Abstract
Liprins are a conserved family of scaffolding proteins important for the proper regulation and development of neuronal synapses. Humans have four liprin-αs and two liprin-βs which all contain long coiled-coil domains followed by three tandem SAM domains. Complex interactions between the coiled-coil and SAM domains are thought to create liprin scaffolds, but the structural and biochemical properties of these domains remain largely uncharacterized. In this study we find that the human liprin-β2 coiled-coil forms an extended dimer. Several protease-resistant sub-domains within the liprin-β1 and liprin-β2 coiled-coils were also identified. A 2.0 Å crystal structure of the central, protease-resistant core of the liprin-β2 coiled-coil reveals a parallel helix orientation. These studies represent an initial step towards determining the overall architecture of liprin scaffolds and understanding the molecular basis for their synaptic functions.
LAR family transmembrane protein-tyrosine phosphatase interacting proteins, or liprins, are a conserved family of proteins involved in the formation of the presynaptic cytomatrix of the active zone.(1),(2) The human genome encodes four liprin-αs and two liprin-βs, but only liprin-αs bind to LAR.(2) Liprin-α1 has also been found to interact with numerous other proteins including CASK,(3) ERC,(4) GIT,(5) KIF1A,(6) RIM,(7) and RSY-1(8). Liprin-α1 was first directly linked to the neural synapse with the discovery that a mutation in the C. elegans liprin-α1 homolog, syd-2, led to the development of abnormally large synaptic active zones and impaired synaptic transmission.(9) Subsequently, liprin-α1 has been shown to be important for photoreceptor targeting and selection in Drosophila,(10),(11) axon guidance in C. elegans,(12) and synapse development in rat hippocampal neurons.(13) At post-synaptic sites, liprin-α1 is involved in the trafficking of AMPA receptors through its interaction with GIT.(5) Recently, the human protein kazrin, previously known for its involvement in the regulation of desmosome assembly, was found to be a member of the liprin family.(14) Specifically, the kazrinE isoform has sequence similarity to liprins and also binds to LAR. In Drosophila, the kazrinE homologue, termed liprin-γ, was found to work in concert with liprin-α and liprin-β to regulate the development of photoreceptor and neuromuscular synapses.(15)
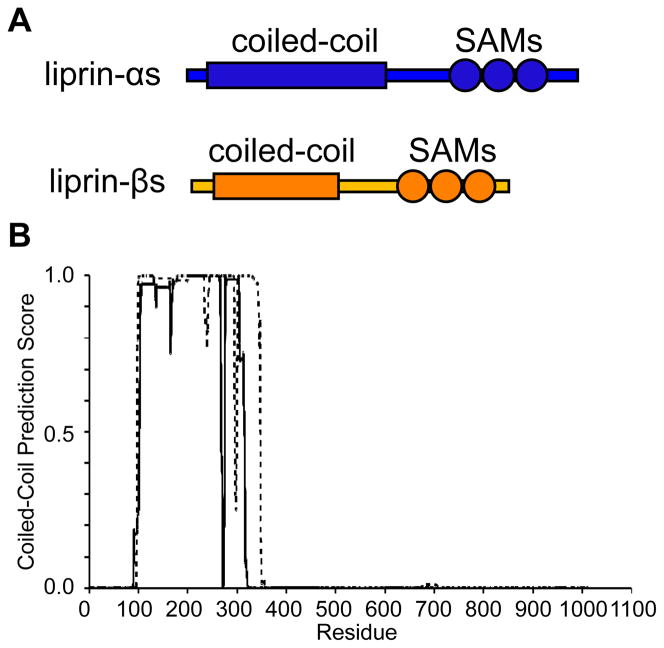
All liprins possess the same overall domain composition comprised of an N-terminal predicted coiled-coil region followed by three tandem SAM1 domains. The coiled-coil domains of liprin-βs are significantly shorter than liprin-αs (Figure 1A).(2) Immunoprecipitation experiments have shown that liprin-αs interact with other liprin-αs through their coiled-coil domains, but not liprin-βs.(2) Similarly, liprin-β coiled-coils interact with other liprin-β coiled-coil domains, but not liprin-αs. The C-terminal SAM domains appear to mediate interactions between liprin-αs and liprin-βs. The extensive interactions between liprins, coupled with their high expression in brain tissue, suggested early on that they possessed the ability to make complex neural scaffolding structures.(2)
Figure 1. Domain Organization of Liprin-β1 and -β2.
(A) The diagram shows the relative sizes and domain composition of liprin-αs (blue) to liprin-βs (orange). (B) A COILS (16) prediction using a 24-residue window for the coiled-coil domains from human liprin-β1 (dashed line) and liprin-β2 (solid line).
The overall architectures of the presumed liprin scaffolds are unknown since there is little structural information for the coiled-coil or SAM domains from any of the liprin proteins. Though there are many reliable algorithms to determine the presence of coiled-coil domains in proteins directly from the primary amino acid sequence,(16)(17)(18) it remains challenging to determine the number and orientations of the coiled-coils in each oligomer.(19)(20)(21)(22) SAM domains are also readily identified by their amino acid sequence and usually function as protein-protein interaction motifs, but they can have many different functions, including binding to RNA, lipids, and forming homo- and heteropolymers.(23) In the case of the liprin SAM domains, they have been predicted to form polymers, but this has not been verified experimentally.(24) Understanding the individual structures of these coiled-coil and SAM domains from liprins should help explain how they form the scaffolds necessary to execute their function in synapse development.
