Abstract
Communication between Mre11 and Rad50 in the MR complex is critical for the sensing, damage signaling, and repair of DNA double-strand breaks. To understand the basis for interregulation between Mre11 and Rad50, we determined the crystal structure of the Mre11–Rad50–ATPγS complex. Mre11 brings the two Rad50 molecules into close proximity and promotes ATPase activity by (1) holding the coiled-coil arm of Rad50 through its C-terminal domain, (2) stabilizing the signature motif and P loop of Rad50 via its capping domain, and (3) forming a dimer through the nuclease domain. ATP-bound Rad50 negatively regulates the nuclease activity of Mre11 by blocking the active site of Mre11. Hydrolysis of ATP disengages Rad50 molecules, and, concomitantly, the flexible linker that connects the C-terminal domain and the capping domain of Mre11 undergoes substantial conformational change to relocate Rad50 and unmask the active site of Mre11. Our structural and biochemical data provide insights into understanding the interplay between Mre11 and Rad50 to facilitate efficient DNA damage repair.
Keywords: DNA damage signaling, DNA repair, genomic stability, Mre11–Rad50, structural biology
DNA double-strand breaks (DSBs) are generated during normal DNA metabolism and are also produced by endogenous and exogenous genotoxic stresses (Kanaar et al. 2008; Jackson and Bartek 2009). DSBs are one of the most detrimental DNA damages, which can be lethal or cause chromosomal abnormalities. To maintain genomic stability, cells must repair the DSB sites, and do this by nonhomologous end-joining (NHEJ) or homologous recombination (HR) (Khanna and Jackson 2001; Rass et al. 2009). Although these two pathways use different mechanisms to repair the damage, they both employ the MRN complex, which consists of Mre11, Rad50, and Nbs1 (or functional homolog Xrs2 in Saccharomyces cerevisiae) (Paull and Gellert 1999; Chen et al. 2001; Yang et al. 2006).
In the MRN complex, Mre11 and Rad50 are highly conserved among all species, whereas much less of the NBS1 (Xrs2) sequence is conserved and is evidently found only in eukaryotes (Carney et al. 1998; Aravind et al. 1999). The MRN complex possesses several important biochemical features that are essential for DNA damage repair and signaling. One of these is the nuclease activity of Mre11, which is essential for the initial processing of DNA ends containing adducts that could interfere with further processing and for resolving the secondary structure of DNA ends. Mre11 is responsible for DNA binding and 3′-to-5′ dsDNA exonuclease, ssDNA structure-specific endonuclease, hairpin nuclease, and strand-annealing activities (Connelly et al. 1998; Paull and Gellert 1998; Trujillo et al. 1998; Hopfner et al. 2000a). Cells devoid of Mre11 endonuclease activity are highly sensitive to DNA-damaging agents (Buis et al. 2008; Williams et al. 2008). Another important feature is ATP-binding and hydrolysis activity by Rad50, which is critical for MRN function (Hopfner et al. 2001; Lobachev et al. 2004). ATP or nonhydrolyzable ATP induces dimerization of Rad50 and promotes DNA-binding activities of the MR complex (Hopfner et al. 2000b; Moncalian et al. 2004). In addition, ATP hydrolysis regulates endonuclease activity of Mre11 and promotes the DNA-unwinding activity of MRN, which illustrates the functional significance of the communication between Mre11 and Rad50 (Trujillo and Sung 2001; Moncalian et al. 2004; Chen et al. 2005; Lee and Paull 2005).
Perturbation of the MRN complex activity significantly affects genomic stability: Aberrant reduction of the MRN complex is associated with some types of sporadic tumors (Giannini et al. 2002; Bartkova et al. 2008). In mice and humans, null mutations in any components of the MRN complex cause early embryonic lethality or cell inviability (Xiao and Weaver 1997; Luo et al. 1999; Zhu et al. 2001; Dumon-Jones et al. 2003; Buis et al. 2008). Hypomorphic mutations in Mre11 and Nbs1 result in the cancer-predisposing genome instability syndromes ATLD and NBS1, respectively. Together, these data illustrate the role of MRN as a keystone complex in the cellular response to DNA damage in mammals (Taylor et al. 2004; Wang et al. 2004).
Atomic force microscopy (AFM) and electron microscopy (EM) studies of MR complexes from various species have revealed that the MR complex exists as a heterotetrameric or hetero-oligomeric assembly, which can be further divided into head, coil, and hinge domains (Connelly et al. 1998; Anderson et al. 2001; Chen et al. 2001; de Jager et al. 2001, 2004). Within the head of the MR complex, which interacts with DNA, the N-terminal and C-terminal domains of Rad50 are connected by the central coiled-coils of Rad50, and each Rad50 subunit is assembled to form an ATP-dependent dimer through intermolecular head-to-tail interactions between the N-terminal and C-terminal domains (de Jager et al. 2001; Hopfner et al. 2001). The crystal structures of PfRad50CD have revealed that ATP functions as a molecular switch that triggers rotation of lobes I and II by 30° and the dimerization of the PfRad50CD (Hopfner et al. 2000b). Mre11, another component of the head, consists of nuclease, capping, and DNA-binding domains in eukaryotic Mre11 (D'Amours and Jackson 2002). The crystal structures of the PfMre11 and TmMre11 core domains have shown that the nuclease domain consists of five phosphoesterase motifs and an active site with two Mn2+ ions (Hopfner et al. 2001; Das et al. 2010). The capping domains of PfMre11 and TmMre11 are relatively less conserved and are believed to provide the specificity for recognizing the DNA substrates.
Although recent studies made important progress toward understanding the structural and biochemical properties of the MRN complex, we have little knowledge on the communication between Mre11 and Rad50 because the structural information at the atomic level is limited to individual domains of Rad50 and Mre11. It is poorly understood how Mre11 recognizes Rad50, which is critical to an understanding of MR function. Furthermore, it is unknown how the ATPase activity of Rad50 is coordinated with conformational changes and functions of Mre11. The mechanism underlying DNA binding to the MR complex is also unclear, as the relative positions of Mre11 and Rad50 are ambiguous. We now report on the crystal structure of the Mre11–Rad50–ATPγS complex from Methanococcus jannaschii, and provide a basis for the regulation between Mre11 and Rad50 in the MR complex and their biochemical and cellular activities. We show that Mre11 stabilizes the dimerization of Rad50, locks residues around the ATP-binding site within Rad50, and promotes efficient Rad50 ATP hydrolysis. The structure of the MR complex in conjunction with biochemical analysis reveals that ATP binding and hydrolysis trigger substantial conformational changes of Rad50 and Mre11 in the MR complexthat ultimately lead to the dislocation of Rad50 from the nuclease domain of Mre11 to unmask its active site. The data provide insights into the understanding of the allosteric effects of ATP binding and hydrolysis on the biochemical activities of the MR complex.
Results
MjMre11 is essential for the ATP hydrolysis activity of MjRad50
To obtain the crystals of the Mre11–Rad50 complex, we made a large number of constructs of various eukaryotic and archaeal Mre11 and Rad50 proteins. However, our initial attempts to crystallize full-length Mre11 and Rad50 from various species were not successful, largely due to the insolubility or aggregation properties of the Mre11–Rad50 complex proteins. To circumvent these problems, we removed the portion of the central coiled-coil region of Rad50 based on sequence alignment, solubility prediction, and available structural information. After extensive trials, we obtained diffraction-quality crystals using full-length Mre11 and Rad50 (residues 1–190 and 825–1005; designated as Rad50CC throughout the text) from M. jannaschii.
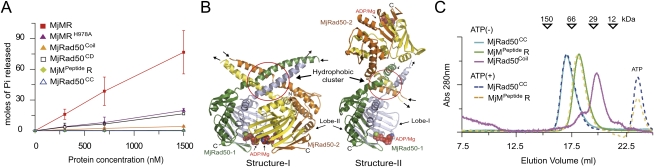
We characterized the biochemical features of the MjMre11–Rad50CC (MjMR) complex (Supplemental Fig. S1). Initially, we observed that free MjRad50CC is deficient in ATP hydrolysis activity (Fig. 1A). To understand the basis for the lack of ATP hydrolysis activity of MjRad50CC, we determined the crystal structure of ADP–MjRad50CC. We obtained two different forms of the MjRad50CC crystal (Supplemental Table S1; Supplemental Material). The two crystal structures of ADP–MjRad50CC showed very similar ATPase domain arrangements with root-mean-square deviations (RMSDs) of 0.57 Å for 295 Cα atoms. However, they showed significantly different dimeric MjRad50CC arrangements in that the two MjRad50CC subunits in each structure had notable variations in relative orientation (Fig. 1B; Supplemental Fig. S2). Furthermore, the conformations of the coiled-coil arm are significantly different in the two ADP–MjRad50CC structures, which demonstrate the flexibility of the coiled-coil arm region. The coiled-coil arm region near the MjRad50 catalytic domain has a cluster of conserved hydrophobic residues, consistent with that of PfRad50, which has been proposed to interact with PfMre11 (Supplemental Figs. S2B,D, S3; Hopfner et al. 2001). Nearly 25 residues in the hydrophobic patch in the MjRad50CC coiled-coil arm interact extensively with same residues from another MjRad50CC to form an ATP-independent dimer in both structures. Such coiled-coil-mediated dimerization of MjRad50CC would interfere with ATP-induced engagement of MjRad50CC molecules and ATP hydrolysis (Fig. 1A–C; Supplemental Material).
