depletion in human cancer cell lines, cells were seeded into 96-well plates at 1500–2500 cells per well. At 24 h post-seeding, cells were transfected with 10 nM siRNA using Lipofectamine. CPT1C RNA knockdown efficiency was determined as follows: Four CPT1C siRNAs (Dharmacon) were transfected either individually into seeded cells at 40 nM each or as a pool of 10 nM each. Transfection of 40 nM siRNA targeting luciferase was used as the negative control (sicontrol). Quantitative RT–PCR was performed as described above at 72 h post-transfection. The sequence of CPT1C siRNA1 was 5′-GAAAUCCGCUGAUGGUGAA-3′, and CPT1C siRNA2 was 5′-GACAAAUCCUUCACCCUAA-3′.
Metabolic stress stimuli, rapamycin, and metformin
Cells were subjected to metabolic stress and control conditions as follows. Transfected cells were treated at 24 h post-transfection. Unless otherwise stated, cells were incubated at 37°C in atmospheric oxygen. For hypoxia, cells were incubated in 0.2% O2 in a hypoxia incubator (ESBE Scientific) for 1, 2, or 3 d before being transferred back to normoxia for 4, 3, or 2 d, respectively. For glucose deprivation, cells were incubated in medium without glucose, or this medium supplemented with a stock glucose solution to the concentrations indicated in the figures. 2-DG (Sigma) and metformin (Sigma) were prepared as stock solutions in PBS and diluted to the required concentrations in standard medium. Rapamycin (Chemietek) was prepared as a stock solution in DMSO and diluted to the required concentrations in standard medium. The appropriate vehicle controls were used for all experiments involving chemical treatments.
FAO
MCF-7 cells (2 × 106) were incubated for 1 h in Krebs Ringer buffer with 5 mM glucose. Washed cells were resuspended in buffer containing 0.5% BSA and 1 μCi [1-14C] palmitic acid (GE Healthcare/Amersham) but no glucose. Cells were seeded in the center wells of organ culture dishes (Falcon 353037), the lids were sealed with vacuum grease, and the dishes were incubated for 4 h at 37°C in 5% CO2. Upon removal of the dishes from the incubator, 1 mL of 1 M NaOH was pipetted through a hole in the lid into the outer ring of each dish, and 300 μL of 2 N HCl was pipetted into each center well. The holes were resealed with Scotch tape, and the radioactive CO2 released from the medium in the center well was collected overnight at room temperature. The following morning, 800 μL of NaOH from the outer ring of the dish was added to 5 mL of Ecoscint to determine counts per minute (cpm) attributable to 14CO2 production.
ATP production
Cells were cultured in low glucose (5.6 mM) DMEM containing 10% FBS and seeded into 96-well plates. For CPTC1 loss of function, cells were transfected at 24 h post-seeding with 40 nM siRNAs as above. At 48 h post-transfection, the medium was changed to PBS and ATP levels were measured from time 0 to 24 h using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Cell numbers were measured by direct cell counting for each time point and used to normalize ATP production levels. Luminescence was measured using a SpectraMax M5 (Molecular Devices).
In situ hybridization
In situ hybridization was performed as described (Hui and Joyner 1993; Skinnider et al. 2001). The full-length mouse Cpt1c ORF was used as the probe.
In vivo tumor cell exposure to hypoxia
MMTV-PyMT634Mul mice were obtained from the Animal Resource Center of the Ontario Cancer Institute. At age 3 mo, tumor-bearing females were i.p. injected with 0.01 mL/g 10 mM EF5 (2-[2-nitro-1H-imidazol-1-yl]-N-2,2,3,3,3-pentafluoropropyl acetamide), which was kindly provided by Dr. Cameron Koch (University of Pennsylvania). The injected mice were randomly allocated to the normoxia (n = 4) or chronic hypoxia (n = 5) groups and sealed into air-tight chambers (Billups-Rothenberg) that were filled with either outside atmosphere or a humidified mixture of 7% oxygen in nitrogen, respectively. At ∼3.5 h post-gassing, mice were sacrificed and tumors of <10 mm in diameter were excised and snap-frozen in liquid nitrogen for subsequent analysis. Levels of EF5 within tumors were quantified by IHC using anti-ELK3-51 antibody (provided by Dr. C. Koch). The total area of EF5+ staining in a tumor section (excluding areas of necrosis and connective tissue) was quantified using the positive pixel algorithm of Aperio ImageScope (Aperio Technologies).