In this study we have focused on determining the structure of the coiled-coil domains of human liprin-βs. Specifically, we cloned the complete coiled-coil domains of human liprin-β1 and -β2 and focused the majority of our efforts studying liprin-β2. The coiled-coil domain of liprin-β2 was found to be predominantly dimeric by equilibrium sedimentation and chemical cross-linking experiments. Three protease-resistant sub-domains of the liprin-β1 and -β2 coiled-coils were also identified including the central third of the liprin-β2 coiled-coil, for which we were able to determine a crystal structure.
Materials and Methods
Cloning and Mutagenesis
The template DNA for cloning all human liprin-β1 (Image ID 5276255; Accession # BC050281) and liprin-β2 (Image ID 4826610; Accession # BC021714) constructs was obtained from Open Biosystems. The complete coiled-coil construct for liprin-β1 was created by PCR amplifying DNA corresponding to residues G97 through T342 and inserting the fragment into a modified pET-3a vector between MluI and SalI restriction sites containing an N-terminal sequence (MEKTR) for efficient expression and a C-terminal His-tag (RRHHHHHH) for purification. The complete coiled-coil construct for liprin-β2 was similarly cloned by inserting residues S100 through E315 into the same pET-3a plasmid restriction sites. The 21 kDa liprin-β1 protease-resistant fragment from E110 to K290 was cloned into a pQE2 vector (Qiagen) between restriction sites SphI and SalI containing a cleavable N-terminal His-tag (MKHHHHHHHMKAK). The 19 kDa and 9 kDa liprin-β2 protease-resistant fragment from A102 to A265 and K185 to A265, respectively, were cloned into the same pQE2 vector restriction sites. Note that an additional lysine residue (underlined) was inserted between the His-tag and the first protein residue to provide a stopping point for the exoprotease. This additional lysine was unnecessary for the 9 kDa construct which already contained an initial lysine. The C145S mutant of the liprin-β2 construct was created by QuickChange site-directed mutagenesis (Stratagene). All plasmid constructs were verified by DNA sequencing (Genewiz).
Protein Expression and Purification
Complete liprin-β1 and liprin-β2 coiled-coils were expressed in BL-21 (DE3) cells. Two separate 2 L cultures of LB were incubated at 37 °C until OD600 ≈ 0.6 and induced with 0.2 μg/mL (final concentration) IPTG for 3–4 hours at 37 °C. Cells were harvested by centrifugation, flash frozen in liquid nitrogen, and stored at −80 °C. Cells (7.22 g for liprin-β1 wild-type, 8.23 g for liprin-β2 wild-type, and 10.4 g liprin-β2 C145S) were lysed by sonication in 40–50 mL of lysis buffer (50 mM HEPES-NaOH, 500 mM NaCl, 1 mM TCEP or 5 mM BME, 0.5–1 mg/mL lysozyme, 0.1 mM PMSF, and DNase, pH 7.5). The lysate was then centrifuged for 10 min at 15 krpm in a Sorvall SS-34 rotor at 4 °C. The desired protein was found in the soluble and insoluble fractions, but the soluble protein did not bind efficiently to the Ni column under native conditions, perhaps due to proteolysis of the affinity. The protein could be purified from the insoluble fraction, however. The insoluble pellet was dissolved in 40 mL denaturing binding/wash buffer (20 mM Tris-HCl, 300 mM NaCl, 6 M urea, 10 mM BME, and 10 mM imidazole pH 8.0) and incubated with turning at 4 °C for 1 hour. Insoluble material was removed by centrifuging for 10 min at 15 krpm as above and the supernatant was incubated with gentle stirring with 5 mL (bed volume) Ni-NTA resin for 1 hour at 4 °C. The Ni beads were collected over a column and extensively washed with more than 50 mL of binding/wash buffer. The protein was eluted with 25 mL denaturing elution buffer (20 mM Tris-HCl, 300 mM NaCl, 200 mM imidazole, 6 M urea, pH 8.0). After diluting the protein to 50 mL with 8 M urea, the protein was refolded by dialyzing into refolding buffer at 4 °C (20 mM HEPES-NaOH, 500 mM NaCl, 1 mM EDTA, 1 mM TCEP, pH 7.5). The C145S mutant was refolded into the same buffer containing 1 M ammonium sulfate. Solid ammonium sulfate was added to the refolded wild-type proteins (1 M final concentration) which were then purified over a 5 mL HiTrap HP phenylsepharose hydrophobic interaction column, washing first with 45 mL buffer A (0.5 M ammonium sulfate, 20 mM Tris-HCl, pH 7.5) and eluting using a gradient to 100% buffer B (20 mM Tris-HCl, pH 7.5) over 45 mL. All proteins were further purified on a Superdex 200 column in 20 mM HEPES-NaOH, 500 mM NaCl, pH 7.5. Wild-type proteins were first reduced with excess DTT before gel filtration.
The 21, 19, and 9 kDa liprin β1 and β2 fragments were expressed and stored in frozen M15 cells (Qiagen) as described above for BL21(DE3). Cells were lysed by sonication in lysis buffer as above and the protein was purified directly from the soluble fraction which was bound directly to Ni resin for 1 hour at 4 °C. Unbound protein was washed from the resin using non-denaturing wash buffer (20 mM HEPES-NaOH, 500 mM NaCl, 5 mM imidazole, pH 7.5) and the bound protein was eluted with non-denaturing elution buffer (20 mM HEPES-NaOH, 500 mM NaCl, 200 mM imidazole, pH 7.5). Proteins were dialyzed overnight at 4 °C into phosphate buffer (20 mM PBS, 500 mM NaCl, pH 7.2). To remove the N-terminal His-tag, 15 mL aliquots of 21, 19, and 9 kDa fragments (1 mg/mL, 1 mg/mL, and 3 mg/mL) were treated with 50 μL DAPase (10 U/mL, Qiagen) pre-activated with 50 μL of 20 mM cystamine-HCl for 5 min at room temperature. The cleavage reaction was allowed to proceed at 37 °C for 3 hours and the DAPase and un-cleaved protein was then removed using subtractive Ni affinity chromatography.