Figure 1.
Biochemical properties of the MjMR complex. (A) The ATPase activities of various MjRad50CC proteins were analyzed. The wild-type MjMR (red square), MjRad50CC (open triangle), MjMre11 C-terminal peptide-bound MjRad50CC (light-green diamond), MjRad50CD (open square), MjMRH978A (MjRad50) mutant (green triangle), and MjRad50coil mutant (yellow triangle) (see the Supplemental Material) are shown. The reaction was performed for 1 h at 37°C. (B) The two crystal structures of MjRad50CC exhibit different dimeric arrangements. In both structures, the dimer is formed through the same hydrophobic residues (highlighted in a red circle) in the coiled-coil near MjRad50CD. However, the orientation of lobes I and II is the same in the two structures. The N-terminal (orange and green) and C-terminal (yellow and cyan) segments are shown.. (C) Gel filtration analyses of free MjRad50CC (light blue or blue), MjMpeptideR (MjMre11 C-terminal peptide-MjRad50CC; green), and MjRad50coil (magenta) (Supplemental Material). Protein samples (20 μM) were injected into the Superdex200 (10/300) column equilibrated with the buffer containing 20 mM bis-tris propane-HCl (BTP), 200 mM NaCl, 5 mM dithiothreitol (DTT), 0.1 mM EDTA, and 5% glycerol (pH 7.4).
To eliminate the negative effect on ATP-dependent dimerization by the hydrophobic patch within the coiled-coil arm of MjRad50CC, we mixed MjRad50CC with the C-terminal peptide (residues 310–366) of MjMre11, which is predicted to bind the coiled-coil arm of MjRad50CC (Supplemental Fig. S4). Gel filtration analysis confirmed that the MjMre11 peptide dissociated the ATP-independent MjRad50CC dimer into a monomer (Fig. 1C). Nevertheless, the MjMre11 peptide-bound MjRad50CC did not form a dimer in the presence of ATP and did not exhibit any ATP hydrolysis activity (Fig. 1A,C). MjRad50CC exhibited strong ATP hydrolysis activity only in the presence of full-length MjMre11, which suggests that full-length MjMre11 is required for the ATP-mediated MjRad50 dimerization and the stimulation of ATP hydrolysis (Fig. 1A).
Overall structure of the MjMR complex
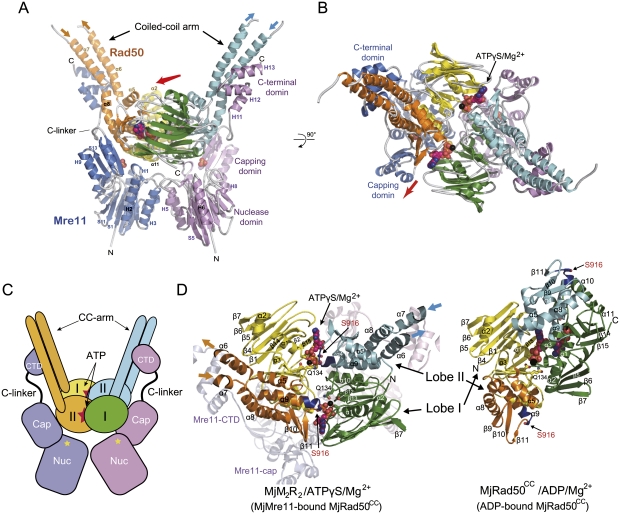
Each asymmetric unit contains one MjMR complex, and the MjMR dimer is generated by a crystallographic twofold axis. The MjMR complex consists of a head and two arms that are extended in a V shape (Fig. 2A–D). The head is composed of both core domains of MjMre11 and MjRad50CC, and the arms are formed by the coiled-coil of MjRad50CC and the C-terminal domain of MjMre11. The overall structure of the MjMR complex reveals that MjMre11 embraces MjRad50CC in such a manner that its C-terminal domain holds the coiled-coil arm of MjRad50CC, and the core domain (nuclease and capping domain) of MjMre11 interacts with the ATPase domain of MjRad50CC. The coiled-coil arm of MjRad50CC is located opposite to the interface of MjRad50CC that interacts with MjMre11 core domain. The overall dimension of the head of the complex is 84 × 97 × 85 Å, similar to the size of the reported EM structure of the head region of the PfMR complex (radius of 46 Å) (Hopfner et al. 2001).
Figure 2.
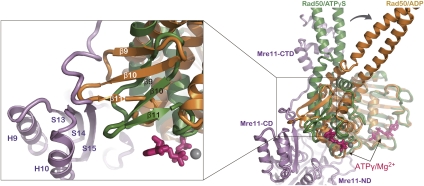
Overall structure of the MjMR–ATPγS complex. (A) Overall structure of the MjMR–ATPγS complex viewed from the side. The structure shows that the MjRad50 dimer sits on top of the MjMre11 dimer (blue and magenta). In each MjRad50 subunit, lobe I (yellow and green) and lobe II (orange and cyan) are shown in different colors for clarity. The axis (shown in a red arrow, also in B) that runs across the central groove of the MjRad50CC dimer is directed toward the capping domain of MjMre11. (B) Overall structure of the MjMR–ATPγS complex viewed from the top (looking down the vertical axis of A). The two ATPγS molecules are shown in a space-filling model. (C) Overall scheme of the MjMR–ATPγS complex. The active site of MjMre11 is shown as a yellow star. The MjMR–ATPγS complex is shown in same orientation and color scheme as that in A. (D) Structural comparison between the ATPγS–MjMre11-bound (left) and ADP-bound MjRad50 (right) dimer. Each lobe is colored in the same scheme as in A. Several key residues in ATP (or ADP) binding are labeled, and the signature motif is colored in dark blue.
The structure of MjRad50CC in the MjMR consists of lobe I (residues 1–137 and 939–1005) with an α/β fold, a smaller lobe II (residues 138–158 and 859–938), and two coiled-coils that protrude away from the base (Fig. 2A–C). MjRad50CC molecules are assembled into a head-to-tail dimer, in which ATPγS and Mg2+ ions are sandwiched between the two subunits, and the catalytic residues are directed from both MjRad50CC molecules, similar to those of PfRad50CD (Hopfner et al. 2000b). MjMre11 can be divided into an N-terminal core domain and a C-terminal domain linked by an extended connecting loop (C-linker). The MjMre11 nuclease domain is composed of eight helices and 12 strands forming a carcineurine-like fold, and the capping (or specificity) domain consists of three β strands packed by two helices on one side (Fig. 2A; Supplemental Fig. S5A–D). The C-terminal domain of MjMre11 is composed of three helices (H11 to H13), each 12 residues long, forming an S-shaped structure connected to the capping domain through an extended linker with 16 residues. The C-terminal three helices of MjMre11, which interact with each other through hydrophobic interactions, bind perpendicularly to the coiled-coil arm of MjRad50CC (Figs. 2A, 3A). The C-linker that connects the capping domain and the C-terminal domain runs parallel to the C-terminal part of the MjRad50CC coiled-coil.
When viewed toward the center of MjMre11, the MjRad50CC dimer is rotated toward each subunit of MjMre11 such that the axis (Fig. 2A,B, red arrow) across the groove at the center of the MjRad50CC dimer interface is directed to the capping domain of each MjMre11 (Fig. 2A,B). The formation of the interface between MjMre11 and MjRad50CC buries 3800 Å2 of the total surface area. Although not extensive, both lobe I and lobe II of MjRad50CC are involved in the interaction with the nuclease and capping domain of MjMre11, while the coiled-coil arm of MjRad50CC tightly interacts with the C-terminal region of MjMre11 (Fig. 3A–D; Supplemental Fig. S6A–C).
