Abstract
Essential messenger RNA (mRNA) export factors execute critical steps to mediate directional transport through nuclear pore complexes (NPCs). At cytoplasmic NPC filaments, the ATPase activity of DEAD-box protein Dbp5 is activated by inositol hexakisphosphate (IP6)-bound Gle1 to mediate remodeling of mRNA–protein (mRNP) complexes. Whether a single Dbp5 executes multiple remodeling events and how Dbp5 is recycled are unknown. Evidence suggests that Dbp5 binding to Nup159 is required for controlling interactions with Gle1 and the mRNP. Using in vitro reconstitution assays, we found here that Nup159 is specifically required for ADP release from Dbp5. Moreover, Gle1-IP6 stimulates ATP binding, thus priming Dbp5 for RNA loading. In vivo, a dbp5-R256D/R259D mutant with reduced ADP binding bypasses the need for Nup159 interaction. However, NPC spatial control is important, as a dbp5-R256D/R259D nup42Δ double mutant is temperature-sensitive for mRNA export. Further analysis reveals that remodeling requires a conformational shift to the Dbp5–ADP form. ADP release factors for DEAD-box proteins have not been reported previously and reflect a new paradigm for regulation. We propose a model wherein Nup159 and Gle1-IP6 regulate Dbp5 cycles by controlling its nucleotide-bound state, allowing multiple cycles of mRNP remodeling by a single Dbp5 at the NPC.
Keywords: nucleocytoplasmic transport, DEAD-box proteins, nucleotide exchange factors, ADP
Dynamic changes in the composition and assembly of messenger RNA (mRNA)–protein interactions underlie many of the central events in eukaryotic gene expression and regulation (Moore 2005). These events include mRNA synthesis, processing, nuclear export, trafficking, translation, and turnover. Members of a family of conserved enzymes, the DEAD-box proteins (Dbps) numbering >20 in Saccharomyces cerevisiae and human cells, are critical for multiple steps in the mRNA life cycle (Rocak and Linder 2004; Cordin et al. 2006). The Dbps are RNA-dependent ATPases that act via helicase activity for unwinding RNA duplexes and/or remodeling activity for altering the protein composition of mRNA–protein (mRNP) complexes (Fairman et al. 2004; Jankowsky and Bowers 2006). The function of these specific Dbp activities must be controlled both spatially and temporally to ensure proper gene expression. However, the mechanisms for regulating Dbps are not fully defined, leaving a gap in our understanding of gene expression.
One of the key steps in the gene expression pathway is the exit of mRNPs from the nucleus (Kelly and Corbett 2009; Stewart 2010; Rodriguez-Navarro and Hurt 2011). This occurs via nuclear pore complexes (NPCs), >60-MDa structures that form channels spanning the nuclear envelope (Brohawn et al. 2009). Each export-competent mRNP has many associated proteins, including those that directly bind mRNA and a heterodimeric transport receptor (Mex67–Mtr2 in S. cerevisiae; TAP/NXF1–p15/NXT1 in metazoan) that mediates docking of mRNPs to NPCs and their translocation (Stewart 2007b; Kelly and Corbett 2009). During mRNA export, protein–protein and protein–mRNA interactions in the mRNP are altered through a remodeling mechanism (Daneholt 2001). For example, in S. cerevisiae, the export receptor Mex67–Mtr2 and the nuclear poly(A)-binding protein Nab2 are removed from the mRNP (Lund and Guthrie 2005; Tran et al. 2007) by Dbp5, an essential Dbp (hDbp5/DDX19B in human cells) (Snay-Hodge et al. 1998; Tseng et al. 1998; Schmitt et al. 1999). At steady state in S. cerevisiae cells, Dbp5 is localized at the NPC through a direct interaction with Nup159/Rat7 (Nup214 in human cells) (Hodge et al. 1999; Schmitt et al. 1999). Nup159 resides exclusively in filaments extending from the NPC cytoplasmic face, and thus it allows localization of Dbp5 for action during terminal steps in mRNP export (Kraemer et al. 1995; Hurwitz et al. 1998; Hodge et al. 1999; Schmitt et al. 1999). However, Dbp5 alone is a weak ATPase and requires inositol hexakisphosphate (IP6)-bound Gle1 for activation (Alcazar-Roman et al. 2006; Weirich et al. 2006). Spatial control of Dbp5 activation is thought to be achieved by Gle1 docking at Nup42 (hCG1 in human), which is adjacent to Nup159 in the NPC cytoplasmic filaments (Strahm et al. 1999; Miller et al. 2004; Kendirgi et al. 2005; Alber et al. 2007). It is speculated that multiple cycles of Gle1-IP6/Dbp5 action contribute to the translocation of a single mRNP through the NPC (Stewart 2007). However, the order of events in mRNP export at the NPC cytoplasmic filaments has not been resolved.
Several recent structural studies give insight into the mechanism by which Dbp5 cycles between nucleotide, RNA, and Nup159 binding. First, our previous studies suggest that Dbp5ATP and Dbp5ADP have distinct conformations, as detected by limited trypsin digestion (Tran et al. 2007). In addition, structural studies of hDbp5 and the N-terminal domain (NTD) of yeast Dbp5 support this conclusion and indicate that Dbp5 undergoes dramatic nucleotide-dependent conformational changes between the AMP-PNP (a nonhydrolyzable ATP analog) and ADP-bound states (Collins et al. 2009; Fan et al. 2009). Intriguingly, in a reconstituted in vitro system, Dbp5 remodels a Nab2-RNP in the presence of ADP alone. This suggests that ADP binding or the Dbp5–ADP-bound (Dbp5ADP) state is functional (Tran et al. 2007). However, both ATP hydrolysis and nucleotide binding are required for in vivo function, as dbp5 mutants lacking either function are recessive lethal (Hodge et al. 2011). It is unknown how the in vitro ADP-dependent remodeling activity is linked to the Dbp5 mechanism for ATP hydrolysis in a cell.
Second, crystallographic analysis of hDbp5 has also revealed that the Nup214- and RNA-binding sites overlap and are mutually exclusive (von Moeller et al. 2009). Moreover, ATP-bound Dbp5 (Dbp5ATP), ADP-bound Dbp5 (Dbp5ADP), and nucleotide-free Dbp5 (apo-Dbp5) states are predicted to have differential Nup159- and RNA-binding affinities. This is based on the Nup214 affinity for apo-hDbp5 being greater than that for hDbp5ATP (von Moeller et al. 2009), and Dbp5ATP affinity for RNA being greater than Dbp5ADP (Weirich et al. 2006; Tran et al. 2007). These results have raised the question of whether Nup159 removes RNA from Dbp5, or whether RNA dissociates Dbp5 from Nup159. Some evidence suggests that Nup159 binding is a first step for positioning Dbp5 to interact with Gle1 and the mRNP at the NPC. The NTD (residues 1–387) of Nup159 is necessary and sufficient for Dbp5 binding, and nup159 mutants lacking Dbp5 binding are defective for mRNA export (Hodge et al. 1999; Schmitt et al. 1999). However, there is also evidence that Dbp5 can potentially associate with mRNA in the nucleus, and also that it acts in the cytoplasm during translation. Immunoelectron microscopy studies of the large Balbiani ring mRNP in the salivary glands of Chironomus tentans found Dbp5 present in this mRNP both cotranscriptionally and throughout the export process (Zhao et al. 2002). Genetic and physical interaction studies have also linked Dbp5 to transcription factor IIH function (Estruch and Cole 2003). During translation in S. cerevisiae, Dbp5 and Gle1-IP6 are required for efficient translation termination (Bolger et al. 2008). Thus, control of Dbp5 activity is likely compartment/site-specific. As Dbp5 binding to Nup159 is unique to the cytoplasmic NPC filaments, understanding its role is critical for determining the specificity and order of steps during mRNP export.
To determine how Dbp5 cycles between remodeling events at the NPC, we directly analyzed Dbp5 in an in vitro reconstituted system and investigated the functional properties of specific dbp5 mutants. We found the Nup159-NTD is specifically required to stimulate the release of ADP from Dbp5. As such, we speculate that Nup159 executes a step following ATP hydrolysis and directly facilitates continued remodeling cycles per Dbp5 at the NPC. This model is supported by the phenotype of a dbp5RR mutant that bypasses the requirement for Nup159 docking and results in a dbp5RR protein with altered ADP binding. However, Gle1 binding to Nup42 in dbp5RR mutant cells is required, as a dbp5RR nup42Δ double mutant is temperature-sensitive and defective for mRNA export. Moreover, in vitro, Gle1-IP6 promotes ATP-binding to Dbp5. Analysis of the dbp5RR protein and a catalytically dead dbp5 (dbp5EQ) in the in vitro mRNP remodeling assay further suggests that the in vitro activity is linked to an apo-Dbp5-to-Dbp5ADP transition that presumably structurally mimics the in vivo Dbp5ATP-to-Dbp5ADP conversion. Taken together, we propose a novel mechanism whereby the Dbp5 nucleotide cycle is regulated and the system favors recycling of Dbp5 at the NPC.