Equilibrium Sedimentation
Equilibrium sedimentation was performed at 20 °C using a Beckman Optima XL-A analytical centrifuge. From the amino acid sequence of liprin-β2 coiled-coil C145S mutant the following were calculated: a monomeric molecular weight of 26992.4 g/mol, an extinction coefficient of 9970 M−1cm−1, and a partial specific volume of 0.7327 at 20 °C.(25)(26)(27) Initial solutions of liprin-β2 coiled-coil C145S were 0.18, 0.12, and 0.059 mg/mL which were centrifuged at 10, 12, 14, and 17 krpm. The equilibrium profiles were measured at 228 nm. A cell with a 1.2 mm path length was used for all experiments and the buffer for all samples was 20 mM Tris-HCl, 500 mM NaCl, 100 mM ammonium sulfate, 1 mM EDTA, pH 7.5 (1.0269 calculated density at 20 °C). Samples were centrifuged at each speed until the absorbance profile difference did not change significantly over approximately 4 hours. Baseline measurements were obtained to perform experimental background correction by spinning at high speed (40 krpm) to completely pellet the protein. The data were globally fit using a non-linear least-squares exponential fitting algorithm for a single non-associating species with Beckman Origin software. Attempts to fit the data to a monomer-dimer equilibrium model yielded a large association constant suggesting the system effectively behaved as a dimer at the concentrations investigated. Sedimentation experiments using higher protein concentrations (0.5–2.0 mg/mL) was consistent with a mixture low molecular species (i.e. mostly dimer and some tetramer) with a small amount of higher molecular weight oligomers that complicated fitting to a simple dimer-tetramer equilibrium model (GOF > 14). Size exclusion chromatography experiments in the buffer used for equilibrium sedimentation were compared to a lower salt buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, pH 7.5) which yielded essentially the same elution volumes, indicating no significant effects of salt concentration on the oligomer size.
Chemical Cross-Linking
The Liprin-β2 coiled-coil mutant C145S was dialyzed into 20 mM phosphate, 500 mM NaCl, pH 7.2 to give a stock solution of 21 μM. The BS3 cross-linking agent was dissolved in 20 mM phosphate, pH 7.2 to give a stock solution of 1 mM. Protein dilutions were performed in the same phosphate buffer containing 500 mM NaCl and BS3 dilutions were made in straight phosphate buffer. For the BS3 titration and time series, cross-linking was initiated by mixing 5 μL of protein solution with 5 μL of cross-linker solution and allowed to proceed at room temperature. Reactions were quenched at different times with 10 μL of 100 mM Tris-HCl, pH 8.0 and 20 μL SDS loading buffer was added to each sample before boiling 10 min and running on an SDS-PAGE gel. For the protein dilution series, cross-linking was initiated by mixing 18 μL of protein solution with 2 μL of BS3 stock solution and allowed to proceed at room temperature for 5 minutes. Reactions were quenched with 20 μL of 100 mM Tris-HCl, pH 8.4. A total of 40 μL of 2X SDS loading buffer was added to each reaction before boiling for 10 min and running on an SDS-PAGE gel.
Limited Proteolysis
Wild-type liprin-β1 and liprin-β2 coiled-coils were concentrated using Amicon Ultra 10k MWCO centrifugal concentrators to 2 mg/mL in 20 mM HEPES-NaOH, 500 mM NaCl, 2 mM DTT, pH 7.5. Lyophilized proteases α-chymotrypsin (type II from bovine pancreas), proteinase K (from Tritirachium album), and trypsin (TPCK treated from bovine pancreas) were obtained from Sigma and dissolved in water to give 1 mg/mL stock solutions that were used immediately. Dilutions of proteases were also made in water. A master reaction mix was first made containing 187.5 μL of the 2.5X reaction buffer (500 mM Tris-HCl, 50 μM CaCl2, pH 7.5), 120 μL of protein stocks, and 67.5 μL of fresh 10 mM DTT from which 16 μL aliquots were mixed with 4 μL of protease solutions. Final concentrations of proteases in the reactions were 100 μg/mL, 20 μg/mL, 4 μg/mL, 800 ng/mL, and 160 ng/mL. Reactions were initiated by addition of protease and quenched after exactly 20 minutes with 20 μL 2X stop buffer (125 mM Tris-HCl, 5 mM PMSF, 4% (w/v) SDS, 5% (v/v) BME, 20% (v/v) glycerol, pH 6.75 and trace bromophenol blue). All reactions were boiled immediately for 10 min before running on an SDS-PAGE gel.
Large-scale reactions with proteinase K were scaled accordingly, quenched with PMSF, and purified by size-exclusion chromatography using a Superdex 200 10/300 GL column with 20 mM HEPES-NaOH, 500 mM NaCl, pH 7.5. Aliquots from fractions containing protein fragments were desalted and analyzed by mass spectrometry (Applied Biosystems/MDS Sciex QSTAR XL Mass Spectrometer). All fractions were also run on an SDS-PAGE gel with 0.1 mM thioglycolate in the upper running buffer and blotted for 2 hours at 120 mA onto a Sequi-Blot PVDF membrane (Bio-Rad) using 10 mM CAPS, pH 11, 10 % methanol as the transfer buffer. The PVDF membrane was stained with 0.025% Coomassie Blue in 40% methanol and destained with 50% methanol, i.e. in the absence of acetic acid to avoid N-terminal acetylation. Bands corresponding to each protein fragment were cut from the membrane and analyzed by three cycles of Edman degradation. The software PAWS (i.e. Protein Analysis Work Sheet, Genomic Solutions Inc.) was used to identify sequences within each coiled-coil domain consistent with the observed masses and N-termini.