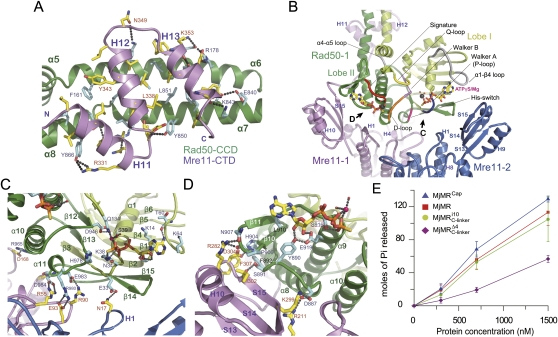
Figure 3.
Recognition specificity at the MjMre11–Rad50CC interfaces. (A) The primary interface between MjMre11 (CTD: C-terminal domain; magenta) and MjRad50 (CCD: coiled-coil arm domain; green). Interface residues from the two molecules are highlighted in yellow (MjMre11) and cyan (MjRad50CC). H bonds and ion pairs are shown in a dotted line. For close-up views, see Supplemental Figure S6. (B) The two MjMre11 (magenta and blue) and one MjRad50 (green) are shown to highlight the interfaces between MjMre11 and MjRad50. Another MjRad50 (Rad50-2) is omitted for clarity. Two ATPγS molecules are shown in one MjRad50CC molecule to represent both sides of interactions between ATPγS and MjRad50; one side shows the signature motif (orange)–ATPγS interaction, and another side shows the P loop (blue)–ATPγS interaction. Regions marked with C and D are shown in close-up views in separate figures. (C) A close-up view of the interface between the nuclease domain of MjMre11-1 (magenta) and the P loop and His switch (green). (D) The signature motif of MjRad50 is stabilized by loops S14–H10 and S15–H11 and a strand, S15 (magenta), of capping domain through the interaction with strand β10 and loop β10–β11 of MjRad50CC (green). (E) ATPase activities of wild-type MjMR and three mutant proteins.
Recognition specificity between MjMre11 and the coiled-coil arm of MjRad50
The MjMre11–MjRad50CC interface can be largely divided into three parts: the C-terminal domain of MjMre11 and the coiled-coil arm of MjRad50CC (Fig. 3A), the capping domain of MjMre11 and lobe II of MjRad50CC (Fig. 3B,D), and the nuclease domain of MjMre11 and lobe I of MjRad50CC (Fig. 3B,C). Each MjRad50CC subunit interacts with both MjMre11 subunits, as does each MjMre11 subunit. However, as shown in Figure 3, B–D, the interaction between the green-colored Rad50-1 and the magenta-colored Mre11-1 is predominant, whereas the green-colored Rad50-1 only weakly associates with the blue-colored Mre11-2.
The primary interface is formed between the C-terminal domain of MjMre11 and the coiled-coil arm of MjRad50CC, which covers ∼70% of the total buried surface. In this interface, three helices, H11 to H13 of MjMre11, bind perpendicularly to the coiled-coil arm of MjRad50CC (Fig. 3A). The core interface is predominantly hydrophobic, with 16 residues from MjMre11 interacting with 19 residues from MjRad50CC, with additional specificity provided by 10 intermolecular hydrogen (H) bonds and two ion pairs at the center of the interface. The details of the molecular interactions are described fully in Supplemental Figure S6. The importance of this interface is that hydrophobicity is highly conserved in both Mre11 and Rad50 from other species and tight interaction between MjMre11 and MjRad50CC that can be resistant to 6 M GuHCl is achieved, presumably through this hydrophobic interface (Hopfner et al. 2001).
In addition to these extensive interactions between the C-terminal domain of MjMre11 and the coiled-coil arm of MjRad50CC, the C-linker connecting helix H11 and the capping domain of MjMre11 also interacts with lobe II of MjRad50CC (Supplemental Fig. S7A). For example, Leu319 from MjMre11 packs against Tyr871 and Leu874 from MjRad50CC, and Glu317 (MjMre11) interacts with Lys877 (MjRad50CC) through an ion pair. Importantly, despite these interactions, residues forming the C-linker exhibit the highest temperature factors in MjMre11 and are expected to be highly flexible (Supplemental Fig. S7B).
The capping domain locks the P loop and the signature motif of MjRad50 and suppresses ATP hydrolysis
Previous structural studies of PfRad50CD–ATP recognition have established that ATP-dependent dimerization of Rad50 is achieved primarily through two major interactions between the ATP γ-phosphate and P loop from one Rad50 subunit, and between the ATP γ-phosphate and the signature motif from another Rad50 subunit (Hopfner et al. 2000b; Moncalian et al. 2004). In addition, another region such as the His switch is proposed to play a critical role in ATP hydrolysis (Fig. 1A). We observed similar interactions between ATPγS and MjRad50 in the MjMR–ATPγS structure (Figs. 2D, 3B). Importantly, the MjMR–ATPγS complex structure reveals that the N-terminal core domain (nuclease and capping domains) of MjMre11 interacts with regions near the ATP-binding site of MjRad50CC that include the P loop, signature motif, and His switch of MjRad50CC (Fig. 3B). Since these interactions are near the ATP-binding site, they are expected to control the ATP hydrolysis and also contribute to the dimerization of MjRad50CC. The differences between the arrangements of MjRad50 subunits in ADP-bound and ATPγS–Mre11-bound MjRad50 are illustrated in Figure 2D.
In Figure 3B, we omitted one MjRad50CC (MjRad50-2) subunit to highlight the interaction between ATPγS and MjRad50CC and the interaction between MjMre11 and MjRad50CC. Thus, the two ATPγS molecules that interact with Rad50-1 represent both faces of an ATPγS molecule that interact with the two MjRad50 subunits. As shown in Figure 3, B and C, on one face of ATPγS, the P loop (residues 32–38) of the green-colored Rad50-1 surrounds the β-phosphate and γ-phosphate of ATPγS and interacts extensively with these atoms. Asn17 from the blue-colored MjMre11 (MjMre11-2) forms an H bond with Glu33 of the P loop of MjRad50CC (MjRad50-1). In addition, loop β13–α11, containing the His switch (His978), is stabilized through three ion pairs between MjMre11 (MjMre11-1) and MjRad50CC (MjRad50-1; Glu93–Arg980; Arg55–Glu984; Arg90–Glu983) and two H bonds (Fig. 3C).
On another face of ATPγS, the MjRad50CC (MjRad50-1) regions that contact the γ-phosphate as well as the base and ribose ring are stabilized by MjMre11 (MjMre11-1: loops S14–H10 and S15–H11, and a strand, S15) (Fig. 3D): First, strand β10 and loop β10–β11, which surround the adenine base, are stabilized by strand S15 and loop S14–H10 of MjMre11 through a number of H bonds and van der Waals interactions. In particular, the main chain oxygen of Ile302 of MjMre11 forms an H bond with Ser891 that is next to Tyr890 in loop α8–β9 of MjRad50CC. Tyr890 not only interacts with the 2′ and 3′ hydroxyl groups of the ribose ring, it also forms an H bond with Glu919 in the signature motif of MjRad50 (Fig. 3D). The interaction between strand S15 of MjMre11 and β10 of MjRad50CC may contribute to the stability of the base, sugar, and phosphate portions of ATP. Together, a number of interactions between the capping domain of MjMre11 and lobe II of MjRad50CC stabilize the MjRad50CC dimer and active site, thereby affecting the ATP hydrolysis activity of MjRad50CC.
To examine the effect of the interaction described above on ATP hydrolysis, we mutated the capping domain and its interacting region in MjRad50CC and analyzed its ATP hydrolysis activity (Fig. 3E). We simultaneously replaced the residues at the capping domain (Arg282, Asp304, and Phe307) of MjMre11 and lobe II (Ser891, Phe892, Pro906, and Asn907) of the MjRad50CC interface with alanine or serine (designated as the capping domain mutant) (Fig. 3D; Supplemental Figs. S3, S4; Supplemental Table S2). Interestingly, the capping domain mutant increased the ATP hydrolysis activity by 15% compared with that of the wild-type MjMR complex, which suggests that the interaction between the capping domain and lobe II (loops α8–β9 and β10–β11) locks the movement of lobe II required for ATP hydrolysis activity.