In vivo shRNA inhibition of tumor growth
The pRS shRNA expression cassettes (Origene) encoding shRNA directed against CPT1C (sense 5′-CGGACTATGTTTCCTCAGGCGGTGGATTC-3′) or GFP (catalog no. TR30001; control) were packaged into amphotropic retroviruses using transient transfection of Phoenix cells via FuGENE 6 reagent (Roche). Culture supernatants were collected 2 d after transfection and filtered (0.45 μm). For retroviral infection, MDA-MB-468 cells or HCT116 cells were cultured for 24 h in 1:1 Phoenix conditioned medium (DMEM, 10% FBS, 8 μg/mL polybrene; Sigma-Aldrich). Transfection was repeated three times to increase efficiency. Infected cells were trypsinized, counted, and injected s.c. into nude mice at 5 × 106 (MDA-MB-468) or 3 × 106 (HCT116) cells per mouse (five mice per group). Tumor volumes were measured in situ twice weekly, and viable tumor volumes were calculated assuming ellipsoid growth patterns.
Flow cytometry analyses
Cell cycle analysis of ES cells was performed using the BrdU Flow kit (BD Bioscience). Shifts in mitochondrial membrane potential were detected using a standard protocol and JC-1 (Stratagene). Cell fluorescence was detected and analyzed using a flow cytometer (FACSCalibur, Becton Dickinson), CellQuest, and FlowJo software according to standard protocols. Active caspase-3 was detected by flow cytometry using the BD Bioscience kit (BD PharMingen). Cleaved caspase-9 was detected using carboxy-fluorescein-labeled caspase inhibitors (B-Bridge International, Inc.). Apoptosis was measured by standard Annexin V (BD Bioscience) and propidium iodide staining.
Electron microscopy
Cpt1cgt/gt and Cpt1c+/gt ES cells were fixed in 0.1 M phosphate buffer containing 4% formaldehyde and 0.5% glutaraldehyde, and treated with 1% osmium tetraoxide. After dehydration in ethanol gradients and a polymerization step, tissue sections of 70 nm were examined using standard protocols.
Statistics
The paired t-test or unpaired t-test was used for comparisons where appropriate. For statistical interpretation, P < 0.05 (*) is considered significant, P < 0.01 (**) is considered highly significant, and P < 0.005 (***) is considered very highly significant.
Ethical statements
The use of human samples and their associated clinical information was approved by the UHN Research Ethics Board. All mice were maintained in compliance with the guidelines of the Canadian Council on Animal Care.
Acknowledgments
We thank members of the Mak and Pan laboratories for insightful comments, M. Saunders for scientific editing, and S. McCracken, R. Cairns, and B. Wouters for helpful suggestions. This work was supported by grants from the Forschungskredit of the University of Zurich and Oncosuisse (to K.Z.), the Canadian Cancer Society (to M.T.), the Princess Margaret Hospital Foundation (to J.G.P. and T.W.M.), and the Canadian Institutes of Health Research (to T.W.M).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1987211.
Supplemental material is available for this article.