Crystal Structure Determination
The recombinant 9 kDa liprin-β2 coiled-coil fragment was dialyzed into 20 mM Tris, 5% glycerol, pH 8.0 and concentrated to 14.9 mg/mL using a 5,000 MWCO Amicon Ultra centrifugal concentrator. Crystals were grown by the hanging drop method using 190 mM ammonium iodide, 23% PEG 3350 as the well solution and mixing 1 μL of the protein sample with 0.5 μL of the well solution. Crystals grew overnight at room temperature (~21 °C) and were cryoprotected using the well solution supplemented with 30% glycerol before data collection. Three datasets, collected from different sections of a single large crystal at Advanced Photon Source (APS, Argonne National Labs), were processed and merged with Denzo/Scalepack.(28) The crystals belonged to space group C2221 with cell dimensions of a = 29.965 Å, b = 162.653 Å, c = 34.387 Å, and α = β = γ = 90° with a single molecule in the asymmetric unit. The molecular replacement search model was generated by submitting the complete sequence of the crystallized fragment from K185 through A265 of liprin-β2 to the Phyre (29) (Protein Homology/analogY Recognition Engine) web server which generated a model corresponding to residues L212 through T264 of liprin-β2 based on the coiled-coil structure of tropomyosin. An initial molecular replacement solution was found by Phaser (30) using this homology model. The amino acid register was set automatically using ARP/wARP (31) building into a side-chain omit map. The initial solution from Phaser was refined by iterative cycles of manual building with COOT (32) and REFMAC5 (33). During refinement the anisotropy was corrected by performing ellipsoidal truncation and anisotropic scaling using the UCLA MBI Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/).(34) TLS refinement was used toward the end of refinement using a single TLS group for the entire protein molecule in the asymmetric unit. Residues with clear density for more than one side-chain conformation were refined with partial occupancy in dual conformations.
Structure Analysis
The packing of the liprin-β2 coiled-coil dimer was analyzed using the SOCKET (35) web server and the coiled-coil parameters were analyzed using TWISTER. (36) In order to be analyzed by both programs, a dimeric coiled-coil structure was first generated by applying a crystallographic two-fold symmetry operation to create a coordinate file for the dimer model. As alternate conformations are not allowed by these programs, a single conformation was assigned to each residue. Graphics were prepared in PyMOL (Schrödinger, LLC).
Results
Design of β1 and β2 Coiled-Coil Expression Constructs
To define the extent of the coiled-coil domains for both liprin β1 and β2 (1011 and 876 amino acids respectively) we employed the COILS (16) coiled-coil prediction server. As shown in Fig. 1B, residues 97–342 of liprin β1 and 100–315 of liprin β2 were strongly predicted to be coiled-coils. Both regions were expressed in E. coli and purified.
Both β1 and β2 coiled-coil constructs were prone to intermolecular oxidative disulfide bond formation as evidenced by peak shifts in gel filtration experiments in the presence and absence of a reducing agent (data not shown). To eliminate complications due to disulfide bond formation during further biochemical analysis of the liprin β2 construct, a C145S mutant was made by site-directed mutagenesis to remove its single Cys. The C145S mutant eluted from the gel filtration column at the same volume as the reduced wild-type construct (data not shown), indicating that this mutant was the same size and shape as the reduced wild-type protein, but not prone to oxidation.
The Liprin β2 Coiled-Coil Forms a Dimer
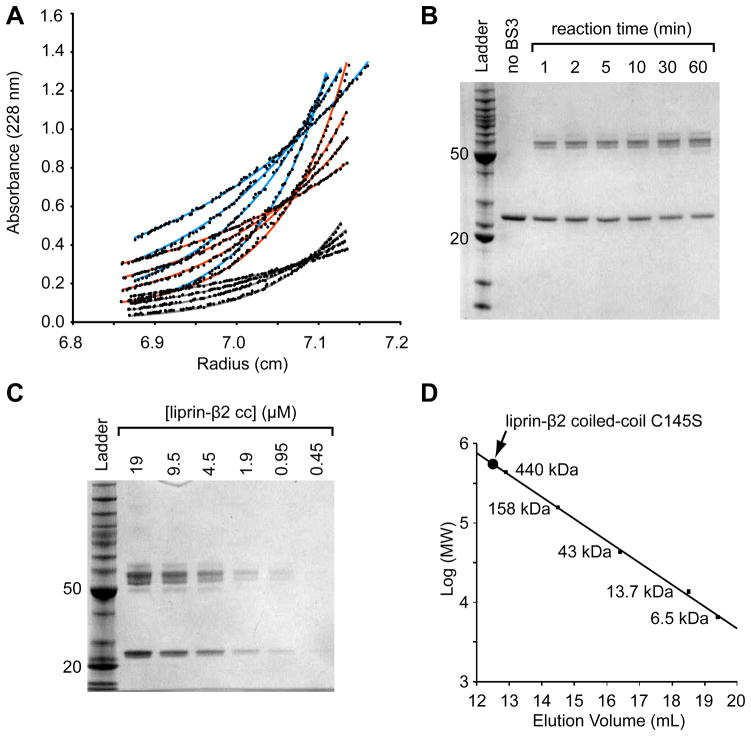
As the coiled-coil domains were expected to have an unusually extended architecture, we employed equilibrium sedimentation to determine the molecular weight of the oligomer because the method measures mass independent of shape (Figure 2A). Sedimentation experiments were performed at three concentrations (6.8, 4.5, and 2.2μM) and four different speeds (10, 12, 14, and 17 krpm) using the liprin β2 C145S coiled-coil. As shown in Fig. 2A, the data was globally fit well by a single exponential for a simple dimeric species (GOF = 1.17).