Results
Nup159 promotes Dbp5 release of ADP but not ATP
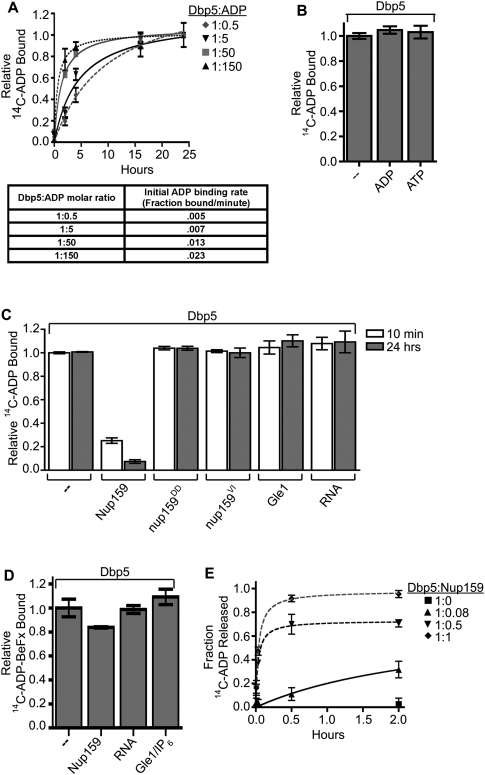
We showed previously that ADP is sufficient for Dbp5-dependent mRNP remodeling in vitro (Tran et al. 2007). To investigate this further, we initiated studies to analyze Dbp5 nucleotide-binding states. First, we characterized ADP binding using equilibrium binding assays. Purified, recombinant Dbp5 was incubated with increasing concentrations of radiolabeled 14C-ADP over a time course of 24 h and binding was measured in a filter-binding assay (Fig. 1A). Initial ADP-binding rates increased with increasing molar ratios of ADP to Dbp5 (Fig. 1A, bottom). However, binding did not plateau for several hours at any ratio tested. This long equilibration time for Dbp5–ADP binding had not been reported previously, and we initially tested for specificity by competition assays with unlabeled nucleotides. Using 14C-ADP–Dbp5 samples (molar ratio of 5:1) that were equilibrated for 24 h, filter-binding assays were performed after addition of excess unlabeled ADP or ATP (Fig. 1B). Surprisingly, neither nucleotide diminished the level of bound 14C-ADP over the course of an additional 24 h. We showed previously that excess unlabeled ADP and ATP can equally compete for binding when added coincidentally with radiolabeled ADP (Tran et al. 2007). Taken together, this indicates that, once bound to ADP in vitro, nucleotide release from Dbp5 is slow.
Figure 1.
Nup159 promotes Dbp5 release of ADP. (A) Equilibrium binding assays show an increase in initial rate of Dbp5–14C-ADP binding: Dbp5 (1 μM) was incubated with increasing concentrations of 14C-ADP (0.5–150 μM) over the course of 24 h. Samples were removed at several time points and used in filter-binding assays. Background integrated density signals of 14C-ADP at each concentration were subtracted from the Dbp5-bound ADP signal. As listed in the table, initial velocities of binding (fraction 14C-ADP bound per minute) were calculated during the linear phase of the binding curve at each Dbp5:14C-ADP ratio tested. (B) Dbp5 does not release bound ADP after equilibration: After 24-h equilibration of 1 μM Dbp5 and 5 μM 14C-ADP:MgCl2, excess unlabeled ATP:MgCl2 or ADP:MgCl2 (10 mM of each) was added, and filter-binding assays were conducted after an additional 24 h incubation. (C) Nup159-NTD does not promote release of the nonhydrolyzable ATP analog ADP-BeFx: After 26-h equilibration of 1 μM Dbp5 with 5 μM 14C-ADP-BeFx, either 1 μM Nup159NTD, 500 nM Gle1 with 200 nM IP6, or 2 μM poly(A)25 RNA was added, and filter-binding assays were performed at 24 h after addition. (D) Nup159-NTD specifically promotes Dbp5 release of bound ADP: After 24-h equilibration of 1 μM Dbp5 with 5 μM 14C-ADP:MgCl2, either 1 μM Nup159-NTD, 500 nM Gle1 with 200 nM IP6, or 2 μM RNA was added, and filter-binding assays were performed at 10 min and 24 h after addition. Two altered nup159 proteins (nup159DD and nup159VI) that do not bind Dbp5 in vitro were used as negative controls. (E) Increasing concentration of Nup159 promotes faster Dbp5 release of bound 14C-ADP. Dbp5 (1 μM) was incubated with 5 μM 14C-ADP for 24 h. Increasing concentrations of Nup159 were added and filter-binding assays were performed over the course of 2 h. All data points and bars represent at least three independent experiments with standard error bars.
To test whether known Dbp5-interacting factors stimulate ADP release, we added either bacterially expressed, purified recombinant Nup159-NTD (amino acids 2–387), full-length Gle1 with IP6, or RNA to the 14C-ADP-binding assay after the initial 24-h equilibration period (Fig. 1D). Nup159-NTD promoted significant release of ADP within minutes, whereas Gle1-IP6 and RNA did not have any effect at any time point tested. We further characterized this Nup159 release activity by testing increasing molar ratios of Nup159 to Dbp5 in the 14C-ADP-binding assay again, after the initial 24-h equilibration period. We found that the initial rates of release are dependent on the concentration of Nup159 relative to Dbp5 (Fig. 1E). Interestingly, full release was achieved at a 1:1 Dbp5:Nup159 molar ratio, indicating that Nup159 promotes release by direct binding. To test for the effect of direct Dbp5–Nup159 binding, we generated two different altered versions of Nup159 that lack Dbp5 binding: nup159-D331R/D333R (nup159DD) and nup159-V323E/I326E (nup159VI) (Supplemental Fig. 1A–C). Neither nup159DD-NTD nor nup159VI-NTD promoted ADP release from Dbp5 (Fig. 1D). We also tested the effect of Nup159-NTD, Gle1-IP6, and RNA on the ability to promote the release of 14C-ADP-BeFx, which mimics an ATP in the active site prior to hydrolysis. After a 24-h binding incubation, no significant release of 14C-ADP-BeFx was observed (Fig. 1C). We conclude that the Nup159-NTD function is specific to the release of ADP from Dbp5.
Gle1-IP6 promotes Dbp5 ATP binding
The ADP equilibrium binding data for Dbp5 was surprising because we and others have shown that Dbp5 has robust ATPase activity in vitro without the addition of Nup159-NTD to promote ADP release (Alcazar-Roman et al. 2006; Weirich et al. 2006). However, the action of Nup159 during the ATPase cycle is not fully resolved. Others have also reported that Nup159-NTD has no effect on the RNA-dependent ATPase activity of Dbp5 (Weirich et al. 2006), whereas, under different assay conditions, the Nup214-NTD is inhibitory for hDbp5 activity (von Moeller et al. 2009). As noted above, the predicted overlapping RNA- and Nup159-binding sites complicate the interpretation of how Nup159 might impact in vitro ATPase activity. In an in vivo ATPase cycle, access to Nup159, Gle1, and RNA is likely spatially distinct compared with the reconstituted in vitro reactions. We hypothesized that there could be a factor that promotes ATP loading in vitro to facilitate Dbp5 nucleotide cycling and allow continued hydrolysis.