Rad50 binding alters the dimer interface of Mre11
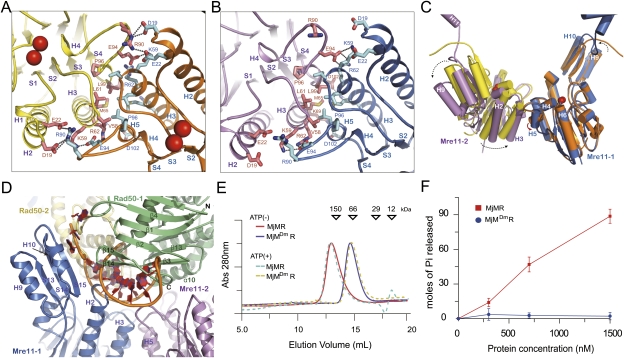
To understand whether MjMre11 undergoes conformational changes upon binding to MjRad50CC, we determined the crystal structure of free MjMre11 and compared it with the structure of MjMre11 in the MjMR complex (Fig. 4A–C; Supplemental Table S1). Although the overall structures of the individual domains are similar in free and complex MjMre11, significant differences are observed at the dimeric interface of MjMre11. The free MjMre11 dimerizes through continuous and extensive surface regions, which can be divided into two parts (Fig. 4A). One part is formed between loop H4–H5 and helix H2 of MjMre11, in which Arg90 from one MjMre11 forms ion pairs with Asp19 and Glu22 from another MjMre11. Another part is formed between helices H3 and H5. This four-helical bundle dimer interface is comprised of hydrophobic interactions formed by Val58, Leu61, Met65, and Leu99 from each MjMre11 and an ion pair between Lys59 and Glu94. In the dimer interface of free MjMre11, the angle between helix H5 of one MjMre11 and helix H3 of another MjMre11 is ∼60° (Fig. 4C; Supplemental Fig. S5B).
Figure 4.
Conformational changes of MjMre11 and MjRad50CC in the MjMR–ATPγS complex. (A) A close-up view of the free MjMre11 dimer interface. Compared with MjMre11 that bound to ATPγS–MjRad50, a new interface is formed between loop H4–H5 and helix H2 through four ion pairs and two H-bonds. Red spheres represent Mn2+ ions. (B) A close-up view of the MjMre11 dimer interface in the MjMR complex. Leu61, Met65, Pro96, and Leu99 from the two helices H3 and H5 interact with the same residues from another MjMre11 to form a four-helical bundle interface. (C) Structural superposition of the free MjMre11 (yellow and orange) and ATPγS–MjRad50-bound MjMre11 (magenta and blue) dimer. A dotted arrow illustrates the rotation of MjMre11. (D) ATP-bound MjRad50 blocks the active site of MjMre11. A branched DNA (orange) from PfMre11 (PDB ID: 3DSD) is modeled on the structure of the MjMre11 (blue and violet)–MjRad50 (green and light brown) complex. (E) Size exclusion chromatography of the MjMre11 dimer interface mutant (MjMDmR). The MjMR complex (20 μM) containing wild-type or mutant MjMre11 was loaded onto Superdex200 (10/300) equilibrated with the buffer containing 20 mM BTP, 200 mM NaCl, 5 mM DTT, 0.1 mM EDTA, 5% glycerol, and 2 mM ATP (pH 7.4) at 4°C. Prior to injection, the MjMR complex was incubated with 10 mM ATP for 30 min on ice. The MjMR complex incubated with 10 mM ATPγS showed a result identical to that with 10 mM ATP and is omitted for clarity. (F) MjMDmR (blue circle) exhibits impaired ATP hydrolysis activity.
The binding of MjRad50CC induces substantial conformational changes in the MjMre11 dimer (Fig. 4A–C). First, MjMre11 subunits rotate toward each other such that the interactions between loop H4–H5 and helix H2 are perturbed. Concomitantly, the four-helical bundle dimeric interface becomes more tightly packed. Here, helix H5 is rotated by >60° upon MjRad50CC binding. Both hydrophobic and ion pair interactions in the dimeric interface are further stabilized in the MjMR complex. If MjMre11 does not experience rigid body rotation upon MjRad50CC binding, MjMre11 (in particular, the active site region) would collide with MjRad50CC. Movement of MjMre11 also produces new interactions between MjMre11 and another MjRad50CC, such as an ion pair between Asn17 and Glu33 (MjRad50CC), which could contribute to the MjMR complex formation (Fig. 3C).
Mre11 dimerization is crucial for ATP hydrolysis of Rad50
In the MjMR complex, the MjRad50CC dimer sits on the core domain of the MjMre11 dimer. In particular, the regions including loops H4–H5, S3–H4, S6–H6, H10–S15, S7–H7, and S15–H11 and helix H1 of MjMre11 are covered by the MjRad50CC dimer (Figs. 3B, 4D). The MjMre11 interface to which MjRad50CC binds is continuous and conserved (Supplemental Fig. S8). The dimeric MjMre11 structure forms a frame that promotes the ATP-mediated engagement of MjRad50CC. In the MjMR complex, MjMre11 dimerizes through a four-helical bundle formed by the two helices H3 and H5. In addition to interactions between Leu61, Met65, Pro96, and Leu99 at the center of the four-helical bundle dimeric interface, Arg62 and Lys69 from one MjMre11 form ion pairs with Glu94 from another MjMre11, and Arg69 forms an ion pair with Asp102 to further support the dimer interface (Fig. 4B).
To examine whether the four-helical bundle interface of MjMre11 is important for the dimerization and ATPase activity of MjRad50CC molecules, we perturbed the dimeric interface of MjMre11 and analyzed the ATPase activity of the MjMR complex. We generated a mutant with Met65Arg, Glu94Ala, and Leu99Trp in which all resides were replaced simultaneously (Supplemental Table S2). This mutant eluted as a heterodimeric MjMR complex in size exclusion chromatography in both the absence and presence of ATP (or ATPγS) (Fig. 4E). As we expected, this mutant failed to exhibit any ATP hydrolysis activity, demonstrating that dimerization of Mre11 is critical for the ATPase activity of Rad50CC (Fig. 4F). Together, these data suggest that dimerization of MjMre11 is required for ATP-mediated engagement of MjRad50CC and ATP hydrolysis.
Structural comparison between ADP–MjRad50 and ATPγS–MjMre11-bound MjRad50
In the MjMR–ATPγS complex structure, the angle between the coiled-coil arms and the longitudinal axis of the MjMR complex is ∼34° (Figs. 2A,C, 5). This observation is consistent with the AFM study of the human MR complex, in which the angle between coiled-coils and the longitudinal axis of the complex is 24° ± 8° and 27° ± 8° in the absence and presence of AMP-PNP, respectively (Moreno-Herrero et al. 2005).
Figure 5.
ATP hydrolysis results in large changes in the structure of MjRad50CC in the MjMR complex. (Right) The structure of MjRad50 (green) from the complex is shown superimposed on the structure of ADP–MjRad50 (orange) by aligning the lobe Is. (Left) Rotation of lobe II (orange) is expected to cause steric clash with the capping domain (magenta).
Structural comparison between ADP-bound (structure I in Fig. 1B) and ATPγS–MjMre11-bound MjRad50CC reveals that lobe I or lobe II of MjRad50 is rotated by 30° relative to each other, similar to the structures of ATP-free and ATP-bound PfRad50CD (Hopfner et al. 2000b). In the superimposed structures, this rotation relocates the coiled-coil arm as much as 40 Å, positions the two coiled-coil arms nearly parallel, and disengages the MjRad50CC dimer (Fig. 5). While the movement of lobe I does not affect the overall structure of the MjMR complex, the rotation of lobe II of MjRad50CC would be predicted to cause a steric clash between loop α8–β9, strands β9 and β10 of MjRad50CC and helix H10, and strand S15 and loop S15–H11 of the capping domain (Fig. 5). These altered positions of the loop and strands in MjRad50CC are no longer compatible with the interaction with MjMre11. Thus, the movement of MjRad50 lobe II can push the capping domain of MjMre11 away from the nuclease domain.
DNA-binding site within the MjMR–ATPγS complex
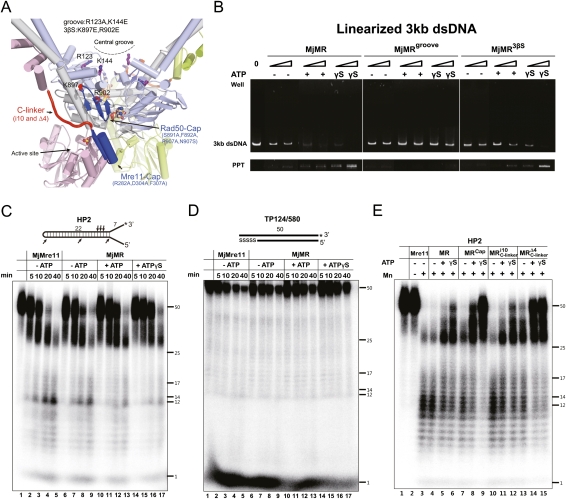
Both Mre11 and Rad50 bind to DNA (Paull and Gellert 1998; Hopfner et al. 2000b; Trujillo and Sung 2001; Williams et al. 2008). Previously, the central groove has been proposed to be a DNA-binding site in Rad50 and other SMC family members (Hopfner et al. 2000b; Lammens et al. 2004; Koroleva et al. 2007; Woo et al. 2009). We tested this possibility by replacing Arg123 and Lys144 at the groove of MjRad50 (MjMRgroove, which points directly toward the surface) with glutamate and analyzing its DNA-binding activity (Fig. 6A). We used two short DNA molecules (HP2 and TP 124/580) and a linearized 3-kb DNA for electrophoretic mobility shift assays (EMSAs) (Fig. 6B; Supplemental Fig. S9A,B). Although slight differences are observed between the short DNA and the long DNA binding to MjMR, presumably because of the differences in DNA length and the ratio between protein and DNA, these DNA molecules bind to the MjMR complex in a similar pattern. Figure 6B and Supplemental Figure S9 show that the wild-type MjMR complex stably interacts with two short DNA molecules and a long duplex DNA in the presence of ATP or ATPγS (Fig. 6B; Supplemental Fig. S9). In contrast, the MjMRgroove mutant did not interact (or interacted weakly) with both DNA molecules in the presence of ATP (or ATPγS).