Figure 2. The Liprin-β2 Coiled-Coil Forms an Extended Dimer.
(A) The liprin-β2 coiled-coil domain concentration distributions after sedimentation to equilibrium at various speeds and protein concentrations. The three different concentrations (light blue - 0.18 mg/mL, orange - 0.12 mg/mL, and grey - 0.059 mg/mL) were equilibrated at four different speeds (10, 12, 14, and 17 krpm, bottom to top) and globally fit to a single exponential (shown as lines) with a variable molecular weight. The data was best fit with the molecular weight equal to the dimer (52,598 Da measured from fit; 53,984.8 calculated from sequence). (B) An SDS-PAGE gel showing the results of a liprin-β2 coiled-coil domain chemical cross-linking time-series experiment performed using a constant concentration of protein (5 μM) and BS3 cross-linker (50 μM). (C) An SDS-PAGE gel showing the results of a chemical cross-linking of the liprin-β2 coiled-coil domain at different protein concentrations, at a constant BS3 cross-linker concentration (100 μM) for a total time of 5 min. The ladder used for the gels in B and C contains standard proteins of 10, 15, 20, 25, 30, 40, 50, 60, 70, 80, 90, 100, 120, 160, and 220 kDa (BenchMark Protein Ladder, Invitrogen). For reference the 20 and 50 kDa bands of the ladder are indicated on each gel. (D) Superdex S-200 gel filtration chromatography of the liprin-β2 coiled-coil domain C145S. The elution volume of a set of globular protein standards is also indicated. The protein standards from largest to smallest are ferritin, aldolase, ovalbumin, ribonuclease A, and aprotinin (GE Healthcare). The void volume of the column was determined using blue dextran 2000 to be approximately 8.3 mL. The liprin-β2 coiled-coil (concentration < 0.5 mg/mL) elutes at 12.6 mL (~525 kDa) or approximately 20-fold higher than its monomeric molecular weight.
Chemical cross-linking using BS3 was performed to further establish the oligomer size of the β2 coiled-coil (Figure 2B and 2C). BS3 contains two amine-reactive succinimidyl ester moieties approximately 11 Å apart that should covalently couple exposed primary amines (e.g. from lysine residues) in close proximity. First, the C145S β2 coiled-coil concentration was held constant at 5 μM in the presence of a 10-fold stoichiometric excess of BS3 (50 μM) (Figure 2B). The cross-linking reaction was quenched at different time points to monitor the extent of cross-linking as a function of time. The reaction is essentially complete within 1 min and only monomeric and dimeric species are observed despite the large excess of cross-linker. A set of bands centered at the dimer molecular weight, rather than a single band, is observed, presumably due to different cross-linking points that affect migration through the gel. The cross-linking experiment was then performed for 5 min at a constant BS3 concentration (100 μM) using a dilution series of the C145S β2 coiled-coil (Figure 2C). This experiment shows that the relative ratio of dimer to monomer remains constant regardless of the protein concentration. These experiments suggest that the β2 coiled-coil is primarily dimeric.
The size of the liprin-β2 coiled-coil domain was also estimated by size exclusion chromatography. As shown in Fig. 2D, it migrates at a position corresponding to a 525 kDa globular protein or the size of a globular 20mer. The large hydrodynamic radius suggests that the protein is highly extended as might be expected for a coiled-coil structure.
Identification of Stable, Protease-Resistant β1 and β2 Coiled-Coil Sub-Domains
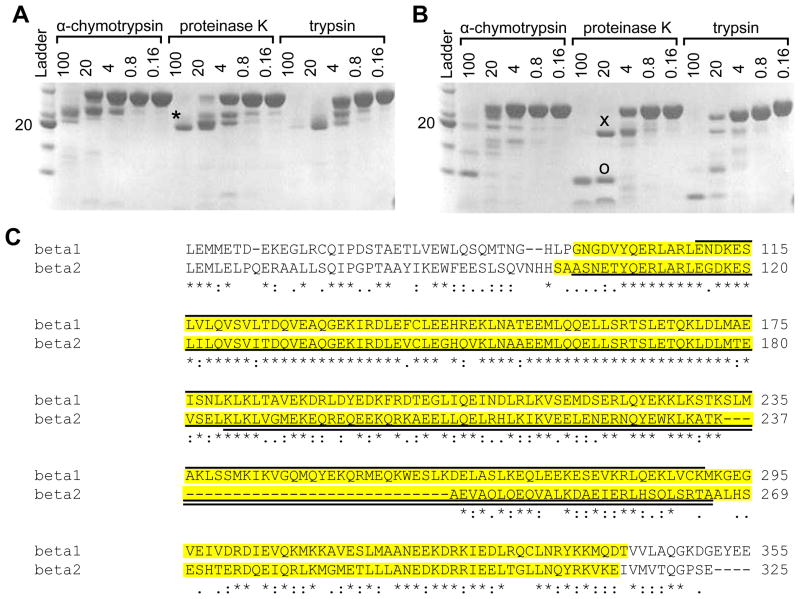
Despite extensive efforts, we were unable to obtain high quality crystals of the full length coiled-coil domains. We therefore searched for smaller, stable sub-domains. To identify smaller and presumably more rigid sub-domains, the wild-type liprin β1 and β2 coiled-coil constructs were subjected to limited proteolysis using α-chymotrypsin, proteinase K, and trypsin under reducing conditions (Figure 3A and 3B). Limited proteoylsis of the liprin β1 coiled-coil yielded a single, approximately 21 kDa fragment in the presence of all three enzymes as assessed by SDS-PAGE analysis. Limited proteolysis of the liprin β2 coiled-coil yielded two fragments, approximately 9 kDa and 19 kDa, as assessed by SDS-PAGE analysis. The consistent size of these fragments regardless of the different sequence specificities of these proteases suggests that these fragments are likely to represent common, stable subdomains within the entire coiled-coil domains.