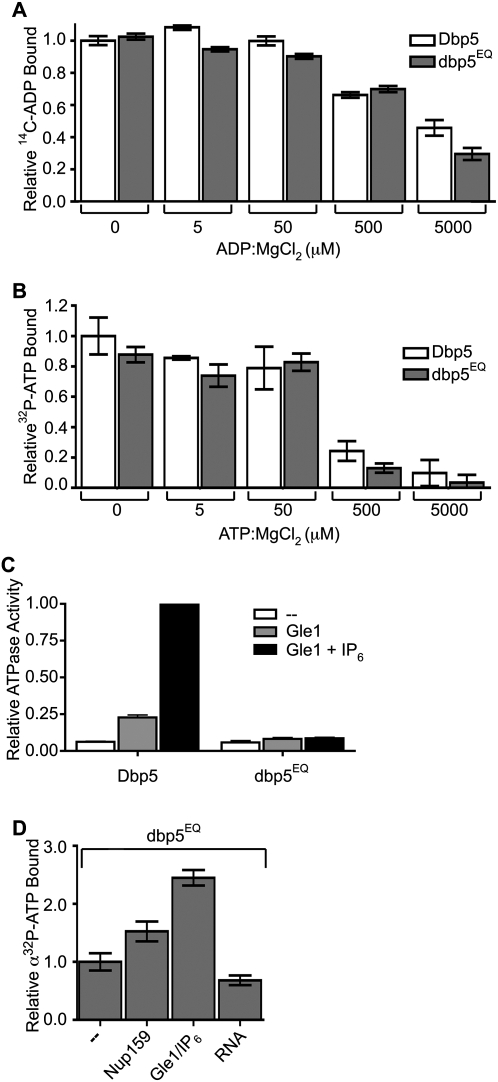
To identify this factor, we generated an altered dbp5 protein with a glutamate-to-glutamine change (E240Q) in the eponymous DEAD sequence of motif II (designated dbp5EQ) (Fig. 3A, below). Motif II is one of several highly conserved motifs present in all Dbps and is required for ATP hydrolysis (Linder 2006). Binding assays showed that the dbp5EQ protein bound ATP and ADP (Fig. 2A,B). However, using in vitro colorimetric ATPase assays, purified recombinant dbp5EQ had only a low basal level of activity that was not stimulated by Gle1-IP6 (Fig. 2C), indicating a predominant defect in the ability to hydrolyze ATP. Thus, using the dbp5EQ protein eliminated effects of ATP hydrolysis and allowed measurement of ATP binding in the presence or absence of known Dbp5 interaction partners or cofactors (Fig. 2D). We found that, in the presence of limiting ATP, Gle1-IP6 enhanced the relative level of dbp5EQ–ATP binding in vitro more than twofold. The effect of Nup159-NTD or RNA was not as robust. This correlates with data showing that Gle1-IP6 lowers the Km of Dbp5 for ATP (Weirich et al. 2006), and indicates that full access to such factors might overcome the requirement for ADP release by Nup159 in vitro. In addition, these results support a potential mechanism with Gle1-IP6 stimulating both ATP loading on Dbp5 and ATP hydrolysis.
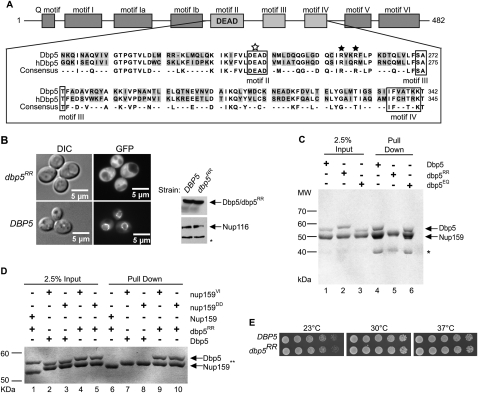
Figure 3.
dbp5RR mutant does not bind Nup159 and displays no in vivo growth defects. (A) Amino acid residue alignment for a portion of S. cerevisiae Dbp5 and Homo sapiens hDBP5. Black stars designate conserved arginine residues that were changed to aspartic acid residues (R256D and R259D) in Dbp5 to yield dbp5RR. The open white star represents a conserved glutamate residue that was changed to glutamine (E240Q) to yield dbp5EQ. Conserved DEAD-box family sequence motifs are boxed and identified below alignment. (B) Cytoplasmic mislocalization of GFP-dbp5RR by live-cell microscopy: S. cerevisiae strains with DBP5 chromosomally deleted, expressing either GFP-DBP5 or GFP-dbp5RR on plasmids, were used in live-cell fluorescent microscopy experiments to visualize subcellular localization of each protein. (Left) DIC image. (Right) GFP. Western blot analysis of yeast whole-cell extracts was conducted to test for relatively equal levels of expression of GFP-Dbp5 and GFP-dbp5RR. Nup116 protein level was used as a loading control. (*) Degradation products of Nup116. (C) Nup159-NTD interaction in vitro with dbp5RR is not detected: Soluble binding assays were conducted with 200 pmol of bacterially expressed recombinant His6-Nup159-NTD and immobilized on Ni-NTA agarose resin and either recombinant Dbp5, dbp5RR, or dbp5EQ. Input and bound fractions were analyzed by SDS-PAGE and Coomassie staining. (*) Nup159-NTD degradation product. (D) Swapping the salt bridge interaction allows nup159DD and dbp5RR interaction: Soluble binding assays were conducted as in C with His6-Nup159-NTD, His6-nup159VI-NTD, His6-nup159DD-NTD, Dbp5, or dbp5RR. (E) dbp5RR strain does not have growth defects: S. cerevisiae strains with DBP5 chromosomally deleted, expressing either wild-type DBP5 or dbp5RR, were used to perform serial dilution growth assays at 23°C, 30°C, and 37°C.
Figure 2.
Gle1-IP6 enhances Dbp5 binding to ATP. (A,B) Adenine nucleotide binding of Dbp5 versus dbp5EQ: Filter-binding assays were conducted to assay for relative binding to ATP and ADP. Purified recombinant Dbp5 or dbp5EQ (1 μM) was incubated with 1 μM 32P-ATP:MgCl2 or 14C-ADP:MgCl2 and increasing concentrations of unlabeled nucleotide ranging from 0 nM to 5000 nM. Bars represent the averages of three independent experiments with standard error shown. (C) dbp5EQ is inhibited for ATPase activity: Colorimetric ATPase-coupled enzyme assays were performed with purified recombinant 500 nM Dbp5 or dbp5EQ, 1 mM ATP:MgCl2, and 1 μM poly(A)25 RNA. Gle1 (250 nM) was added with or without 100 nM IP6. (D) Gle1-IP6 promotes ATP loading of dbp5E240Q: dbp5E240Q (2 μM) was incubated with 10 nM α32P-ATP in the presence of 2 μM Nup159-NTD, 1 μM Gle1 with 400 nM IP6, or 4 μM poly(A)25 RNA for 4 h. Filter-binding assays were performed. In all panels, bars represent the averages of three independent experiments with standard error bars.
A dbp5RR mutant bypasses the need for Nup159 interaction in vivo by affecting ADP binding
Although nup159 mutants with defects in Dbp5 binding have strong defects in mRNA export (Hodge et al. 1999; Weirich et al. 2004), to date, yeast dbp5 mutants with diminished Nup159 binding have not been characterized. The recent crystal structure of the NTD of hDbp5 in complex with the Nup214-NTD identified several surface-accessible amino acids of Dbp5 and Nup159 that form critical salt bridges as part of their interaction (Napetschnig et al. 2009; von Moeller et al. 2009). Each of these amino acids is conserved in the yeast Dbp5 protein, including Arg 256 and Arg 259 (R256 and R259) in Dbp5. Based on the hDbp5–Nup214 structure, the yeast Dbp5 R256 and R259 residues are predicted to interact with the D331 and D333 residues in Nup159-NTD. As described above, when these aspartic acids in Nup159 were changed to arginine, the resulting nup159DD protein lacked Dbp5 binding (Supplemental Fig. 1B). We also found that a nup159DD mutant strain was temperature-sensitive for growth (Supplemental Fig. 1C). This is completely analogous to the phenotypes for the previously published rat7ΔN and nup159VI mutants that lack Dbp5 binding (Hodge et al. 1999; Weirich et al. 2004) (Supplemental Fig. 1B,C).