Figure 6.
DNA-binding and nuclease activities of the MjMR complex. (A) Overall structure of the MjMR–ATPγS complex showing the positions of mutations used in this study. (B) EMSAs with the wild-type and two MjMR mutant proteins (MjMRgroove and MjMR3βS) using 4 nM linearized 3-kb dsDNA. The molar ratio of protein:DNA was 50:1 and 200:1. Reactions containing the buffer (10 mM BTP-HCl, 50 mM NaCl, 5 mM DTT, 5% glycerol at pH 7.5) with 5 mM MgCl2 in the absence or presence of 1 mM ATP (or ATPγS) were incubated for 30 min at 26°C. Reaction products were analyzed on 0.5% agarose gel. (Bottom lane) Unlike the MjMre11–3-kb DNA complex (or the MjMR–short DNA complex), which is soluble, the MjMR complex precipitated in the presence of a 3-kb DNA. Thus, we did not compare the 3-kb DNA binding by MjMre11 to the binding by MjMR complex. It is possible that the long duplex DNA binding induces the conformational change of the MjMR complex to form oligomeric aggregation states. (C) Kinetic analysis of the endonuclease activities of free MjMre11 (lanes 2–5), free MjMR (lanes 6–9), MjMR–ATP (lanes 10–13), and MjMR–ATPγS (lanes 14–17). Nuclease assays were performed with 200 nM protein in 5 mM MnCl2 on a fully paired hairpin substrate with a 32P-labeled 3′ end (20 nM). A schematic of the oligonucleotide substrate is shown at the top. The asterisk indicates the location of the 32P label in the diagram of the substrates, and the arrows indicate the cleavage sites. Reactions were incubated for 5, 10, 20, or 40 min at 55°C before separation on a denaturing polyacrylamide gel. ssDNA markers are indicated. (D) Exonuclease activity analysis performed using a 50-bp substrate. he 32P-label is indicated by an asterisk, and the length of the oligonucleotide is indicated. Five nucleotides in one 3′ end are connected through phosphorothioate bonds, which are shown as “SSSSS.” The reaction buffer and conditions were same as those in C. (E) Nuclease assays performed with wild-type or mutant MjMR (200 nM) and 20 nM HP2 for 60 min at 55°C.
If the DNA-binding sites are connected in MjMre11 and MjRad50, the DNA bound to the central groove could pass the three-stranded sheet formed by β9–β11 to reach the active site of MjMre11 (Fig. 6A). Thus, we made a second mutant (MjMR3βS) by replacing Lys897 and Arg902 of MjRad50 with glutamate. The MjMR3βS mutant exhibited decreased DNA binding in the presence of ATP or ATPγS compared with the wild-type MjMR complex (Fig. 6B; Supplemental Fig. S9). Biochemical analyses including ATPase activities, gel filtration chromatography, and circular dichroism studies of MjMRgroove and MjMR3βS imply that the surface mutations that we introduced did not affect the overall architecture of the MjMR complex (Supplemental Figs. S10, S11A,B). These results suggest that at least parts of the central groove and the β10 and β11 strands of MjRad50 subunits are involved in DNA binding.
An active site of MjMre11 is largely masked by MjRad50 in the ATPγS-bound MjMR complex
The crystal structures of PfMre11 bound to synaptic and branched DNA have shown that 17 residues in the six DNA recognition loops are involved in DNA binding (Williams et al. 2008). The structure of the MjMR complex showed that the DNA-binding region observed in PfMre11–DNA structures is largely blocked by MjRad50CC in the MjMR–ATPγS complex (Fig. 4D). The only way in which DNA could access the active site in this complex is through the channel formed by loops S7–H7, S8–S9, and S10–H8 from MjMre11 and helices α10 and α11 from MjRad50CC, which can accommodate an ssDNA (Supplemental Fig. S12). This channel is located close to the capping domain and strands β9 to β11 from lobe II of MjRad50. Alternatively, large conformational changes of MjRad50 could unmask the active site of MjMre11, and DNA binds to the active site similarly to those bound to PfMre11.
We examined the DNA-binding activities of MjMre11 and MjMR using the HP2 and TP124/580 substrates to understand the effect of ATP (ATPγS)-bound MjRad50 on DNA binding by MjMre11 (Supplemental Fig. S9A,B). While some fraction of free MjMre11 binds to DNA, a smaller fraction of free MjMR interacts with DNA. The ATP–MjMR complex exhibits slightly increased DNA binding compared with the free MjMR complex, and more MjMR binds stably to the DNA molecules in the presence of ATPγS. However, as we show in Figure 6B and Supplemental Figure S9, the stable interaction between wild-type ATPγS (or ATP)–MjMR and DNA is due primarily to DNA binding to MjRad50. The MjMR mutant protein (MjMRgroove or MjMR3βS) that contains the perturbed DNA-binding site within MjRad50 interacts with DNA relatively weakly (or similarly) in the presence of a nucleotide compared with the free MjMR mutant protein (Fig. 6B; Supplemental Fig. S9). This effect is more apparent in MjMR–TP124/580 binding. These data suggest that nucleotide-bound MjRad50 interferes with DNA binding by MjMre11.
ATP-bound MjRad50 negatively regulates the nuclease activity of MjMre11
To understand the role of ATP and MjRad50 in the nuclease activity of MjMre11, we performed a kinetic analysis on free MjMre11, free MjMR, and ATP (or ATPγS)-bound MjMR complex using a substrate with a paired hairpin structure, HP2 (25 base pairs [bp]), with 7-nucleotide (nt) 3′ overhangs and 6-nt 5′ overhangs (Trujillo and Sung 2001). Free MjMre11 initially generated an ∼35-nt product as a major form, which gradually degraded to an ∼30-nt product and then multicut products (5–22 nt) (Fig. 6C, lanes 2,3). Further incubation of free MjMre11 with a DNA substrate led to the formation of 5- to 22-nt fragments as major products, whereas the ∼35-nt product almost disappeared and only a small amount of an ∼30-nt DNA remained (Fig. 6C, lanes 4,5).
The free MjMR complex exhibited a cleavage pattern similar to that of free MjMre11 (Figs. 6C, lanes 6–9; Supplemental Fig. S13A). However, in each stage, the endonuclease activity was slightly attenuated, which suggests that free MjMR recognized a DNA substrate in a manner similar to that of free MjMre11, but that MjRad50CC interfered weakly with the nuclease activity of MjMre11. Although the ATP-bound MjMR complex produced an ∼30-nt DNA and multicut products (5–22 nt) in a pattern similar to that of free MjMre11 or free MjMR, it cleaved the substrates in a much delayed manner (Fig. 6C, lanes 2–5,10–13; Supplemental Fig. S13). The ATPγS-bound MjMR complex also delayed the product generation compared with the free MjMR complex (Fig. 6C, lanes 14–17; Supplemental Fig. S13). While the ATPγS-bound MjMR complex failed to generate most of the small fragments, it did produce a notably decreased amount of the ∼30-nt DNA at 40 min (Fig. 6C, lane 17). We also analyzed exonuclease activities of free MjMre11 and MjMR using a 50-bp substrate in the absence or presence of ATP or ATPγS. We observed delayed patterns of product generation in the presence of ATP and ATPγS similar to those in endonuclease reactions (Fig. 6D). We further examined the effects of ATP (and ATPγS) on the nuclease activity of MjMR using three different substrates. In the different substrates, the MjMR proteins exhibited nucleolytic patterns similar to those we observed on the HP2 substrate, which suggest that the nucleotide-bound MjRad50 interferes with the nuclease activities of MjMre11 (Supplemental Fig. S14).