Figure 3. Limited Proteolysis of Liprin-β1 and -β2 Coiled-Coils.
(A) An SDS-PAGE gel showing the results of limited proteolysis of the liprin-β1 coiled-coil (30.8 kDa) by several proteases at a series of concentrations (μg/mL). (B) An SDS-PAGE gel showing the results of limited proteolysis of the liprin-β2 coiled-coil (27.0 kDa) by several proteases at a series of concentrations (μg/mL). The 21 kDa(*), 19 kDa (x), and 9 kDa (o) bands were identified by mass spectrometry and Edman degradation. The ladder is the same as in Figure 3. (C) ClustalW (41) sequence alignment of human liprin-β1 and -β2 coiled-coils. The regions cloned in the full-length coiled-coil constructs are highlighted in yellow. The 21 kDa liprin-β1 fragment is indicated by a solid line above the alignment and the 19 kDa and 9 kDa fragments of liprin-β2 are indicated by solid lines below the alignment. The extent of sequence conservation is indicated below the alignment.
To identify the sequences of the protease stable sub-domains, limited proteolysis experiments of both liprin β1 and β2 coiled-coils using proteinase K were scaled up and the resulting components were purified by size-exclusion chromatography. Fractions containing the purified fragments were analyzed by ESI mass spectrometry and N-terminal sequencing by Edman degradation (Figure 3C). A single consistent sequence was identified for the 21 kDa liprin β1 coiled-coil sub-domain (calculated mass 21,385.5 kDa; measured mass 21,389.0 kDa; N-terminal sequence: END) from E110 through K290. A single consistent sequence was also identified for the 19 kDa liprin β2 coiled-coil sub-domain (calculated mass 19,120.6 kDa; measured mass 19,121.0 kDa; N-terminal sequence: ASN) from A102 through A265. The smaller 9 kDa sub-domain of the liprin β2 coiled-coil appeared to be heterogeneous by both mass spectrometry and Edman degradation analysis. Nevertheless, a likely component of this mixture was identified from the largest species (calculated mass 9723.1 kDa; measured mass 9722.2 kDa; N-terminal sequence: KLK) that encompassed amino acids K185 through A265. The common C-terminus of this 9 kDa and the 19 kDa fragment further suggests that K185-A265 is the dominant component of this mixture.
Three constructs encompassing these protease-resistant cores of the liprin β1 and β2 coiled-coils were cloned into a pQE2 bacterial expression vector containing a cleavable, N-terminal His-tag for purification purposes. All constructs expressed into the soluble fraction and were purified under native conditions. The N-terminal His-tag was removed using an exo-peptidase and the resulting tag-free constructs were purified by a subtractive Ni affinity column. The 21 kDa β1 or 19 kDa β2 fragments did not yield crystals, but we were able to obtain crystals of the 9 kDa β2 fragment of liprin-β2.
X-ray Crystallography of the Central β2 Coiled-Coil Core
The 9 kDa β2 coiled-coil crystals belonged to space group C2221 with a unit cell of a = 29.965 Å, b = 162.653 Å, c = 34.387 Å, and α = β = γ = 90° (Table 1). The crystals diffracted to approximately 2.0 Å, but the diffraction was strongly anisotropic with significantly reduced diffraction intensity along the a* reciprocal axis (Supplemental Figure 1). The structure was solved by molecular replacement using a homology model as the search model. Remarkably, the homology model generated by the Phyre server (29) was based on a tropomyosin structure that bore only 16% identity to the liprin β2 construct. The search model corresponded to residues L212 through T264, but electron density for residues preceding L212 was clearly visible in the initial molecular replacement solution, lending confidence that the correct solution had been obtained. To further validate this initial solution, all residues were converted to alanine to create an omit map that clearly showed positive side-chain electron density throughout the entire structure. The amino acid register was determined automatically by allowing ARP/wARP (31) to build into this omit map. It became clear during refinement that the strongly anisotropic data needed to be corrected by performing ellipsoidal truncation and scaling.(34) Before this anisotropy correction the refinement stalled (R/Rfree approximately 0.35–0.40) and the electron density was less well-defined than would be expected from a 2.0 angstrom electron density map (Supplemental Figure 2). Immediately following the anisotropy correction, the electron density became much better defined and the R/Rfree dropped accordingly (final R/Rfree of 0.233/0.291). Attempts to refine the structure in a lower symmetry space group did not improve the R-factor or the quality of the electron density map.
Table 1.
