Abstract
Chromatin-modifying enzymes are known to be critical components for the correct differentiation of embryonic stem cells into specific lineages, such as neurons. Recently, the role of Polycomb group proteins has been studied in the specification and differentiation of muscle stem cells. In this perspective, we review a recent study by Juan and colleagues (pp. 789–794) in Genes & Development of the role of the polycomb group protein Ezh2 in muscle stem cells, and discuss the implications for general lineage restriction.
Keywords: Polycomb, Ezh2, satellite cells, muscle growth, muscle regeneration
Polycomb-mediated gene repression has been shown to be critical for the differentiation potential of embryonic stem cells (ESCs). However, because the catalytic subunit Ezh2 is an essential gene in mice, the role of the polycomb-repressive complex in tissue specification and differentiation has been difficult to study in mammals. To circumvent this problem, Juan et al. (2011) present data in the April 15, 2011, issue of Genes & Development from experiments using genetically engineered mice that lack Ezh2 specifically in muscle satellite cells, a type of tissue stem cell required for postnatal muscle growth and repair. In this perspective, we discuss muscle development and polycomb-mediated repression of extraneous gene programs, and how this might reflect a mechanism to prevent nonlineage gene activation by members of transcription factor families that are capable of binding widely across the genome and participating in many different differentiation programs.
Transcriptional control in skeletal muscle myogenesis
In vertebrates, cells that will become the skeletal muscles of the body are formed through a stepwise process of cell fate restriction and commitment, beginning with an epithelial-to-mesenchymal cell state transition (EMT). In the developing embryo, groups of epithelial somite cells will express the EMT-promoting transcription factors TWIST and/or SNAIL in order to generate early mesenchymal cells that are capable of becoming cartilage, bone, dermal, or skeletal muscle tissues (Delfini et al. 2009). Specifically, subsets of these somitic cells are restricted in their lineage potential by expression of a paired box transcription factor, PAX3, and/or expression of the basic helix–loop–helix (bHLH) factor MYF5, which specifies them as muscle precursors (Maroto et al. 1997; Tajbakhsh et al. 1997).
During development, PAX3+ cells undergo a wave of proliferation and, in the case of the cells destined to contribute to limb muscles, migrate to the terminal limb buds (Bober et al. 1994; Daston et al. 1996). The initial number of muscle progenitor cells in either the somites or limb buds has been suggested to be insufficient to account for the mass of muscle produced during embryogenesis, suggesting that these muscle progenitors are capable of substantial subsequent proliferation (Maina et al. 1996). To induce this population of PAX3+ cells to undergo terminal myogenic differentiation and form myotubes, extrinsic signals from the surrounding tissue(s) activate the myogenic transcription factor genes Myf5 and Myod (Rudnicki et al. 1993; Tajbakhsh et al. 1998). In mice lacking both Myf5 and Myod, PAX3+ precursor cells are capable of both proliferation and migration to the limb buds but, once in place, adopt nonmuscle cell fates (Kablar et al. 1999). In particular, these cells differentiate into cells with chondrocyte-like characteristics (Kablar et al. 1999). Similar to the double-knockout mice, in Myf5-null mice, the PAX3+ cells in the myotome—the portion of the somite that will give rise to the trunk muscles—migrate aberrantly and adopt alternative cell fates (Tajbakhsh et al. 1996). For the cells that migrate to the limb buds and adopt nonmuscle fates, it is unclear whether this occurs as a result of stochastic misexpression of lineage-specific transcription factors or results from competing extrinsic signals that can operate on these cells in the absence of the Myod/Myf5-driven myogenic program that would presumably be dominant. However, experiments tracing the lineage of Myf5-null cells that remain in the myotome found that muscle precursor cells on the dorsal side began to express Dermo-1, a gene normally expressed in dermal tissue, while those on the ventral side began to express scleraxis, a marker of collagenous tissues such as tendons (Tajbakhsh et al. 1996). This difference in alternative cell fates suggests that, in the absence of an overriding muscle program, extrinsic signals produced by the surrounding tissues drive these cells to adopt specific fates, and would thus suggest that the results in the Myf5-null mice are not merely a result of stochastic expression events.