The capping domain and C-linker of MjMre11 and lobe II of MjRad50 are important for the allosteric regulation in the MR complex
Previous studies have established that ATP binding and hydrolysis of the Rad50 subunits result in the allosteric effect that controls the several biochemical activities of the MR complex, including DNA binding and nuclease activities. Our structural and biochemical results showed that the capping domain of MjMre11 interacts with lobe II of MjRad50, and implied that the flexible C-linker, which connects the C-terminal domain of MjMre11 and MjRad50 (indirectly through the coiled-coils) to the capping domain, may be involved in conformational changes of the MR complex. Thus, we hypothesized that lobe II of MjRad50 and the capping domain and C-linker of MjMre11 may be involved in mediating the allosteric signal of ATP hydrolysis to nuclease activities of the MjMR complex. We tested this idea by using two different classes of mutant proteins. First, we examined nuclease activities of the capping domain mutant (MjMRcap), which destabilizes the capping domain of MjMre11 and lobe II of MjRad50, and compared them with those of the wild-type MjMR complex (Fig. 6A,E, lanes 7–9). The endonuclease analysis revealed that MjMRcap consumed less substrate and generated a notable amount of the ∼35-nt and 30-nt products in the presence of ATP and ATPγS (Fig. 6E, lanes 7–9; Supplemental Fig. S15). However, this mutant generated a relatively small amount of 12- to 14-nt fragments with ATPγS in the reaction. These results suggest that the interaction between lobe II of Rad50 and the capping domain of Mre11 is linked to the endonuclease activity of MjMre11.
We then examined the role of the C-linker of MjMre11 in DNA binding using two mutant proteins. In one mutant (MjMRi10), we increased the C-linker length by adding 10 residues (GSTGSGSTGS) between residues 314 and 315 of MjMre11, whereas, in another mutant (MjMRΔ4), we reduced the C-linker length by deleting four residues (308–311). Since the flexible C-linker may play an important role in the movement of the C-terminal domain of MjMre11 and MjRad50, we hypothesized that altering the flexibility of the C-linker may affect the nuclease activity of the MR complex. The ATP hydrolysis activity of the i10 mutant was similar to that of the wild-type MR complex (Fig. 3E). While this mutant generated a similar amount of 30-nt products, it generated slightly increased 5- to22-nt products in either the ATP-free or ATP-bound state (Fig. 6E, lanes 10,11). Deletion of four residues of the C-linker is expected to restrain the motion of the C-terminal domain and the capping domain. Presumably because of these reasons, the Δ4 mutation attenuated ATPase activity of the MjMR complex by 40% (Fig. 3E). Only limited amounts of the substrate were consumed by this mutant; both ATP-bound and ATPγS-bound MjMRΔ4 exhibited attenuated endonuclease activities in a product generation pattern, similar to that of ATPγS-bound MjMRcap (Fig. 6E, lanes 13–15; Supplemental Fig. S15). This result suggests that the flexibility of the C-linker is associated with the endonuclease activity of MjMR.
Discussion
So far, little is known of how Mre11 and Rad50 assemble into the head of the MR complex and affect the DNA-binding and nuclease activities of the complex, since structural information at the atomic resolution is limited to each component. In the present study, we provide the biochemical and structural basis for the allosteric regulation between Mre11 and Rad50 in the head of the MR complex. MjMR possesses several unique features, such as the endonuclease activity of free MjMre11, and also shares many common features with the eukaryotic MR complex, including the ATP-mediated control of the interaction and regulatory functions between MjMre11 and MjRad50. Thus, the structure of MjMR–ATPγS in conjunction with the biochemical studies clearly provides the fundamental principles to understand the regulation mechanism between Mre11 and Rad50.
Communication between Mre11 and Rad50
Several groups have performed ATP hydrolysis analyses using Rad50 molecules from various species. In each case, some variations were observed. For instance, free gp46 and ScRad50 exhibit relatively weak or negligible ATPase activity (Trujillo et al. 2003; Herdendorf et al. 2010). An independent study from another group (Ghosal and Muniyappa 2007) reported that ScRad50 possesses some ATPase activity that can be stimulated in the presence of Mre11. PfRad50CD shows some ATPase activity, and PfMre11 increases the ATPase activity twofold (Hopfner et al. 2000b). Thus, it appears that Rad50s from different species possess different levels of the basal ATPase activity, and the requirement of Mre11 for ATP-mediated engagement of Rad50CDs and stimulation of ATP hydrolysis is a conserved feature from phage to eukaryotes. In other SMC family members, the ATPase activity of the eukaryotic SMC1/3 complex in cohesion is stimulated by the C-terminal winged helix domain of an additional factor, Scc1, which interacts with the ATPase domains of SMC1/3 (Arumugam et al. 2006). Therefore, ATPase stimulation of the SMC protein by an additional protein may be a common feature in SMC family members.
The MjMR–ATPγS structure reveals that the two MjMre11–Rad50 interfaces are critical in regulating the ATPase activity of MjRad50. While the interaction between the C-terminal domain of MjMre11 and the coiled-coil arm of MjRad50 allows the MjMre11 dimer to bring the two MjRad50 subunits within the proximal distance, interaction between the capping domain of Mre11 and a three-stranded β sheet of Rad50 stabilizes the engagement of the ATPase domains and suppresses ATPase activity. The capping domain mutant increased the ATP hydrolysis activity by ∼15% compared with that of the wild-type MjMR complex (Fig. 3E). This increased ATP activity of the capping domain mutant suggests that the interactions between the capping domain of MjMre11 and loops α8–β9 and β10–β11 of MjRad50 lock the movement of lobe II, stabilize engagement of MjRad50, and suppress ATPase activity. By perturbing the interactions between the capping domain and loops α8–β9 and β10–β11 in lobe II, lobe I and lobe II of MjRad50 could be rotated with enhanced freedom. This feature resembles the non-SMC proteins ScpA and ScpB, which bind to the ATPase domain of Bacillus subtilis SMC and suppresses its ATPase activity (Hirano and Hirano 2004; Hirano 2005).
Perturbation of the Schizosaccharomyces pombe Mre11 dimer interface dramatically decreases the resistance against DNA-damaging agents (Williams et al. 2008). Based on the crystal structures of PfMre11–DNA and yeast genetics analysis, it has been concluded that dimerization of Mre11 is required for DNA binding. However, we showed that disruption of the MjMre11 dimer interface prevents ATP-mediated engagement of MjRad50 molecules and abolishes ATP hydrolysis (Fig. 4E,F). Therefore, we propose that another crucial role for the dimerization of Mre11 is to stimulate Rad50-mediated ATP hydrolysis.
Coordinated conformational changes of MjRad50 and MjMre11
Structural analysis of ADP-bound and ATPγS–MjMre11-bound MjRad50CC reveals that lobe II of MjRad50CC and the capping domain of MjMre11 undergo conformational changes upon ATP hydrolysis (Fig. 5). What would be the consequence of these conformational changes? Since the movement of lobe II of MjRad50CC leads to the collision with the MjMre11 capping domain, lobe II could push away and rotate the capping domain from the nuclease domain, which has been proposed to be important for the selection of DNA substrates and possibly be involved in the partial unwinding of DNA molecules (Williams et al. 2008). Flexibility of the capping domain is supported by (1) the open conformation of the capping domain relative to the nuclease domain in the free MjMre11 dimer (compared with the capping domain in the MjMR complex); (2) the PfMre11–DNA structure, which demonstrates the capping domain movement in the presence of DNA (Williams et al. 2008); and (3) the differences in the orientation of the capping domains in TmMre11, PfMre11, and MjMre11 (Supplemental Fig. S5A–D; Das et al. 2010). Such relocation of the capping domain might be accompanied by the conformational changes in the C-terminal domain and the C-linker of MjMre11. Concomitantly, the flexible C-linker of MjMre11 would induce large movement of MjRad50CC, and ultimately may lead to the exposure of the MjMre11 active site in the MjMR complex.
Implications of ATP hydrolysis on the MjMR nuclease activities
We showed that ATP-bound MjRad50 negatively regulates the nuclease activity of MjMre11. Both the endonuclease and exonuclease activities of the MjMR complex were decreased in the presence of ATP or ATPγS in a time-dependent manner (Fig. 6C,D). The decreased nuclease and DNA-binding activities of the MjMR complex in the presence of a nucleotide compared with those of free MjMR suggest that ATP-bound MjRad50CC blocks the access of a substrate DNA. Compared with free MjRad50, nucleotide-bound MjRad50 inhibits nuclease activities of MjMre11 more efficiently on various substrates, which suggests that the ATPase-induced conformational change of MjRad50CC could unmask the MjMre11 active site (Fig. 6C; Supplemental Fig. S14).