Based on the salt bridge interaction between hDbp5 and Nup214 (von Moeller et al. 2009), we predicted that the dbp5RR mutant should have a phenotype identical to the nup159DD mutant. Thus, we generated a dbp5-R256D/R259D mutant (designated dbp5RR) by changing the sequence to encode aspartic acid residues rather than arginines (R256D and R259D) (Fig. 3A). To test for localization at the nuclear rim (e.g., at Nup159), we performed live-cell fluorescence microscopy with strains expressing either GFP-Dbp5 or GFP-dbp5RR (Fig. 3B). GFP-Dbp5 displayed characteristic nuclear rim/NPC localization, whereas GFP-dbp5RR was mislocalized to the cytoplasm. Western blotting of whole yeast cell lysates confirmed that GFP-Dbp5 and GFP-dbp5RR had similar protein expression levels (Fig. 3B, right panel). We also performed immunoprecipitation experiments from whole yeast cell lysates with anti-Dbp5 antibodies. Immunoblotting for Nup159 showed coisolation with wild-type Dbp5; however, Nup159 was not isolated with dbp5RR protein (Supplemental Fig. 1D). To directly confirm this lack of interaction, soluble binding assays were conducted with bacterially expressed, purified recombinant His6-Nup159-NTD, Dbp5, and dbp5RR. When His6-Nup159-NTD was immobilized on Ni-NTA agarose, wild-type Dbp5 was isolated (Fig. 3C, lane 4). However, no binding was detected for dbp5RR (Fig. 3C, lane 5). As a control, another altered protein (dbp5EQ) was also tested and showed binding to Nup159-NTD (Fig. 3C, lane 6). To test whether the salt bridge interaction was swapped by the corresponding changes in the dbp5RR and nup159DD proteins, direct binding experiments were conducted. The dbp5RR and nup159DD proteins showed restored in vitro interaction (Fig. 3D). Thus, the dbp5RR protein behaved as predicted with regard to Nup159 and nup159DD binding. However, surprisingly, serial dilution growth analysis of the dbp5RR mutant strain showed no growth defects as compared with wild-type (Fig. 3E). This was striking given the temperature-sensitive growth of the rat7ΔN, nup159DD, and nup159VI mutants (Supplemental Fig. 1C; Hodge et al. 1999; Weirich et al. 2004).
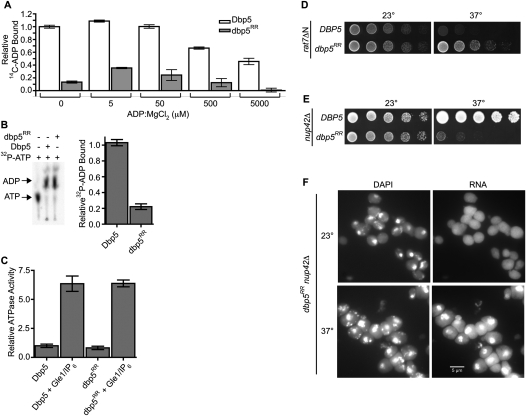
If the lack of a growth defect in the dbp5RR mutant reflected no requirement for Nup159 binding, this could mean that the growth defect of the rat7ΔN, nup159DD, and nup159VI mutants reflects pleiotropic perturbations and defects in more than just Dbp5 binding. Alternatively, the dbp5RR protein might be altered such that it specifically does not require Nup159 interaction. To examine these possibilities, we measured the relative ADP binding to dbp5RR as compared with wild-type Dbp5 (Fig. 4A). Using a filter-binding nucleotide competition assay (Tran et al. 2007), we found that dbp5RR protein had significantly lower binding to ADP. This suggests that dbp5RR does not require Nup159 function in vivo because its decreased ADP binding allows efficient Nup159-independent ADP release after hydrolysis. To directly test for this possibility in vitro, we compared the ability of Dbp5 and dbp5RR to release ADP after hydrolysis. To do this, we performed an ATPase assay using α32P-ATP and allowed both Dbp5 and dbp5RR to incubate until hydrolysis of α32P-ATP to α32P-ADP was fully complete (as assessed by thin layer chromatography) (Fig. 4B, left panel). Samples were then removed from each hydrolysis reaction and assayed by filter binding to measure the relative ADP levels still bound to each protein (Fig. 4B, right panel). Compared with wild type, dbp5RR protein retained significantly less bound ADP after hydrolysis, reflecting increased release independent of Nup159-NTD binding. We speculated that the dbp5RR mutant is viable because the ADP release phenotype bypasses the requirement for Nup159 binding.
Figure 4.
dbp5RR does not require Nup159 for ADP release. (A) dbp5RR has reduced binding affinity to ADP as compared with wild-type Dbp5: Dbp5 (1 μM) or dbp5RR (1 μM) was incubated with 5 μM 14C-ADP and increasing concentrations of unlabeled ADP:MgCl2. Filter-binding assays were performed to measure the relative fraction of ADP bound to each protein. Bars represent the averages of three independent experiments with standard error bars. (B) dbp5RR does not require Nup159 for ADP release: α32P-ATP was incubated with either Dbp5 or dbp5RR and 400 μM poly(A)25 RNA for 24 h to allow complete hydrolysis to α32P-ADP to occur. After 24 h, samples for filter-binding assays were removed and the remaining reaction was stopped with proteinase K digestion. (Left) The digested samples were spotted onto thin-layer chromatography plates to monitor ATP hydrolysis. (Right) The portion of the samples removed for filter-binding assays were used to monitor the relative amount of ADP still bound to either Dbp5 or dbp5RR. Bars represent the averages of three independent experiments with standard error shown. (C) dbp5RR is active for ATP hydrolysis and Gle1-IP6 stimulation: Colorimetric ATPase-coupled enzyme assays were performed as described in Figure 1B with 500 nM Dbp5 or 500 nM dbp5RR. Gle1 (250 nM) with 100 nM IP6 was added to measure relative levels of stimulation. The ATPase activity of each sample was normalized to the maximally stimulated Dbp5 with Gle1-IP6 sample. Bars represent the averages of three independent experiments with standard error shown. (D) The dbp5RR mutant complements the rat7ΔN temperature sensitivity. Yeast strains were grown at the permissive temperature and serially diluted on plates for growth at 23°C and 37°C. (E) The dbp5RR nup42Δ double mutant has a synthetic fitness growth defect. Yeast strains were grown at the permissive temperature and serially diluted on plate for growth at 23°C and 37°C. (F) Nuclear poly(A) RNA accumulates in arrested dbp5RR nup42Δ cells. In situ hybridization with an oligo(dT) probe for poly(A) RNA localization was conducted on dbp5RR nup42Δ cells grown at 23°C and shifted to growth for 1 h at 37°C. DAPI staining marks the nucleus (left) and fluorescent signal for the poly(A) RNA (right).
To directly test for the ability of the dbp5RR mutant to bypass the Nup159 requirement, we constructed rat7ΔN dbp5RR double mutants. The rat7ΔN mutant is temperature-sensitive at 37°C (Fig. 4D; Hodge et al. 1999) and Dbp5 localization at the NPC is diminished, as in the nup159VI mutant (Hodge et al. 1999; Weirich et al. 2004). Remarkably, the dbp5RR mutant complemented the rat7ΔN temperature sensitivity, as evidenced by the growth of the rat7ΔN dbp5RR double mutant at 37°C (Fig. 4D). We conclude that the altered ADP binding by the dbp5RR protein eliminates the requirement for Nup159 interaction. Moreover, a primary role for Nup159 in the Dbp5 cycle is to facilitate ADP release.
The dbp5RR mutant requires activation by Gle1 at the NPC
Gle1-IP6 enhances the ATP binding of wild-type Dbp5 (Fig. 2D) and stimulates its ATPase activity (Alcazar-Roman et al. 2006; Weirich et al. 2006). Compared with wild type, the dbp5RR protein had a similar basal level of ATPase activity and was fully stimulated by Gle1-IP6 (Fig. 4C). This indicates that the Nup159- and ADP-binding perturbations in dbp5RR are specific and do not alter its catalytic activity or regulation by Gle1-IP6.
Extensive genetic studies have linked Dbp5, Gle1, and IP6 functions to Nup42 at the NPC (Miller et al. 2004; Alcazar-Roman et al. 2006), with Nup42 directly binding Gle1 (Murphy and Wente 1996; Strahm et al. 1999). To test whether the dbp5RR mutant is dependent on efficient Gle1 function at the NPC, we generated a dbp5RR nup42Δ double mutant. Both single mutants were not temperature-sensitive; however, the dbp5RR nup42Δ double mutant was lethal at 37°C (Fig. 4E). To assay for mRNA export, in situ hybridization for poly(A) RNA localization was conducted. Cells were grown at the permissive temperature (23°C) and shifted for 1 h to 37°C. The dbp5RR nup42Δ double-mutant cells showed nuclear accumulation of poly(A) RNA (Fig. 4F), whereas the single mutants did not (Saavedra et al. 1997; Stutz et al. 1997). Thus, in the absence of Nup159 binding, Nup42 function is required for dbp5RR activity in mRNA export at elevated temperatures. We conclude that, even if Nup159-catalyzed ADP release is not essential, spatial control of Dbp5 at the NPC cytoplasmic filaments is still necessary for proper mRNA export.