X-ray Diffraction Data and Refinement Statistics
| Data Collection | Liprin-β2 (K185 to A265 (PDB code 3QH9) |
|---|---|
| Wavelength (Å) | 1.4938 |
| Space Group | C2221 |
| Cell Dimensions | |
| a, b, c (Å) | 30.0, 162.7, 34.4 |
| α, β, γ (°) | 90, 90, 90 |
| Matthews Coefficient (Å3/Da) | 2.15 |
| % Solvent | 42.95 |
| Resolution (Å) (highest shell) | 80.00–2.00 (2.07–2.00) |
| Rsym | 0.099 (0.348) |
| I/Iσ | 20.7 (5.0) |
| % Completeness | 97.0 (84.1) |
| Redundancy | 10.2 (7.2) |
|
| |
|
Refinement
| |
| No. Reflections | 5774 |
| Rwork | 0.232 |
| Rfree* | 0.298 |
| No. Atoms | 638 |
| B-factor (Å2) all atoms | 37.0 |
| protein | 38.0 |
| water | 19.6 |
| Rmsd Bond Lengths (Å) | 0.0234 |
| Rmsd Bond Angle (°) | 2.0 |
| Ramachandran Statistics (%) | |
| most favored | 100 |
| additionally allowed | 0 |
| generously allowed | 0 |
| disallowed | 0 |
Calculated from 5% random sampling of data excluded from refinement
Analysis of the β2 Coiled-Coil Structure
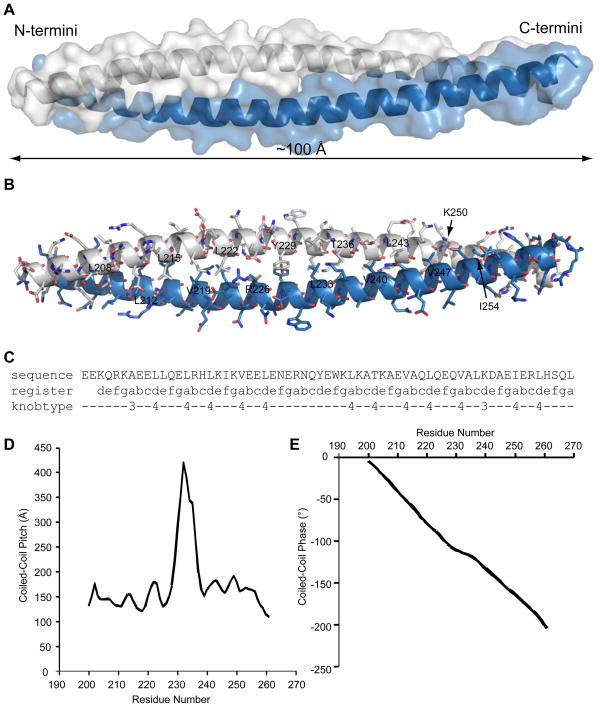
Although the construct used for crystallization included residues K185 through A265 of liprin-β2, the electron density was only interpretable for residues Q198 through R263. A single protein molecule was found in the asymmetric unit that formed a single, continuous α-helix which interacted with a symmetry related copy to form a parallel, homodimeric coiled-coil approximately 100 Å long (Figure 4A and 4B). Throughout the crystal lattice the dimeric coiled-coils pack with their long axes parallel to the long b-axis of the unit cell. Analysis by SOCKET (35) reveals typical knobs-into-holes packing throughout most of the dimer with typical Leu and Val residues occupying the majority of the “a” and “d” residues of the heptad repeat involved in packing between the two molecules of the coiled-coil (Figure 4C). A notable exception are residues R226 and Y229 which occupy successive “a” and “d” residues near the center of the coiled-coil, but do not exhibit classical knobs-into-holes type packing. Analysis by TWISTER (36) reveals a sharp increase in the local pitch of the coiled-coil surrounding these residues indicating that the coiled-coil unwinds slightly at this location, but the register of the heptad pattern does not change (Figure 4D and 4E). Overall, the two-stranded coiled-coil forms a typical left-handed supercoil with an average coiled-coil pitch of 161 ± 38 Å.
Figure 4. Structural Analysis of the Core of the Liprin-β2 Coiled-Coil.
(A) An x-ray crystal structure model of the proteolytically-stable coiled-coil sub-domain of liprin-β2. Each molecule of the dimer is shown in separate colors (light blue and white) with a transparent surface representation and ribbon helices to illustrate the overall packing of the two-stranded, parallel coiled-coil. (B) A model showing all side chain atoms in stick representation and the backbone traced with a ribbon. Carbon atoms are shown in light blue and white as in panel A, oxygen in red, and nitrogen in dark blue. All the visible “a” and “d” residues from the heptad repeat that form the core packing interactions are labeled alternately on opposing helices. (C) A coiled-coil packing analysis by SOCKET.(35) Only one sequence is shown since the register of the heptad repeat and knob types are symmetric. Type 3 and 4 knobs are both defined as complementary in that they also make part of another hole. Type 3 knobs are positioned across from a hole, whereas type 4 knobs pack directly inside holes.(35) (D) A graph of the local pitch of the coiled-coil at each residue as measured by TWISTER.(36) (E) A graph of the change in phase of the coiled-coil also measured by TWISTER which shows that the coiled-coil unwinds slightly at the center of the structure.
Discussion
Synaptic vesicles are organized by a complex protein assembly that ensures the rapid release of neurotransmitters in response to incoming action potentials.(1)(37) The liprin protein family plays an important part in regulating and maintaining this synaptic architecture,(38) but the molecular basis for these functions is unknown. Previous immunoprecipitation experiments have shown that liprin-β coiled-coils interact with themselves,(2) but the basic stoichiometry and orientation of these interactions has not been defined. The equilibrium sedimentation and cross-linking experiments presented here demonstrate that the coiled-coil of liprin-β2 predominantly forms a dimer. The sequence similarity and similar size-exclusion retention times suggest that liprin-β1 and liprin-β2 are approximately the same size and shape, so it seems probable that liprin-β1 is also dimeric. The orientation of the liprin-β2 coiled-coil oligomer was established from the crystal structure of the central, protease-resistant core. In the context of the entire liprin-β2 protein, this parallel dimer arrangement is predicted to bring all six C-terminal tandem SAMs in close proximity to each other as shown in Fig. 5A, in contrast to a hypothetical antiparallel dimer which would separate the tandem SAM domains.