Once expressed, MYF5 and MYOD induce the expression of additional myogenic bHLH transcription factors, myogenin and Mrf4, as well as many other transcription factors that have necessary roles in both myogenesis and the differentiation of other cell types, such as the Mef2, Runx, and Six families of transcription factors. The myogenic bHLH factors participate in a feed-forward regulatory circuit with these other factors, cooperating with them to regulate transcription of genes expressed in skeletal muscle (Bergstrom et al. 2002; Penn et al. 2004). These differentiation-specific transcription factors will induce the expression of genes necessary for the formation of muscle-specific structures, such as myosin heavy chain (Mhc) and muscle-specific creatine kinase (Ckm) (Arnold and Braun 1996; Bergstrom et al. 2002).
As mentioned above, although the initial muscle mass is created during embryogenesis, substantial muscle growth occurs during postnatal development. This growth depends on subpopulations of somite-derived cells that undergo a wave of proliferation and then migrate under the myofiber lamina (Mauro 1961; Gros et al. 2005). These cells, known as satellite cells, express the paired-box transcription factor PAX7 and are capable of self-renewal in a PAX7-dependent manner (Oustanina et al. 2004). While Pax7 does not appear to be critical for the specification of satellite cells, Pax7-null cells have limited potential for self-renewal, and the population of cells capable of contributing to muscle repair declines sharply with age (Oustanina et al. 2004). About 10% of the satellite cells express both CD34 and PAX7, but lack expression of Myf5 (Kuang et al. 2007). These cells are thought to represent bona fide adult muscle stem cells, the portion of the PAX7+ satellite cell population capable of self-renewal (Beauchamp et al. 2000; Montarras et al. 2005). The remainder of the satellite cell population likely represents committed myoblasts, capable of a more limited number of divisions. Satellite cells predominantly contribute to muscle fiber growth by fusing with existing myotubes, with the occasional formation of new myotubes (White et al. 2010). In the adult, satellite cells act as a reservoir of PAX7+ stem cells that, when activated by regeneration signals, give rise to PAX7+/MYF5+ myoblasts, which contribute to muscle repair (Kuang et al. 2007). Although satellite cells appear to arise from PAX3+ precursor cells in the somite, it should be noted that embryonic and fetal myoblasts behave differently in culture compared with satellite cells harvested from adult tissues, suggesting differences between the two cell populations (Cossu and Molinaro 1987). However, it is unclear what drives these differences and how they are established.
Cell fate determination and Polycomb group proteins
Ensuring transcriptional specificity across a variety of developmental stages and cellular types requires complex cellular mechanisms to regulate the preservation and alteration of transcriptional information. Although some of this information can be inherited by specifically segregating transcription factors into daughter cells, transcriptional information can also be retained independently of this mechanism. The maintenance of transcription factor-independent gene expression states was first described in the regulation of homeotic genes in Drosophila. During the development of a Drosophila embryo, homeotic genes are activated or repressed in a tissue-specific manner by a number of transcription factors. Remarkably, the expression state of these genes is maintained in subsequent cellular lineages through multiple rounds of mitosis, even after the specifying transcription factors themselves are lost during later stages of development (Mahmoudi and Verrijzer 2001). Polycomb group proteins maintain the cellular identity of these lineages by modifying the chromatin context of homeotic genes. Presumably, these specialized proteins establish a nonpermissive chromatin structure via their ability to modify histones.
Polycomb proteins form two major complexes: Polycomb-repressive complex 1 (PRC1) and PRC2 (Shao et al. 1999; Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). While some of the individual subunits are shared between the two complexes, PRC1 and PRC2 are functionally distinct. PRC1 ubiquitylates histone H2A on Lys 119, leading to chromatin compaction by unknown mechanisms (Francis et al. 2004; Wang et al. 2004). PRC2 predominantly catalyzes the di- and trimethylation of H3 on Lys 27 (H3K27), leading to chromatin-based repression by unknown mechanisms (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). In vitro, addition of PRC2 proteins to purified chromatin results in the compaction of nucleosomes; however, this activity is separable from polycomb-mediated methylation of H3K27, and it remains controversial whether this compaction occurs in vivo (Francis et al. 2004).