Nucleotide-dependent conformational changes are important for the remodeling mechanism
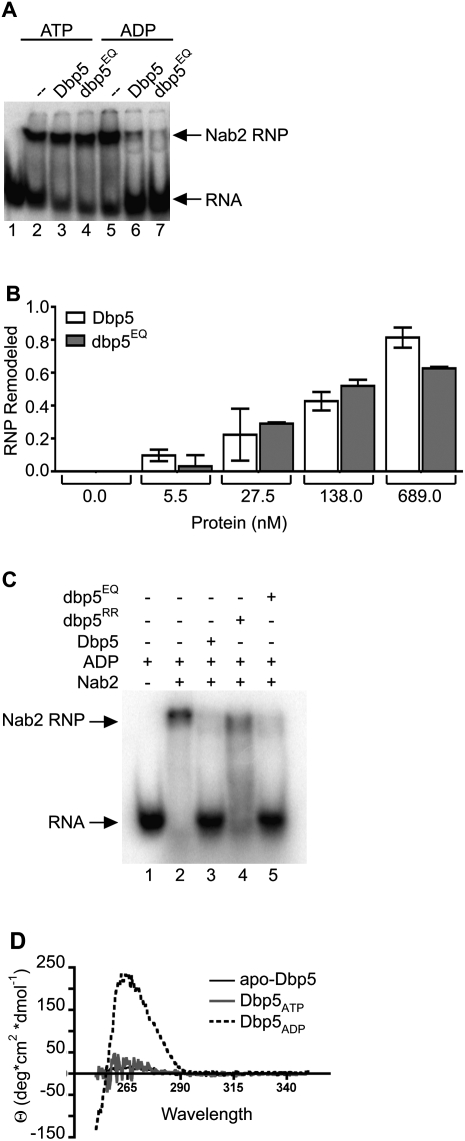
To more directly test for the requirements for ADP binding and ATP hydrolysis during in vitro remodeling, we conducted a series of in vitro remodeling assays with the dbp5EQ and dbp5RR proteins. We speculated that dbp5EQ could be competent for in vitro mRNP remodeling if the mechanism is based on harnessing a conformational change in Dbp5 structure upon ADP binding to the APO form (apo-dbp5EQ → dbp5EQADP). In the presence of ATP alone, neither wild-type Dbp5 nor dbp5EQ showed significant remodeling of the Nab2-RNP (Fig. 5A, lanes 3,4). This supports our prior conclusion that the ATP-bound form is not functional for remodeling (Tran et al. 2007), and suggests that an apo-dbp5EQ → dbp5EQATP conversion is also not sufficient. However, by adding ADP rather than ATP, dbp5EQ remodeled Nab2-RNPs equally as well as wild-type Dbp5, as shown by a gel shift assay (Fig. 5A, lanes 6,7). A titration of Dbp5 protein levels was used to more carefully determine the extent of ADP-dependent remodeling for wild-type Dbp5 and dbp5EQ. As shown in Figure 5B, dbp5EQ was as effective as wild type. This further confirms that ADP binding alone is sufficient to mediate in vitro mRNP remodeling.
Figure 5.
Dbp5 remodeling mechanism is mediated by a conformational change. (A) In vitro remodeling assay demonstrates that dbp5EQ remodels similarly to wild-type Dbp5: In vitro constituted Nab2-RNPs were assembled as described in Tran et al. (2007) with 5′-end labeled 32P-poly(A)25. Nab2 (250 nM) was incubated with 10 nM RNA and 500 nM either wild-type Dbp5 or dbp5EQ in the presence of 1 mM ATP:MgCl2 or 1 mM ADP:MgCl2. Electrophoretic mobility shift assays (EMSAs) were performed and products were visualized by autoradiography. (B) Quantification of Nab2-RNPs remodeling: Increasing concentrations of Dbp5 and dbp5EQ were added to in vitro constituted Nab2-RNPs in the presence of ADP:MgCl2 and filter-binding assays were performed. Densitometry was used to quantify the fraction of Nab2-RNPs that were remodeled at each concentration of protein. Bars represent the averages of three independent experiments with standard error bars. (C) dbp5RR remodels in vitro constituted Nab2-RNPs less efficiently than wild-type Dbp5: EMSAs were performed as described above using 750 nM wild-type Dbp5, dbp5E240Q, or dbp5RR in the presence of 1 mM ADP:MgCl2. The gel was exposed to autoradiography to detect the relative Nab2-RNP and free RNA. (D) Circular dichroism studies demonstrate nucleotide-dependent conformational changes: Near-UV spectral scans of Dbp5 with 1.5 mM ATP:MgCl2, ADP:MgCl2, or MgCl2 alone were collected from 250-nm to 350-nm wavelengths. Spectra of buffer with 1.5 mM ATP:MgCl2, ADP:MgCl2, or MgCl2 were subtracted from the Dbp5 spectra to remove interfering signals from the adenosine moiety. Θ represents molar elipticity, which is calculated as (100 × signal)/(pathlength × amino acid residues × mM concentration).
If efficient ADP binding is required, we predicted that the dbp5RR protein with altered ADP binding would be less effective in mediating in vitro mRNP remodeling with ADP alone. Indeed, dbp5RR remodeling with ADP was markedly reduced as compared with both wild-type Dbp5 and dbp5EQ (Fig. 5C). Thus, altered ADP binding correlated with altered mRNP remodeling in vitro. However, in vivo with ATP and Gle1-IP6, the dbp5RR mutant has no growth or mRNA export defects (Fig. 3E; data not shown). Together, this suggests that the APO-to-ADP transition (with dbp5EQ) and the ATP-to-ADP transition (with dbp5RR) are equivalent mechanistically for mRNP remodeling.
Recent structural studies indicate that Dbp5 exhibits nucleotide-dependent conformational changes (Fan et al. 2009; Napetschnig et al. 2009; von Moeller et al. 2009). Crystallography of hDbp5 with RNA and AMP-PNP showed the two RecA domains in a closed conformation forming a nucleotide-binding cleft between the two domains (von Moeller et al. 2009). In the presence of ADP, however, the two RecA domains are displaced due to the insertion of the N terminus, including an induced α helix (amino acids 1–91), between the domains (Collins et al. 2009). This correlates directly with our limited trypsin digestion studies of full-length Dbp5, wherein the proteolytic products in the presence of ATP are distinct from those observed in the presence of ADP (Tran et al. 2007). In addition, the proteolytic products in the absence of nucleotide (APO) appear the same as with ATP, suggesting similar APO and ATP conformations (Tran et al. 2007). To further investigate the nucleotide-dependent conformations in solution, we performed a series of circular dichroism spectroscopy experiments with full-length recombinant Dbp5 (Fig. 5D). Dbp5 was incubated with ATP or ADP or without nucleotide. Results were plotted as a function of molar ellipticity versus wavelength. Both the Dbp5ATP and apo-Dbp5 samples showed minimal ellipticity across the scan. However, Dbp5ADP displayed a large peak at ∼260–270 nm, likely resulting from changes in aromatic residue accessibility. This indicates that the conformation of Dbp5 in vitro in the presence of ADP (Dbp5ADP) is distinct as compared with either the absence of nucleotide (apo-Dbp5) or the presence of ATP (Dbp5ATP).
Interestingly, from the results shown in Figure 1A, we calculated the approximate on rate of Dbp5–ADP binding for the 1:666 molar ratio used in the in vitro remodeling assay. From this, the resulting initial ADP-binding rate would be ∼7.3% of the total ADP bound per minute. Thus, over a 30-min in vitro remodeling assay, a significant proportion of the apo-Dbp5 would shift to Dbp5ADP by the loading of ADP. It is important to note that, in the in vitro remodeling assays, Dbp5 is present in at least 50-fold to 1500-fold molar excess over the RNP. Therefore, based on the dbp5RR reduced remodeling with ADP, the circular dichroism, and the ADP equilibrium binding results, we suggest that Dbp5 remodeling in vitro is mediated by a conformational change from apo-Dbp5 to Dbp5ADP over the course of the assay. This in vitro mechanism is then effectively equivalent to the conformational change in Dbp5 when ATP has been hydrolyzed to ADP. Taken together, this suggests that the mechanism of Dbp5 remodeling in vitro requires an ATP- or APO-to-ADP conformational change.