References
- Brown JM, Wilson WR 2004. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer 4: 437–447 [DOI] [PubMed] [Google Scholar]
- Buck E, Eyzaguirre A, Brown E, Petti F, McCormack S, Haley JD, Iwata KK, Gibson NW, Griffin G 2006. Rapamycin synergizes with the epidermal growth factor receptor inhibitor erlotinib in non-small-cell lung, pancreatic, colon, and breast tumors. Mol Cancer Ther 5: 2676–2684 [DOI] [PubMed] [Google Scholar]
- Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL 2010. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329: 1201–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497–1501 [DOI] [PubMed] [Google Scholar]
- Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB 2007. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res 67: 6745–6752 [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ 2004. Why do cancers have high aerobic glycolysis?. Nat Rev Cancer 4: 891–899 [DOI] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12: 954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Joshi A, Zhang Y, Jaeger SA, Bulyk M, Tsichlis PN, Shirley Liu X, Struhl K 2010. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell 17: 348–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui CC, Joyner AL 1993. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat Genet 3: 241–246 [DOI] [PubMed] [Google Scholar]
- Ide T, Brown-Endres L, Chu K, Ongusaha PP, Ohtsuka T, El-Deiry WS, Aaronson SA, Lee SW 2009. GAMT, a p53-inducible modulator of apoptosis, is critical for the adaptive response to nutrient stress. Mol Cell 36: 379–392 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18: 283–293 [DOI] [PubMed] [Google Scholar]
- Kerner J, Hoppel C 2000. Fatty acid import into mitochondria. Biochim Biophys Acta 1486: 1–17 [DOI] [PubMed] [Google Scholar]
- Laderoute KR, Amin K, Calaoagan JM, Knapp M, Le T, Orduna J, Foretz M, Viollet B 2006. 5′-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol Cell Biol 26: 5336–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y 2006. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis 9: 230–234 [DOI] [PubMed] [Google Scholar]
- Maglione JE, Moghanaki D, Young LJ, Manner CK, Ellies LG, Joseph SO, Nicholson B, Cardiff RD, MacLeod CL 2001. Transgenic Polyoma middle-T mice model premalignant mammary disease. Cancer Res 61: 8298–8305 [PubMed] [Google Scholar]
- Menendez JA, Lupu R 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 7: 763–777 [DOI] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF 2010. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Suh WK, Jin J, Woo M, Du C, Elia A, Duncan GS, Wakeham A, Itie A, Lowe SW, et al. 2002. Generation and characterization of Smac/DIABLO-deficient mice. Mol Cell Biol 22: 3509–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JG, Mak TW 2007. Metabolic targeting as an anticancer strategy: dawn of a new era?. Sci STKE 2007: pe14 doi: 10.1126/stke.3812007pe14 [DOI] [PubMed] [Google Scholar]
- Pelicano H, Martin DS, Xu RH, Huang P 2006. Glycolysis inhibition for anticancer treatment. Oncogene 25: 4633–4646 [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. 2000. Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Plas DR, Thompson CB 2005. Akt-dependent transformation: there is more to growth than just surviving. Oncogene 24: 7435–7442 [DOI] [PubMed] [Google Scholar]
- Price N, van der Leij F, Jackson V, Corstorphine C, Thomson R, Sorensen A, Zammit V 2002. A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics 80: 433–442 [DOI] [PubMed] [Google Scholar]
- Ramsay RR, Zammit VA 2004. Carnitine acyltransferases and their influence on CoA pools in health and disease. Mol Aspects Med 25: 475–493 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Shaw RJ 2006. Glucose metabolism and cancer. Curr Opin Cell Biol 18: 598–608 [DOI] [PubMed] [Google Scholar]
- Sierra AY, Gratacos E, Carrasco P, Clotet J, Urena J, Serra D, Asins G, Hegardt FG, Casals N 2008. CPT1c is localized in endoplasmic reticulum of neurons and has carnitine palmitoyltransferase activity. J Biol Chem 283: 6878–6885 [DOI] [PubMed] [Google Scholar]
- Skinnider BF, Elia AJ, Gascoyne RD, Trumper LH, von Bonin F, Kapp U, Patterson B, Snow BE, Mak TW 2001. Interleukin 13 and interleukin 13 receptor are frequently expressed by Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 97: 250–255 [DOI] [PubMed] [Google Scholar]
- Swinnen JV, Brusselmans K, Verhoeven G 2006. Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care 9: 358–365 [DOI] [PubMed] [Google Scholar]
- Wolfgang MJ, Kurama T, Dai Y, Suwa A, Asaumi M, Matsumoto S, Cha SH, Shimokawa T, Lane MD 2006. The brain-specific carnitine palmitoyltransferase-1c regulates energy homeostasis. Proc Natl Acad Sci 103: 7282–7287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MJ, Cha SH, Millington DS, Cline G, Shulman GI, Suwa A, Asaumi M, Kurama T, Shimokawa T, Lane MD 2008. Brain-specific carnitine palmitoyl-transferase-1c: role in CNS fatty acid metabolism, food intake, and body weight. J Neurochem 105: 1550–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Kumabe T, Cho YY, Watanabe M, Kawagishi J, Yoshimoto T, Fujino T, Kang MJ, Yamamoto TT 2000. Fatty acid induced glioma cell growth is mediated by the acyl-CoA synthetase 5 gene located on chromosome 10q25.1-q25.2, a region frequently deleted in malignant gliomas. Oncogene 19: 5919–5925 [DOI] [PubMed] [Google Scholar]