In mammals, methylation of H3K27 by PRC2 requires the complex to contain one of the enhancer of zeste proteins: EZH1 or EZH2. EZH2 appears to have greater methylation potential than EZH1, and PRC2 complexes containing EZH1 or EZH2 are at least partially functionally distinct, a fact that likely accounts for the differences that have been observed in their spatial and temporal expression patterns (Margueron et al. 2008). In general, EZH1 is found in quiescent cells, likely serving as a maintenance enzyme to preserve H3K27 methylation levels, while EZH2 is predominantly found in cycling cells, presumably acting to establish new heterochromatin or re-establish pre-existing heterochromatin after cell division (Margueron et al. 2008).
The core PRC2 subunits can associate with a number of nonessential accessory proteins, which may fine-tune the functionality of the complex, likely via targeting of the enzymatic activity to distinct genomic locations. These accessory proteins may directly bind DNA in a sequence-specific manner, allowing recruitment of PRC2 to target loci, or act through intermediaries. In addition, recent research suggests a role for long noncoding RNAs (lncRNAs) in targeting PRC2 to specific genomic locations through either interaction with the core subunit SUZ12 or interactions with accessory subunits (Plath et al. 2003; Rinn et al. 2007; Zhao et al. 2008; Khalil et al. 2009; Maenner et al. 2010).
While H3K27 monomethylation is found in intergenic heterochromatin and over active gene bodies, di- and trimethylation is associated mainly with facultative heterochromatin, a type of silent chromatin that is reversible and is often associated with developmentally controlled gene repression. Indeed, the promoters/enhancers of many lineage-specific genes are associated with H3K27 methylation in ESCs, and some of these elements lose this methylation signature upon differentiation, depending on the specific cell type (Boyer et al. 2006; Cui et al. 2009). During this process, genes that promote cell growth and were active in ESCs become silenced, an event that also appears to be at least partly dependent on PRC2-mediated H3K27 methylation (Bracken et al. 2007). It is possible that this regulatory strategy is widely used in animal development, as ∼50% of the histone H3 in murine ESCs is dimethylated on K27 and an additional 15% is trimethylated (Peters et al. 2003).
In ESCs, potentiation of lineage-specific genes is also dependent on PRC2-dependent activity. While PRC2 represses lineage-specific genes by H3K27 methylation of promoter-associated histones, PRC2 also simultaneously recruits the deposition of the histone variant H2A.Z into these nucleosomes (Creyghton et al. 2008). This histone variant is generally associated with transcriptional activity and is antagonistic to DNA methylation of these promoters (Barski et al. 2007; Zilberman et al. 2008). It is thought that the PRC2-dependent incorporation of H2A.Z protects these genes from being locked into a stable silent state by DNA methylation and allows them to be activated quickly during the differentiation process. Once a specific cell program is established, developmental genes for other tissue types retain H3K27 trimethylation, lose H2A.Z, and acquire DNA methylation over their promoters. Loss of EZH2 in established ESC lines does not impair the expression of stem cell genes such as the Sox and Wnt genes or impair the ability of these cells to self-renew (Boyer et al. 2006). However, ESCs lacking EZH2 have defects in differentiation potential, likely because they cannot repress the stem cell program and prevent the expression of multiple lineage-specific genes. Some research also indicates that ESCs that are lacking Ezh2 and are induced to differentiate down a specific lineage pathway fail to maintain the expression of those genes required for maintaining that cell type identity (Boyer et al. 2006). Because PRC2 is generally considered to be a repressor, this is likely due to the activation of inhibitors or the lack of EZH2-mediated H2A.Z deposition at these sites prior to differentiation, allowing them to become repressed by DNA methylation.
Polycomb is required for specification and commitment of muscle stem cells
In the recent study by Juan et al. (2011) in Genes & Development, the role of PRC2 in myogenic stem cells was examined by deleting the EZH2 subunit of PRC2 in a Pax7-dependent manner. This method used the Cre recombinase driven by the Pax7 promoter to remove Ezh2 from cells coincident with the point of satellite cell specification, allowing Juan et al. (2011) to address the mechanism of satellite cell commitment and maintenance during normal murine muscle development.