Discussion
Here we present evidence defining critical steps in the mRNA export mechanism via facilitated Dbp5 nucleotide cycling and mRNP remodeling at the NPC. Other studies indicate that Dbp5 docking at Nup159 is required for mRNA export (Hodge et al. 1999; Schmitt et al. 1999; Weirich et al. 2004). We show that Nup159 interaction is specifically required for triggering ADP release from Dbp5. Moreover, because the dbp5RR mutant with independent defects in ADP and Nup159 binding is viable and the dbp5RR mutant complements the rat7ΔN temperature sensitivity, the Nup159 interaction is not strictly required for directional mRNA export. However, some level of spatial control at the NPC cytoplasmic face is needed for efficient mRNP transport. This is evidenced by the growth and mRNA export defects in a dbp5RR nup42Δ double mutant wherein Nup42 at the NPC impacts Gle1-IP6 function. We find that Gle1-IP6 acts to mediate both ATP binding to Dbp5 and hydrolysis. Overall, the combined actions of Nup159 and Gle1-IP6 on Dbp5 nucleotide binding provide a mechanism for permitting multiple cycles of mRNP remodeling by a single Dbp5 molecule at the NPC.
We also further investigated the nucleotide-dependent basis for mRNP remodeling. Both circular dichroism (Fig. 5D) and limited trypsin digestion studies (Tran et al. 2007) reveal that the in vitro solution states of Dbp5ATP and apo-Dbp5 are similar. However, Dbp5ADP is distinct. This is supported by published structure studies of different yeast and human Dbp5 complexes. Moreover, from in vitro studies with Dbp5 proteins that have altered ADP binding (dbp5RR) or ATP hydrolysis (dbp5EQ), we found that ADP binding is both necessary and sufficient for mRNA remodeling. As such, transitions in the Dbp5 nucleotide-bound state couple conformational changes to mRNP remodeling. We conclude that in vitro remodeling is facilitated by either (1) a Dbp5ATP-to-Dbp5ADP conformational change induced by in situ ATP hydrolysis and release of Pi, or (2) an apo-Dbp5-to-Dbp5ADP conformational change induced by ADP binding. In vivo, the Dbp5ATP-to-Dbp5ADP mechanism is likely obligatory because of the relatively low level of ADP within cells (van den Brink et al. 2008).
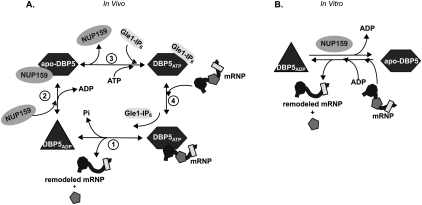
Based on these results, we propose a working model for the in vivo Dbp5 cycle of activity at the NPC (Fig. 6A). Four distinct Dbp5 phases are predicted: Dbp5ATP/mRNP, Dbp5ADP, apo-Dbp5/Nup159, and Dbp5ATP/Gle1-IP6. As shown in Figure 6A, step 1, Dbp5ATP has the highest affinity for RNA, with very weak RNA binding by Dbp5ADP (Weirich et al. 2006; Tran et al. 2007). The conformational change from Dbp5ATP to Dbp5ADP in vivo during ATP hydrolysis triggers mRNA remodeling. In vitro, this can be mimicked by a transition from apo-Dbp5 to Dbp5ADP during nucleotide binding (Fig. 6B). The conformational switch involves the insertion of an N-terminal α helix between the two RecA-like domains in Dbp5 and potentially triggers a corresponding change in the structure of the bound mRNA/mRNP, resulting in the release of RNA-binding proteins (Collins et al. 2009). Next in the cycle (Fig. 6A, step 2), Nup159 directly binds Dbp5ADP and facilitates the ADP release. Of note, apo-hDbp5 has the highest affinity for Nup214 in vitro (von Moeller et al. 2009). This event effectively regenerates Dbp5 for another round of mRNA remodeling. At the NPC (Fig. 6A, step 3), Gle1-IP6 is localized near Nup159–apo-Dbp5 in the NPC cytoplasmic filaments, and Gle1-IP6 enhances ATP binding in vitro and primes Dbp5 for RNA binding. In terms of the in vivo mechanism, the high cellular ATP concentration will likely also facilitate ATP loading. This step does not strictly require Nup159 binding, as evidenced by the dbp5RR mutant phenotype. Considering the full cycle (Fig. 6A, step 4), a previous study reported a potential reduction of the Dbp5/Gle1-IP6 interaction in the presence of RNA (Alcazar-Roman et al. 2010). Thus, when Gle1-IP6–Dbp5ATP bind the mRNA/mRNP, ATP hydrolysis is triggered coincident with Gle1-IP6 release from Dbp5. The steps shown in Figure 6A (steps 4 and 1) together effectively comprise the RNA-dependent and Gle1-IP6-dependent stimulation of Dbp5 ATPase activity. Overall, the model fully accounts for the mutually exclusive binding of RNA and Nup159, and positions Nup159 and Gle1 to regulate nucleotide binding and have direct control over Dbp5 cycling at the NPC.
Figure 6.
Proposed mechanism of Dbp5 nucleotide cycle. The left panel illustrates in vivo steps, and nucleotide binding/hydrolysis are controlled by multiple binding partners. (Step 1) Bound to ATP and an mRNP, Dbp5 is primed for remodeling. Hydrolysis is stimulated by Gle1-IP6, resulting in release of Pi and the remodeled mRNP, and generation of a conformationally changed ADP-bound Dbp5. (Step 2) Bound to ADP, Nup159 promotes ADP release, which confers another conformational change and places Dbp5 in an APO state. (Step 3) Gle1-IP6 promotes the loading of ATP to allow another round of nucleotide cycling and remodeling. (Step 4) Bound to ATP, Dbp5 has the highest affinity for RNA. RNA binding and ATP hydrolysis release Gle1-IP6. The right panel proposes a model for in vitro mimicking via ADP binding to the APO form of Dbp5, allowing mRNP remodeling in the absence of Gle1-IP6. Once bound to ADP in vitro, Nup159 promotes nucleotide release.
If this model is correct, there are several specific predictions for the in vivo behavior of dbp5 mutants with defects in ATP hydrolysis and RNA binding. In the accompanying study (Hodge et al. 2011), we found that such mutants behave as predicted. Notably, the ATP hydrolysis-defective dbp5EQ mutant, which binds to either ATP or ADP (Fig. 2A,B), is a loss-of-function mutant in vivo (Hodge et al. 2011). Further, at steady state in the presence of wild-type Dbp5, dbp5EQ protein is not detected at the nuclear rim/NPC (Hodge et al. 2011). This suggests that the ability of Dbp5 to access Nup159 requires the Dbp5ADP form generated by ATP hydrolysis (Fig. 6A, step 2). As the dbp5EQ protein cannot efficiently enter the mRNP remodeling cycle, this also provides a rationale for why the dbp5EQ mutant is not dominant. In addition, the phenotype and biochemical properties of a dominant dbp5-R369G mutant that lacks RNA binding also directly support the model in Figure 6A, step 4. The dbp5-R369G protein competes with wild-type Dbp5 for Gle1 (Hodge et al. 2011). Since binding sites for Dbp5 at the NPC are limiting, this would reduce the remodeling capacity of wild-type Dbp5. This further indicates that the RNA-binding step follows the interaction between Dbp5 and Gle1-IP6 and is potentially needed to release Gle1 from Dbp5 through ATP hydrolysis.
Although Nup159 and Gle1 impact Dbp5 activity at the NPC, only Gle1 is linked to Dbp5 function during translation termination (Bolger et al. 2008). Prior studies indicate that Dbp5 is executing a RNP remodeling function during translation termination, allowing the recruitment of Sup35 (eRF3) to the termination complex (Gross et al. 2007; Bolger et al. 2008). We predict that this mechanism could also require a Dbp5ATP-to-Dbp5ADP transition and presumably a factor that facilitates ADP release from Dbp5. Possibly, such an ADP release factor is a component of the ribosome or the termination complex. In addition, this factor might play a general role and result in enough basal total cellular regeneration of Dbp5ATP to allow mRNA export and viability at the permissive temperature in strains where Dbp5 cannot bind Nup159; for example, in rat7ΔN, nup159VI, and nup159EE mutants (Supplemental Fig. 1C; Hodge et al. 1999; Weirich et al. 2004). The hypothesized translation termination ADP release factor might also account for the ability of overexpressed DBP5 to suppress completely the temperature sensitivity and RNA export defects of the rat7ΔN mutant (Hodge et al. 1999). As such, mutation of the gene encoding the translation ADP release factor might be lethal in combination with either the rat7ΔN, nup159VI, or nup159EE mutant.