We used two classes of mutant proteins (the capping and C-linker mutants) to understand the regions of the MjMR complex that contribute to the allosteric regulation by ATP (Fig. 6E; Supplemental Table S2). The decreased nuclease activity upon perturbation of the interaction between the Mre11 capping domain and the Rad50 lobe II (loops α8–β9 and β10–β11) region suggests that the stability of lobe II of MjRad50 and the capping domain of MjMre11 is associated with the nuclease activities of MR (Fig. 6E). Mutational analysis of the C-linker showed that the flexibility of the C-linker affected the endonuclease activity of MjMre11 (Fig. 6E). The restricted movement of the C-linker may (1) negatively regulate the disengagement of MjRad50 molecules, (2) limit the movement of the C-terminal domain of MjMre11, and/or (3) restrain the conformational changes in lobe II and the capping domain, and so interfere with the unmasking of the active site. Together, these data support the view that the ATPase-mediated conformational changes of lobe II, the capping domain, and the C-linker play important roles in translating ATP hydrolysis to the DNA processing activities of the MjMR complex.
Why would ATP-bound MjRad50 negatively regulate the nuclease activity of Mre11? One of the roles for Rad50 in the MR complex may be to provide limited digestion to facilitate short or long regions of sequence homology searches in recombining DNA templates. Although it is still unclear how the nuclease is regulated to identify the similar sequence, the regulatory role of ATP-bound MjRad50 shown here provides the possibility that Rad50 is involved in promoting the homology search by restricting nucleolytic degradation.
Models for the ATP-mediated allosteric regulation of the Mre11–Rad50 complex
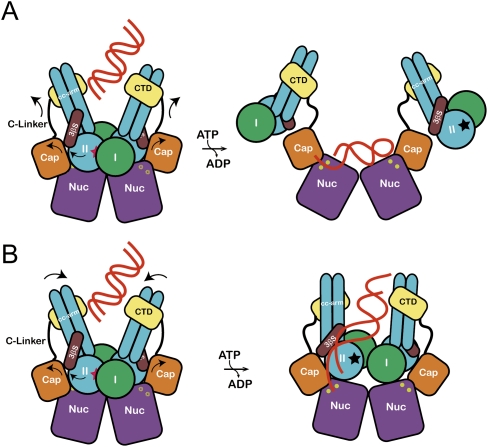
One of the important findings in the present study is that the rotation of each lobe (in particular, lobe II) triggered by ATP hydrolysis might induce gross conformational changes in both MjMre11 and MjRad50, which play a critical role in regulating the nuclease activities of MjMre11. Based on structural and biochemical results, we propose the following models for a nuclease mechanism by the MR complex. In the first model, ATP hydrolysis could disengage and dissociate MjRad50CDs from MjMre11 (while MjRad50 binds to the MjMre11 C-terminal domain through the coiled-coil arm) through the movement of the capping and C-terminal domains and disclose the active site (Fig. 7A). Substrate DNA then binds to the active site of MjMre11 in a mode similar to that observed in the PfMre11–DNA structure. A DNA substrate may bind to the active site in two different directions or bind to two active sites in the MjMre11 dimer simultaneously, and such binding may explain the generation of the ∼30-nt and 12- to 14-nt species (Fig. 6C). However, if DNA binds in this site, it is unclear how DNA binding in this site can be coordinated with the DNA binding in the central groove of MjRad50. One possible scenario is that the DNA molecule initially binds to the groove and stimulates ATP hydrolysis (Trujillo et al. 2003). After disengagement of MjRad50, DNA could subsequently interact with the MjMre11-binding site (Fig. 7A). Although multiple conformational states of the free MjMR complex may exist, we expect that there are restrictions on the conformational freedom of MjRad50 in the MjMR complex. For instance, the interaction between the coiled-coils of the MR complex in various oligomeric states may limit the conformational freedom of MjRad50 in the complex (de Jager et al. 2001, 2004).
Figure 7.
The proposed allosteric regulation models for the MjMR complex. (A) DNA binds to the central groove and stimulates the ATP hydrolysis. Rotation of lobe II (an arrow) triggers the movement of the capping domain and C-terminal domain of MjMre11, which dislocates MjRad50CD and unmasks the active site (yellow circle) of MjMre11. The DNA-binding mode is similar to that of the PfMre11–DNA complex. A dark star represents an ATP-binding site in MjRad50. (B) Rotation of lobe II of MjRad50 is accompanied by movement of the C-terminal domain of MjMre11 and induces the rotation of coiled-coils of MjRad50 such that their positions changed from V shape to parallel (an arrow). This conformational change may open the active site channel to comfortably accommodate the DNA substrate. The DNA molecule bound to the groove passes through a three-stranded sheet (β9–β11) and is guided to the active site channel.
We propose another DNA-binding model in the MjMR complex, which is continuously connected from MjRad50 to MjMre11 (Fig. 7B); a central groove and a three-stranded β sheet appear to be important in MjRad50 from the DNA-binding assays (Fig. 6B), and the capping domain and active site channel of MjMre11 are the only accessible paths to the two Mn2+ sites in the MjMR–ATPγS complex from the crystal structure (Supplemental Fig. S12). In this pathway, in order to reach the MjMre11 active site from the central groove, the DNA substrate must undergo substantial bending near the three-stranded β sheet and capping domain, which is only possible if the capping domain rotates and unwinds the DNA molecule (Fig. 7B). We propose a model in which the rotation of lobe II pushes the capping domain away, which could subsequently partly unwind the duplex part of DNA and direct it to the active site of MjMre11. The conformational changes of the capping domain and the C-linker may open the active site channel more widely. Here, MjRad50CDs may not be completely liberated from MjMre11. Instead, the coiled-coil arm of MjRad50 could rotate according to the movement of lobe II such that the two coiled-coil arms becomes parallel from the V shape (Figs. 5, 7B). Such a movement of MjRad50 might be sufficient to unmask the active site. It has been reported that DNA binding to the MR complex straightens the coiled coils of Rad50 and leads them to parallel orientation (Moreno-Herrero et al. 2005).
Materials and methods
Protein expression and purification
Genes encoding residues 1–190 and 825–1005 of MjRad50 were inserted into pCDFDuet-1 and pETDuet-1, respectively. MjMre11 (residues 1–366) was inserted into the pET28a vector. The Escherichia coli Rosetta (DE3) containing the three vectors was cultured in LB broth media. The complex was first purified by Ni-NTA affinity chromatography using a His tag at the N terminus of MjMre11. The MjMR complex was subsequently eluted with 300 mM imidazole in the same buffer. Fractions containing the MjMR complex were subsequently purified using cation exchange and gel filtration chromatography and concentrated by ultrafiltration. The free MjRad50CC and free MjMre11 (residues 1–313) were also purified by the same procedure as used for the MjMR complex protein. All mutant MjMR complexes were purified in the same procedure as that of the wild-type MjMR complex (Supplemental Fig. S11A,B).
Crystallization and data collection
Crystals of the MjMR complex were grown at 22°C by the hanging-drop vapor diffusion method. The crystallization buffer contained 10% PEG 3350, 0.1 M Tris-HCl (pH 7.2), 0.1 M LiSO4, 1 mM ATPγS, 2% isopropanol, and 5 mM MgCl2. Diffraction data were collected at −170°C using crystals flash-frozen in crystallization buffer containing 30% (w/v) glycerol. Diffraction data from native crystals were collected at 1.000 Å on beamline 4A at the Pohang Advanced Light Source (PAL). The MjMR crystals formed in space group C2221 (with a = 86.96 Å, b = 147.13 Å, and c = 175.91 Å) and contained one complex molecule in an asymmetric unit. Diffraction data integration, scaling, and merging were performed using the HKL2000 package (Supplemental Table S1; Otwinowski and Minor 1997).
Structure determination and refinement
The structures of the complex, free MjRad50CC, and free MjMre11 were determined by the molecular replacement method. We initially determined the structure of MjRad50CC with the PHENIX program using PfRad50CD as a search model (Hopfner et al. 2000b; Adams et al. 2009). For the MjMR complex, MjRad50 was located with the PHENIX program followed by the search of MjMre11 using PfMre11 as a model (Hopfner et al. 2001). After density modification, an electron density map generated at a resolution of 3.5 Å using the PHENIX program showed good quality, which allows most chains to be traced. Successive rounds of model building using COOT (Emsley and Cowtan 2004) and refinement using CNS (Brünger et al. 1998) and PHENIX were performed to build the complete model. The final model consisted of residues 1–366 of MjMre11, residues 1–189 and 830–1005 of MjRad50, two ATPγS molecules, two Mg2+ ions, two SO42− ions, and 20 water molecules. The N-terminal His-tagged residues were not visible and were presumably disordered. We also determined the structure of free MjMre11 using MjMre11 from the complex as a model. The statistics are summarized in Supplemental Table S1.