Using this approach, Juan et al. (2011) found that Ezh2 is required for adult PAX7+ satellite cell homeostasis and proliferation, but is not required for fetal muscle development. This conclusion was drawn from the observations that, at birth, mice with Pax7-driven Ezh2 deletions had a comparable number of similarly sized myofibers as wild-type mice. However, 1-wk-old mice carrying the Ezh2 deletion had substantially smaller myofibers, although a similar number of fibers overall. This suggested that the Ezh2Pax7 transgenic mice had a defect in the ability of PAX7+ satellite cells to contribute to postnatal muscle growth in the absence of Ezh2. To determine whether this defect was due to an inability of PAX7+ cells to self-renew, resulting in a depletion of the pool of satellite cells, Juan et al. (2011) compared the numbers of PAX7+ cells associated with myofibers isolated from mice with or without the Ezh2 deletion. Although fetal muscles had comparable numbers of PAX7+ cells in mice with or without the Ezh2 deletion, there was a marked decrease in the number of PAX7+ cells associated with myofibers isolated from mice with the Pax7-dependent Ezh2 deletion. In addition, when PAX7+ cells isolated from adult myofibers were induced to proliferate in culture for 3 d, cells lacking Ezh2 gave rise to markedly fewer cells. Staining these cells with antibodies to detect either H3 phosphorylated on Ser10 or BrdU incorporation revealed an approximately twofold decrease in the replication potential of the Ezh2-null cells.
To test whether the defect in muscle size was directly related to the inability of PAX7+/Ezh2-null satellite cells to contribute to muscle formation, Juan et al. (2011) used an established model of muscle repair based on muscle damage induced by cardiotoxin. Normally, the damage caused by cardiotoxin induces proliferation of the PAX7+ satellite compartment and results in the differentiation of satellite cells into myofibers. In contrast to the normal situation, Juan et al. (2011) observed a drastic 10-fold reduction in the number of PAX7+ cells and a twofold reduction in H3S10 phosphorylation, leading to the conclusion that EZH2 is required for satellite cell proliferation during muscle regeneration.
Because EZH2 has been implicated in the repression of the cell cycle inhibitor p16 (Cdkn2), Juan et al. (2011) asked whether the inability of Ezh2-null cells to proliferate was due to activation of p16. They found that satellite cells lacking EZH2 did indeed have increased levels of p16. To directly test the idea that the defect in satellite cell proliferation was mediated by p16, Juan et al. (2011) then treated wild-type and Ezh2-null/PAX7+ cells with a siRNA directed against p16. Although the siRNA treatment had no effect on the percentage of wild-type cells positive for H3S10 phosphorylation (a marker of mitosis), cells lacking Ezh2 had a higher percentage of cells positive for H3S10 phosphorylation than those treated with a control siRNA, suggesting that inhibition of p16 could at least partially rescue the proliferation defect in these cells.
Finally, Juan et al. (2011) assessed Ezh2-null cells for the expression of genes associated with other, nonmyogenic, lineages. Comparison of expression profiles from wild-type or Ezh2-null skeletal muscle tissues revealed misexpression of genes normally kept silent by PRC2 in ESCs, such as Zic-1, a gene normally expressed in early somites and the cerebellum. Tissue-specific genes such as Agrp1, normally expressed in hypothalamic neurons, and Opsin1, normally expressed in retinal cones, were also found to be up-regulated in the Ezh2-null muscle cells. The unscheduled expression of these genes and others is likely a direct consequence of EZH2 loss, as these genes are marked by H3K27 methylation in wild-type satellite cells but not in the Ezh2-null cells. Interestingly, up-regulation of other non-skeletal muscle-specific genes was observed in the total muscle lysates from Ezh2-null animals, but not in a sorted PAX7+ population, suggesting that, although the defect originates in the muscle stem cells, it continues to result in aberrant expression in differentiated daughter cells. Surprisingly, these mixed-lineage genes were not up-regulated when Ezh2 was deleted in a MyoD-dependent manner. This suggests that either mixed-lineage genes are already locked into a silent state by the time the EZH2 protein is lost in these committed myoblasts, or some factor, such as PAX7, which is expressed early in satellite cell specification but is lost during the differentiation process, is responsible for the misexpression of these genes in the absence of EZH2-mediated repression.