The roles for both an ATPase-activating factor (Gle1-IP6) and an ADP release factor (Nup159) in spatially regulating Dbp5 further solidify the proposed similarity between directional mRNA export and how the GTPase Ran is controlled for directional karyopherin-mediated transport through the NPC (Tran and Wente 2006). In particular, the control of RanGTP and RanGDP states allows precise regulation of karyopherin–cargo interactions and Ran cycling (for review, see Moore 1998; Gorlich and Kutay 1999), much like we propose for how the Dbp5ATP and Dbp5ADP states are controlled. However, in the case of Ran, the GTPase-activating protein (designated RAN-GAP1/Rna1) and the guanine nucleotide exchange factor (designated RCC1/Prp20) are spatially confined to opposing NPC faces/compartments (the cytoplasm/NPC cytoplasmic filaments and nucleus, respectively) (for review, see Macara 2001). For Dbp5, Nup159 and Gle1 impact mRNP export directionality and Dbp5 remodeling cycles via juxtaposed locations on the NPC cytoplasmic filaments. While there is a variation in localization of the nucleotide release/exchange and activating factors for Dbp5 and Ran, the precise regulation of where each NTPase is stimulated to undergo a conformational change provides directionality for the export of either RNA or proteins.
To our knowledge, this is the first Dbp reported to require a defined ADP release factor. The Dbp5 mechanism does parallel that reported for other ATPases that have nucleotide exchange factors. While nucleotide exchange factors for ATPases are far less studied than guanine nucleotide exchange factors, proteins such as BAP and Hsp110 promote release of ADP by their ATPase-binding partners (Chung et al. 2002; Andreasson et al. 2008). It is possible that other Dbps also use ADP release factors. If so, we speculate that their equivalent DEAD-box motif mutants to that in dbp5EQ will also not be dominant in vivo. However, recent structural studies highlight that the residues involved in ADP binding to Dbp5 and hDbp5 are different from that known for two other Dbps, VASA and eIF4AIII (Collins et al. 2009). Thus, this might also be a novel mechanism for directly controlling Dbp5 and other select Dbps through the need for facilitated ADP release. Future studies will be required to uncover the full range of mechanisms used to differentially regulate Dbps throughout the gene expression pathway.
Materials and methods
Protein expression and purification and yeast growth
All yeast strains were grown in YPD (2% peptone, 2% glucose, 1% yeast extract) at indicated temperatures. All recombinant proteins used (GST-Dbp5, GST-dbp5RR, GST-dbp5EQ, His6-Nup159NTD, His6-nup159NTDDD, His6-nup159NTDVI, and MBP-TEV-Gle1) were bacterially expressed in Escherichia coli Rosetta (DE3) cells (EMD Biosciences) using 0.2 mM IPTG to induce expression for 16–18 h at 16°C. Bacteria were lysed by sonication in Buffer B (150 mM NaCl, 20 mM HEPES at pH 7.5, 20% w/v glycerol) supplemented with 1× protease inhibitor cocktail and 1 mM PMSF. After centrifugation at 10,500 rpm for 30 min at 4°C, soluble fractions were used for affinity chromatography with either glutathione-coupled sepharose (GST-tagged proteins; GE Healthcare), amylose resin (MBP-tagged proteins; New England Biolabs), or Ni+-NTA-coupled agarose (His6-tagged proteins; Qiagen) using the manufacturer's instructions. The GST tag was cleaved during dialysis with Factor Xa (New England Biolabs). The MBP-TEV tag was cleaved during dialysis with ∼15 kU of recombinant, purified His6-TEV protease as described in Kapust et al. (2001). The His6 tag was not cleaved from proteins. All proteins were dialyzed overnight into 1 L of Buffer B at 4°C. All proteins were subsequently aliquoted, snap-frozen in an ethanol and dry ice bath, and then stored at −70°C.
RNP assembly and RNP remodeling assays
Nab2-RNPs were assembled as described in Tran et al. (2007). Briefly, 500 nM His6-Nab2 was incubated with 10 nM 32P-poly(A)+25 for 5 min at 30°C in remodeling buffer (20 mM HEPES at pH 7.5, 100 mM NaCl, 3 mM MgCl2, 10% w/v glycerol). Where appropriate, 1.5 μM Dbp5/dbp5RR/dbp5EQ was added with 1 mM ADP:MgCl2 (Sigma-Aldrich) and incubated for 30 min at 30°C in 20 μL of total reactions. Twenty-microliter samples were resolved in a 4% acrylamide–NaH2PO4 native gel. Each gel was run at 80 V for 1 h at 4°C. The gels were analyzed by autoradiography.
Radiolabeled nucleotide-binding assays
All filter-binding assays were performed with a nylon membrane underneath a Whatman 0.45-μm nitrocellulose membrane (as described in Wong et al. 1992) in a 96-well vacuum dot blotter (Aurogene). Both membranes were soaked for at least 10 min in Binding buffer (20 mM HEPES at pH 7.5, 100 mM NaCl, 3 mM MgCl2, 10% w/v glycerol) prior to dot blotting. All samples in each experiment were done in triplicate, and each experiment was performed three times. For ADP kinetic loading assays, 1 μM Dbp5 was incubated with increasing concentrations from 0.5 μM to 150 μM 14C-ADP:MgCl2 in Binding buffer. Thirty-microliter samples were removed at 30 sec, 10 min, 2 h, 4 h, 8 h, 16 h, and 24 h and spotted onto the nitrocellulose–nylon membrane sandwich using the vacuum dot blotter. Membranes were allowed to dry at room temperature and exposed to Kodak BioMax MS film with a Kodak BioMax TranScreen LE (Eastman Kodak) for 2 h at −70°C. Densitometry was used to quantify the fraction of nucleotide bound as follows: 14C-ADPBOUND/14C-ADPBOUND + 14C-ADPFREE. Initial binding rates were calculated for each Dbp5: 14C-ADP ratio assayed during the linear phase of binding as fraction 14C-ADP bound per minute. These rates were used to extrapolate the initial rate at a Dbp5: 14C-ADP = 1:666, which is the ratio used in the in vitro remodeling assays. For nucleotide competition assays, 1 μM Dbp5, dbp5RR, or dbp5EQ was incubated with 250 nM 14C-ADP:MgCl2 along with increasing amounts of unlabeled ADP:MgCl2 (50 μM, 500 μM, and 5000 μM) for 30 min at room temperature in Binding buffer. Samples were used for filter binding and analyzed by autoradiography as described above. Similarly, assays were performed with 250 nM α-32P-ATP and increasing concentrations of unlabeled ATP:MgCl2 (50 μM, 500 μM, and 5000 μM). Samples were quantified as described above. For nucleotide release assays, 1 μM Dbp5 was incubated with 5 μM 14C-ADP:MgCl2 or 14C-ADP-BeFx in Binding buffer for 24 h at room temperature. His6-Nup159NTD/His6-nup159NTDDD/His6-nup159NTDVI (1 μM), 500 nM Gle1 with 200 nM IP6, or 10 μM RNA was added and 30-μL samples were used for filter binding after 10 min, and then again after 24 h. Nucleotide off-rate assays were performed with 1 μM Dbp5, 5 μM 14C-ADP, and increasing concentrations of His6-Nup159NTD (83 nM, 500 nM, and 1 μM). Thirty-microliter samples were removed 30 sec, 2 min, 10 min, 30 min, and 120 min after addition of His6-Nup159-NTD and used for filter binding as described. 14C-ADP bound was quantified as described above.
Circular dichroism spectroscopy
Experiments were conducted with and without Dbp5 (10 μM) with 1.5 mM ATP:MgCl2,1.5 mM ADP:MgCl2, or 1.5 mM MgCl2 alone in 50 mM sodium phosphate (pH 7.5) and 100 mM NaF with a Jasco J-810 spectrometer using a 1-mm path length at room temperature following a 10-min incubation. Spectral scans were collected in triplicate for each condition in the near-UV wavelength (250–350 nm). Control spectra from buffer alone + MgCl2, 1.5 mM ATP:MgCl2, or 1.5 mM ADP:MgCl2 were subtracted from spectra with Dbp5. Because the ATP and ADP concentrations are in vast excess of Dbp5, the resulting spectra reflect changes in Dbp5 structure only and not differences in the concentration of free adenosine molecules. Molar elipticity (Θ) was calculated as (100 × signal)/(pathlength × amino acid residues × mM concentration).