Nuclease assays
Cleavage reaction mixtures contained 20 nM 32P-labeled substrate DNA molecule(s) and enzyme (as indicated) in reaction buffer (10 mM Bis-Tris propane at pH 7.5, 50 mM NaCl, 2 mM dithiothreitol [DTT], 5% glycerol, 5 mM MnCl2). ATP (1 mM) was also added at the beginning. Reaction mixtures were incubated for 60 min or the specified time (for kinetics) at 55°C, and were stopped by the addition of 1/10 vol of stop mixture (3% sodium dodecyl sufate [SDS], 50 mM EDTA, 0.5 mg/mL proteinase K), followed by 10 min of incubation at 37°C. Reaction products were boiled for 5 min and were resolved on 15% denaturing polyacrylamide gels containing 7 M urea in TBE buffer. Gels were run for 360 min at 13 V cm−1. After electrophoresis, the gels were fixed in fixing buffer (30% methanol, 5% acetic acid, 5% glycerol), dried, and subjected to autoradiography and PhosphorImager analysis. In all analyses, three independent experiments were performed.
ATPase assays
The wild-type MjMRCC complex and its mutant proteins were incubated in 10 μL of reaction buffer (20 mM Tris at pH 8.0, 100 mM NaCl, 5 mM DTT, 10% glycerol) with 5 mM MgCl2 and 50 μM [γ-32P]ATP (PerkinElmer). Reaction mixtures were incubated for 60 min at 37°C, and were stopped by the addition of 1/5 vol of stop mixture (5% SDS, 50 mM EDTA). Reaction product (2 μL) was then spotted onto a polyethyleneimine (PEI) plate (EMD Biosciences) and resolved by thin-layer chromatography for ATP and Pi by using 0.75 M KH2PO4. The plate was dried and analyzed using a BAS 2000 (Fujifilm) bioimage analyzer and was quantified using the Multi Gauge version 3.1 program. In all analyses, three independent experiments were performed to measure the ATPase activities.
EMSAs
DNA-binding activities of the MjMRCC complex and its mutants (MRgroove and MR3βS) were incubated in 10 μL of reactions containing the buffer (10 mM BTP at pH 7.5, 50 mM NaCl, 5 mM DTT, 5% glycerol) with 5 mM MgCl2 or 1 mM ATP or ATPγS. Each sample in increasing amounts (the molar ratio of protein:DNA was 50:1 and 200:1) was incubated with 4 nM blunt-end long-duplex DNA (3 kb) for 30 min at 26°C. After centrifugation at 18,000 rpm for 10 min, the pellet was mixed with 10 μL of loading dye (6 mM Tris-HCl at pH 8.0, 5% glycerol, 0.03% xylene cyanol/bromophenol blue, 0.1% SDS). Supernatant and pellet were resolved by electrophoresis (100 V/20 min) on a 0.5% agarose gel and visualized by ethidium bromide staining. The EMSA for the MR–short DNA complex is described in the Supplemental Material.
Acknowledgments
Coordinates and structure factors have been deposited with the RCSB database with 3AV0 (the MjMR complex), 3AVZ (MjMre11), 3AUY (MjRad50-ADP), and 3AUX (MjRad50-ADP). This work was supported by grants from the National R&D Program for Cancer Control, the Ministry for Health and Welfare (1020280), a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST; nos. 2010-0019706 and 2010-0029766), and the BK21 program (Ministry of Education).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2037811.
Supplemental material is available for this article.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Trujillo KM, Sung P, Erickson HP 2001. Structure of the Rad50–Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J Biol Chem 276: 37027–37033 [DOI] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV 1999. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res 27: 1223–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Nishino T, Haering CH, Gruber S, Nasmyth K 2006. Cohesin's ATPase activity is stimulated by the C-terminal winged-helix domain of its kleisin subunit. Curr Biol 16: 1998–2008 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Tommiska J, Oplustilova L, Aaltonen K, Tamminen A, Heikkinen T, Mistrik M, Aittomäki K, Blomqvist C, Heikkilä P, et al. 2008. Aberrations of the MRE11–RAD50–NBS1 DNA damage sensor complex in human breast cancer: MRE11 as a candidate familial cancer-predisposing gene. Mol Oncol 2: 296–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, Hays L, Morgan WF, Petrini JH 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93: 477–486 [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE 2005. Effect of amino acid substitutions in the Rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem 280: 2620–2627 [DOI] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DRF 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci 95: 7969–7974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol 3: 317–327 [DOI] [PubMed] [Google Scholar]
- Das D, Moiani D, Axelrod HL, Miller MD, McMullan D, Jin KK, Abdubek P, Astakhova T, Burra P, Carlton D, et al. 2010. Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair. J Mol Biol 397: 647–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell 8: 1129–1135 [DOI] [PubMed] [Google Scholar]
- de Jager M, Trujillo KM, Sung P, Hopfner KP, Carney JP, Tainer JA, Connelly JC, Leach DRF, Kanaar R, Wyman C 2004. Differential arrangements of conserved building blocks among homologs of the Rad50/Mre11 DNA repair protein complex. J Mol Biol 339: 937–949 [DOI] [PubMed] [Google Scholar]
- Dumon-Jones V, Frappart PO, Tong WM, Sajithlal G, Hulla W, Schmid G, Herceg Z, Digweed M, Wang ZQ 2003. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res 63: 7263–7269 [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Ghosal G, Muniyappa K 2007. The characterization of Saccharomyces cerevisiae Mre11/Rad50/Xrs2 complex reveals that Rad50 negatively regulates Mre11 endonucleolytic but not the exonucleolytic activity. J Mol Biol 372: 864–882 [DOI] [PubMed] [Google Scholar]
- Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, Ottini L, Crescenzi M, Martinotti S, Bignami M, et al. 2002. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep 3: 248–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdendorf TJ, Albrecht DW, Benkovic SJ, Nelson SW 2010. Biochemical characterization of bacteriophage T4 Mre11/Rad50 complex. J Biol Chem 286: 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T 2005. Condensins: organizing and segregating the genome. Curr Biol 15: R265–R275 doi: 10.1016/j.cub.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Hirano M, Hirano T 2004. Positive and negative regulation of SMC–DNA interactions by ATP and accessory proteins. EMBO J 23: 2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP 2000a. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol 182: 6036–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA 2000b. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC–ATPase superfamily. Cell 101: 789–800 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50–ATPase. Cell 105: 473–485 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Wyman C, Rothstein R 2008. Quality control of DNA break metabolism: in the ‘end’, it's a good thing. EMBO J 27: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27: 247–254 [DOI] [PubMed] [Google Scholar]
- Koroleva O, Makharashvili N, Courcelle CT, Courcelle J, Korolev S 2007. Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J 26: 867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens A, Schele A, Hopfner KP 2004. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr Biol 14: 1778–1782 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT 2005. ATM activation by DNA double-strand breaks through the Mre11–Rad50–Nbs1 complex. Science 308: 551–554 [DOI] [PubMed] [Google Scholar]
- Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K 2004. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol 14: 2107–2112 [DOI] [PubMed] [Google Scholar]
- Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci 96: 7376–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, Alden E, Tainer JA, Paull TT 2004. The Rad50 signature motif: essential to ATP binding and biological function. J Mol Biol 335: 937–951 [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C 2005. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437: 440–443 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979 [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev 13: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 16: 819–824 [DOI] [PubMed] [Google Scholar]
- Taylor AM, Groom A, Byrd PJ 2004. Ataxia-telangiectasia-like disorder (ATLD)—its clinical presentation and molecular basis. DNA Repair (Amst) 3: 1219–1225 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Sung P 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50·Mre11 complex. J Biol Chem 276: 35458–35464 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Yuan SS, Lee EY, Sung P 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem 273: 21447–21450 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Roh DH, Chen L, Van Komen S, Tomkinson A, Sung P 2003. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J Biol Chem 278: 48957–48964 [DOI] [PubMed] [Google Scholar]
- Wang Z, Cummins JM, Shen D, Cahill DP, Jallepalli PV, Wang TL, Parsons DW, Traverso G, Awad M, Silliman N, et al. 2004. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res 64: 2998–3001 [DOI] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, et al. 2009. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell 136: 85–96 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT 1997. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res 25: 2985–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Saidi A, Frappart PO, Min W, Barrucand C, Dumon-Jones V, Michelon J, Herceg Z, Wang ZQ 2006. Conditional deletion of Nbs1 in murine cells reveals its role in branching repair pathways of DNA double-strand breaks. EMBO J 25: 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A 2001. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol 11: 105–109 [DOI] [PubMed] [Google Scholar]