A repressive balancing act: the effects of Polycomb in muscle development
Interestingly, striking differences have been observed in the phenotype of PAX7+ satellite cells, depending on the precise timing of EZH2 loss. As discussed above, loss of EZH2 at the time of satellite cell commitment leads to a proliferation defect due, in part, to the derepression of the cell cycle inhibiter p16. In contrast, when PAX7+ cells were grown for several days in culture and then treated with siRNA against Ezh2, the cells continued to proliferate and were unable to differentiate, a defect linked to the inability of these cells to repress Pax7 via EZH2 during the process of differentiation (Palacios et al. 2010). It is interesting to note that this defect is reversible, as transiently blocking the activation of EZH2 does not permanently inhibit differentiation, but only delays it (Palacios et al. 2010). Surprisingly, when EZH2 was depleted by siRNA in early PAX7+ cell cultures (day 1 in culture), cell growth was impaired, although it is unclear whether this was the result of an inability to self-renew or of unscheduled expression of differentiation-promoting genes (Caretti et al. 2004). EZH2 must also be lost from the genomic loci of differentiation-specific genes to allow for their expression, since constitutive expression of Ezh2 prevented their activation (Caretti et al. 2004). However, it should be noted that not all differentiation-specific genes are repressed in early myoblasts by EZH2. The promoter for myogenin, a muscle-specific regulatory factor that acts in a cooperative manner with MYOD to induce late muscle target gene expression, for example, is not associated with EZH2 or H3K27 methylation in early myoblasts (Caretti et al. 2004). It is possible that differential interaction between EZH2 and accessory factors may account for the ability of EZH2 to be recruited to stem cell genes while simultaneously being lost from lineage-specific genes. However, the ability of accessory proteins such as YY1 to target PRC2 to specific genomic locations remains controversial. In summary, the general picture emerging from experiments in which EZH2 levels in muscle satellite cells have been manipulated is one of a delicate balance between the maintenance of existing epigenetic states and the establishment of new chromatin modifications by polycomb complexes during cell fate specification and differentiation.
Are chromatin modifiers safeguarding cell identity from promiscuous transcription factors?
The finding that the Ezh2-null satellite cells express both myogenic genes and nonmyogenic gene programs is an intriguing one. As noted above, previous work in stem cells has shown that methylation of H3K27 can participate in bivalent chromatin domains that maintain genes in a poised state that can then be either activated or suppressed during lineage commitment and differentiation. The finding that Ezh2 is required to suppress transcription of nonlineage genes in differentiating satellite cells suggests that the process of lineage restriction via epigenetic silencing is also required during terminal differentiation. One avenue of research that might address broader questions about transcriptional control is the elucidation of the mechanism(s) that either permit or preclude activation of these genes.
One possibility is that the activation of the misexpressed genes is a purely stochastic process due to the lack of H3K27 methylation over their promoter elements. Theoretically, the lack of repressive chromatin marks could create a permissive platform for RNA polymerase II (Pol II) to bind and transcribe targets in a nonregulated and relatively random fashion. Another possibility, however, is that the “choice” of genes is actually a result of the widespread binding of transcription factors already present in the cell to additional sites across the genome that lack H3K27 in the absence of EZH2. In this model, genes with binding sequences for myogenic factors such as PAX7, MYF5, MYOD, or MEF2, but that are not normally bound by these factors during myogenesis, may become aberrantly active due to the lack of H3K27 methylation and repressive chromatin structures over these sequences.
Such possibilities bring to mind broader questions about how cells are committed to a given lineage, and (1) how such commitments and restrictions are maintained, and (2) to what extent and how, from a mechanistic standpoint, chromatin modifications are involved in that process. At the crux of such questions is the fact that most transcription factors bind only a small subset of the number of their binding motifs that are present in the genome. The misexpression observed in the Ezh2-null satellite cells brings to the forefront the questions of how much the occlusion of potential transcription factor-binding sites, by chromatin or competing factors, is responsible for keeping cells committed to a single lineage, and how much of the process requires active positive regulation by factors of the appropriate lineage.