ATPase assays
Endpoint ATPase assays using thin-layer chromatography (Alcazar-Roman et al. 2006) and colorimetric enzyme-coupled ATPase rate assays (Kornberg and Pricer 1951) were performed as originally described in respective references. For both assays, 500 nM Dbp5/dbp5RR/dbp5EQ was incubated with 1 μM poly(A)+25, 1 mM DTT, and 1 U of SUPERasin (Ambion) in ATPase buffer (10 mM HEPES at pH 7.5, 45 mM NaCl, 2 mM MgCl2). Where indicated, 250 nM Gle1 and 100 nM IP6 were added. All reactions were started with 1 mM ATP:MgCl2. Each sample in an experiment was done in triplicate, and each experiment was repeated at least three times.
Soluble binding assays
Two-hundred picomoles of recombinant purified His6-Nup159-NTD (or His6-nup159-NTDDD or His6-nup159-NTDVI) was immobilized on 25 μL of packed Ni+-NTA agarose in MHL buffer (50 mM Na2HPO4 at pH 7.4, 10% w/v glycerol, 150 mM NaCl, 20 mM imidazole). Two-hundred picomoles of Dbp5, dbp5RR, or dbp5EQ was added where indicated. Samples were incubated for 45 min at room temperature with gentle rocking. Samples were washed five times with 700 μL of MHL buffer and one final time with 300 μL of MHL buffer. Bound protein was eluted with 1× SDS sample buffer for 30 min at room temperature. Samples were then loaded into 7.5% acrylamide–SDS gels that were run at 30 mA for 2 h at room temperature. Proteins were detected by Coomassie Blue staining.
Live-cell microscopy and in situ hybridization
Yeast strains used for live-cell microscopy to monitor localization of GFP-Dbp5 and GFP-dbp5RR were grown in YPD at 23°C to mid-log phase (OD600nm 0.3–0.6). One-hundred microliters of cell suspension was collected and resuspended in 20 μL of fresh YPD and imaged using a fluorescent microscope (model BX50, Olympus) using an Uplan 100×/1.3 objective and a digital camera (Photometrics Cool Snap HQ, Roper Scientific). Images were processed using the MetaVue software (Universal Imaging), ImageJ (NIH), and Adobe Photoshop 7.0. In situ hybridization was performed on cells that were grown to mid-log phase at 23°C and then shifted for 1 h to a 37°C shaking water bath, following the protocol described in Alcazar-Roman et al. (2010).
Acknowledgments
We thank Elena Conti for sharing data before publication, and members of the Wente laboratory for discussions and comments on the manuscript. This work was supported by grants from the NIH: R37 GM51219 (to S.R.W.), 1F31 HD061181 (to K.N.N.), 1F32 GM075459 (to E.J.T.), and R01 GM33998 (to C.N.N.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2040611.
Supplemental material is available for this article.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701 [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol 8: 711–716 [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman AR, Bolger TA, Wente SR 2010. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J Biol Chem 285: 16683–16692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B 2008. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J Biol Chem 283: 8877–8884 [DOI] [PubMed] [Google Scholar]
- Bolger TA, Folkmann AW, Tran EJ, Wente SR 2008. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134: 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JR, Schwartz TU 2009. The nuclear pore complex has entered the atomic age. Structure 17: 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KT, Shen Y, Hendershot LM 2002. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem 277: 47557–47563 [DOI] [PubMed] [Google Scholar]
- Collins R, Karlberg T, Lehtio L, Schutz P, van den Berg S, Dahlgren LG, Hammarstrom M, Weigelt J, Schuler H 2009. The DEXD/H-box RNA helicase DDX19 is regulated by an α-helical switch. J Biol Chem 284: 10296–10300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P 2006. The DEAD-box protein family of RNA helicases. Gene 367: 17–37 [DOI] [PubMed] [Google Scholar]
- Daneholt B 2001. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci 98: 7012–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F, Cole CN 2003. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol Biol Cell 14: 1664–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E 2004. Protein displacement by DExH/D ‘RNA helicases’ without duplex unwinding. Science 304: 730–734 [DOI] [PubMed] [Google Scholar]
- Fan JS, Cheng Z, Zhang J, Noble C, Zhou Z, Song H, Yang D 2009. Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides. J Mol Biol 388: 1–10 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kutay U 1999. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H 2007. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315: 646–649 [DOI] [PubMed] [Google Scholar]
- Hodge CA, Colot HV, Stafford P, Cole CN 1999. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J 18: 5778–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, Cole CN 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev (this issue). doi: 10.1101/gad.2041611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz ME, Strambio-de-Castillia C, Blobel G 1998. Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically oriented subcomplex. Proc Natl Acad Sci 95: 11241–11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Bowers H 2006. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res 34: 4181–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng 14: 993–1000 [DOI] [PubMed] [Google Scholar]
- Kelly SM, Corbett AH 2009. Messenger RNA export from the nucleus: a series of molecular wardrobe changes. Traffic 10: 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendirgi F, Rexer DJ, Alcazar-Roman AR, Onishko HM, Wente SR 2005. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: a conserved mechanism in the export of Hsp70 mRNA. Mol Biol Cell 16: 4304–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Pricer WE Jr 1951. Enzymatic phosphorylation of adenosine and 2,6-diaminopurine riboside. J Biol Chem 193: 481–495 [PubMed] [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP 1995. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem 270: 19017–19021 [DOI] [PubMed] [Google Scholar]
- Linder P 2006. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res 34: 4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund MK, Guthrie C 2005. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell 20: 645–651 [DOI] [PubMed] [Google Scholar]
- Macara IG 2001. Transport into and out of the nucleus. Microbiol Mol Biol Rev 65: 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AL, Suntharalingam M, Johnson SL, Audhya A, Emr SD, Wente SR 2004. Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. J Biol Chem 279: 51022–51032 [DOI] [PubMed] [Google Scholar]
- Moore MS 1998. Ran and nuclear transport. J Biol Chem 273: 22857–22860 [DOI] [PubMed] [Google Scholar]
- Moore MJ 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309: 1514–1518 [DOI] [PubMed] [Google Scholar]
- Murphy R, Wente SR 1996. An RNA-export mediator with an essential nuclear export signal. Nature 383: 357–360 [DOI] [PubMed] [Google Scholar]
- Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel G, Hoelz A 2009. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci 106: 3089–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S, Linder P 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Hurt E 2011. Linking gene regulation to mRNA production and export. Curr Opin Cell Biol 23: 1–8 [DOI] [PubMed] [Google Scholar]
- Saavedra CA, Hammell CM, Heath CV, Cole CN 1997. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev 11: 2845–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M, et al. 1999. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J 18: 4332–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J 17: 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M 2007. Ratcheting mRNA out of the nucleus. Mol Cell 25: 327–330 [DOI] [PubMed] [Google Scholar]
- Stewart M 2010. Nuclear export of mRNA. Trends Biochem Sci 35: 609–617 [DOI] [PubMed] [Google Scholar]
- Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbach M, Stutz F 1999. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J 18: 5761–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Kantor J, Zhang D, McCarthy T, Neville M, Rosbash M 1997. The yeast nucleoporin rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes Dev 11: 2857–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Wente SR 2006. Dynamic nuclear pore complexes: life on the edge. Cell 125: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell 28: 850–859 [DOI] [PubMed] [Google Scholar]
- Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J 17: 2651–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink J, Canelas AB, van Gulik WM, Pronk JT, Heijnen JJ, de Winde JH, Daran-Lapujade P 2008. Dynamics of glycolytic regulation during adaptation of Saccharomyces cerevisiae to fermentative metabolism. Appl Environ Microbiol 74: 5710–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moeller H, Basquin C, Conti E 2009. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol 16: 247–254 [DOI] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Berger JM, Weis K 2004. The N-terminal domain of Nup159 forms a β-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol Cell 16: 749–760 [DOI] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol 8: 668–676 [DOI] [PubMed] [Google Scholar]
- Wong I, Chao KL, Bujalowski W, Lohman TM 1992. DNA-induced dimerization of the Escherichia coli rep helicase. Allosteric effects of single-stranded and duplex DNA. J Biol Chem 267: 7596–7610 [PubMed] [Google Scholar]
- Zhao J, Jin SB, Bjorkroth B, Wieslander L, Daneholt B 2002. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J 21: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]