Many differentiation programs involve the stepwise activation of genes, often regulated by a feed-forward regulatory circuit involving both lineage-restricted and multilineage transcription factors. Muscle specification begins with the activation of Myf5 and/or Myod, and these muscle-specific factors, in turn, induce the expression of factors that coregulate gene expression in multiple different lineages—such as MEF2, RUNX, and SIX proteins—in a feed-forward circuit (Tapscott 2005). These latter factors also regulate genes expressed in neurons, cartilage, and many other cell types, yet in muscle cells their binding and activity is restricted to the genes of the myogenic program. Because chromatin-modifying enzymes participate in the muscle differentiation process and are recruited to specific loci by myogenic transcription factors, it is possible that the regulation of chromatin structures serves as a mechanism to narrow the differentiation program by restricting the binding sites available to factors involved in other lineage programs (de la Serna et al. 2001; Puri et al. 2001; Mal 2006; Ohkawa et al. 2006). Also, if chromatin accessibility over these sequences is only retained if the site is bound by one of these factors, this may provide a mechanism to prevent cells from going backward toward a stem cell-like state.
This model of restricting the activity of multilineage transcription factors can also be extended to lineage-specific factors. Many lineage-specific transcription factors belong to large families (such as the bHLH and paired-box families of Myod and Pax7, respectively) with multiple members that all recognize the same core sequence motif, yet each family member regulates distinct sets of cell type-specific genes. In humans, the bHLH family of transcription factors has ∼120 members, most of which bind an E-box motif with a common CANNTG core. Despite the common consensus binding sequence, many of the factors mediate distinct lineage-specific functions. For example, MYOD and NEUROD both bind E-boxes, yet specify skeletal muscle and neurons, respectively. The duplication and divergence of transcription factor families is recognized to support the evolution of new functional attributes; however, it is likely that there is substantial selective pressure to maintain some common elements of a differentiation program. For example, most terminally differentiated cell types must express genes that inhibit proliferation, such as p16, thus repressing stem cell characteristics. If there is a need to maintain some commonly regulated genes, then there would be selective pressure on these transcription families to constrain the divergence of sequence recognition. However, this constraint is in conflict with the need for these related transcription factors to activate lineage-specific programs.
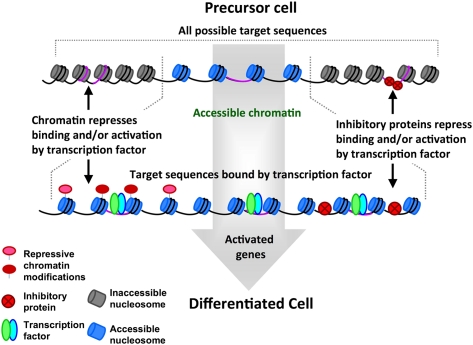
One possible mechanism to resolve the need to both maintain common binding motifs among transcription factor family members and generate lineage-specific transcriptional programs might be through the modulation of chromatin structure during lineage specification that restricts the binding sites available or the transcriptional activity of the bound factor (Fig. 1). In this manner, subsets of binding targets could be made available, or unavailable, to transcription factors in the lineage-committed cell, and the transcriptional activity could be further narrowed by the action of specific cofactors. Chromatin-mediated cell-specific modulation of available binding sites has been reported for the glucorticoid receptor and is likely to be a widespread mechanism to constrain non-lineage-specific transcription (John et al. 2011). This model also fits nicely with the data from experiments testing the role of Ezh2 in muscle specification and differentiation outlined above, which indicate a critical requirement for Ezh2 during initiation of cell type specification to suppress nonrelevant lineage programs. A major question for future research is identifying the molecular mechanisms for selecting which regions to epigenetically repress. Presumably this mechanism would need to distinguish lineage-specific binding regions from binding regions associated with nonlineage programs.
Figure 1.
Model for restriction of transcription factor binding and gene activation during cell lineage commitment. In this model, during cellular lineage commitment and differentiation, the genes activated by transcription factors are restricted by a variety of processes. (Top) We envision that chromatin structure and the occlusion of binding sequences by repressors will dictate the first level of restriction by making subsets of binding targets either accessible or inaccessible to transcription factors (inaccessible nucleosomes are shaded in dark gray, and accessible nucleosomes are shaded in light blue). (Bottom) Sequence motifs located in accessible chromatin domains can be bound by transcription factor complexes, but the silent state of associated genes may be retained by either repressive chromatin marks or the presence of inhibitors. The end result is a carefully modulated pattern of transcription factor activity that leads to the expression pattern of a differentiated cell type.
Acknowledgments
M.L.C. and Z.Y. were both supported by the NIH Interdisciplinary Training Grant in Cancer Research (T32CA080416), S.J.T. was supported by NIH NIAMS (R01AR045113), K.L.M. was supported by a Developmental Biology Predoctoral Training Grant (T32HD007183) from the National Institute of Child Health and Human Development, and A.P.F. was supported by a grant from the University of Washington Child Health Research Center (NIH U5K12HD043376-08).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2054311.
References
- Arnold HH, Braun T 1996. Targeted inactivation of myogenic factor genes reveals their role during mouse myogenesis: a review. Int J Dev Biol 40: 345–353 [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS 2000. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol 151: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell 9: 587–600 [DOI] [PubMed] [Google Scholar]
- Bober E, Franz T, Arnold HH, Gruss P, Tremblay P 1994. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 120: 603–612 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18: 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossu G, Molinaro M 1987. Cell heterogeneity in the myogenic lineage. Curr Top Dev Biol 23: 185–208 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA 2008. H2AZ is enriched at Polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell Mol Life Sci 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Daston G, Lamar E, Olivier M, Goulding M 1996. Pax3 is necessary for migration but not differentiation of limb muscle precursors in the mouse. Development 122: 1017–1027 [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Roy K, Carlson KA, Imbalzano AN 2001. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J Biol Chem 276: 41486–41491 [DOI] [PubMed] [Google Scholar]
- Delfini MC, De La Celle M, Gros J, Serralbo O, Marics I, Seux M, Scaal M, Marcelle C 2009. The timing of emergence of muscle progenitors is controlled by an FGF/ERK/SNAIL1 pathway. Dev Biol 333: 229–237 [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL 2004. Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Gros J, Manceau M, Thome V, Marcelle C 2005. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435: 954–958 [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Derfoul A, Feng X, Ryall JG, Dell'Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V 2011. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev 25: 789–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Tapscott SJ, Goldhamer DJ, Rudnicki MA 1999. Myogenic determination occurs independently in somites and limb buds. Dev Biol 206: 219–231 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA 2007. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2010. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol 8: e1000276 doi: 10.1371/journal.pbio.1000276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Verrijzer CP 2001. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene 20: 3055–3066 [DOI] [PubMed] [Google Scholar]
- Maina F, Casagranda F, Audero E, Simeone A, Comoglio PM, Klein R, Ponzetto C 1996. Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development. Cell 87: 531–542 [DOI] [PubMed] [Google Scholar]
- Mal AK 2006. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J 25: 3323–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D 2008. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32: 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto M, Reshef R, Munsterberg AE, Koester S, Goulding M, Lassar AB 1997. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell 89: 139–148 [DOI] [PubMed] [Google Scholar]
- Mauro A 1961. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol 9: 493–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M 2005. Direct isolation of satellite cells for skeletal muscle regeneration. Science 309: 2064–2067 [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CG, Imbalzano AN 2006. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J 25: 490–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T 2004. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J 23: 3430–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, et al. 2010. TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 7: 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ 2004. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev 18: 2348–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135 [DOI] [PubMed] [Google Scholar]
- Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, Giordano A, Kedes L, Wang JY, Sartorelli V 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell 8: 885–897 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75: 1351–1359 [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Buckingham M 1996. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature 384: 266–270 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M 1997. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89: 127–138 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Borello U, Vivarelli E, Kelly R, Papkoff J, Duprez D, Buckingham M, Cossu G 1998. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development 125: 4155–4162 [DOI] [PubMed] [Google Scholar]
- Tapscott SJ 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132: 2685–2695 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- White RB, Bierinx AS, Gnocchi VF, Zammit PS 2010. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol 10: 21 doi: 10.1186/1471-213X-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S 2008. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]