Figure 5. Possible Organization of Liprin Scaffolds.
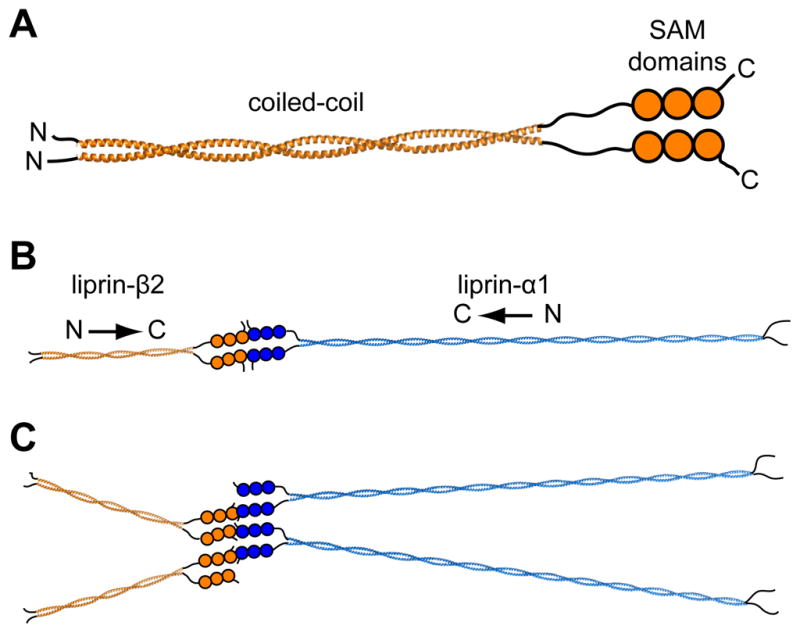
(A) A parallel homodimer model of the complete liprin-β2. The complete coiled-coil (~300 Å) was modeled using a combination of BEAMMOTIF and MAKECCSC.(42) The model is merely intended to convey the orientation and relative sizes of the coiled-coil domains. (B) A speculative scaffold organization is shown consisting of liprin-α1 and liprin-β2 molecules. For simplicity the entire liprin-α1 coiled-coil (~900 Å) was also assumed to form a two-stranded parallel structure which was constructed using MAKECCSC with no attempt to adjust for stutters, stammers, or gaps. The three tandem SAM domains of liprin-αs have been shown in other studies to interact with liprin-βs,(2) which we show here in a “closed dimer” model of α and β dimers. (C) An alternative “open scaffold” model is shown in which multiple α and β liprins stack indefinitely. In both “closed dimer” and “open scaffold” models the coiled-coil domains are predicted to radiate from the central SAM domain interactions.
Importantly, the orientation of the coiled-coil domains in such a dimer dictates the overall model of the predicted liprin scaffold. As mentioned previously, immunoprecipitation experiments have also shown that the liprin-αs interact with liprin-βs through their C-terminal regions containing the SAM domains.(2) Individual SAM domains usually form small α-helix bundles that often form homo- and heteropolymers with interactions between their ML and EH interfaces.(23) The close proximity of three tandem SAM domains on the same molecule in all liprins suggest that they might stack together to form consecutive intramolecular EH-ML interactions as previously observed in the structure of the AIDA-1 tandem SAM dimer.(39) In the context of the parallel coiled-coil dimer, one possibility is that the SAM domains of liprin-αs and liprin-βs might come together to create a “closed dimer” of α and β dimers as shown in Fig. 5B assuming liprin-αs also form parallel coiled-coils (not known). Another possibility is an “open scaffold” in which α and β molecules interact with more than one dimer as shown in Fig 5C. Another more complex possibility is that α and β SAMs form a co-polymer which has been observed before with the SAM domains from Scm and polyhomeotic.(40) Individual liprin SAM domains have also been computationally predicted to form polymeric structures, which could be an important aspect of their scaffolding architecture.(24)
Obviously, concepts of the overall scaffolding structure of liprins remain highly speculative since little is known about the interactions between the SAM domains and the structure of the liprin-α coiled-coils. Regardless, in all of the scaffolding models, the coiled-coil domains are predicted to radiate out from the central axis of SAM domains to interact with other synaptic proteins. At present, the distinct functional roles of the two different liprin-β proteins and four different liprin-α proteins in humans is unknown, but it is known that they can associate. It seems plausible that the coiled-coil domains may form distinct hetero-oligomers that may elicit different functions. Much work remains before we can visualize how liprins organize the complex set of partner proteins that contribute to the construction of the cytomatrix of the active zone.
Supplementary Material
Acknowledgments
We thank Julian Whitelegge and Sara Bassilian (UCLA Pasarow Mass Spectrometry Laboratory) for assistance with mass spectrometry experiments and Kathy Schegg (UNR INBRE Nevada Proteomics Center; NIH Grant Number P20 RR-016464) for Edman sequencing. This work was supported by NIH grant 1R01GM093393 to JUB.
Footnotes
This work was supported by NIH grant R01GM093393 to JUB and a NIH Ruth L. Kirschstein Postdoctoral Fellowship F32GM084615 to RLS.
Coordinates have been deposited in the RCSB Protein Data Bank: PDB code 3QH9 (Liprin-β2 residues K185 to A265).
Abbreviations: SAM: sterile alpha motif; PCR: polymerase chain reaction; BME: β-mercaptoethanol; HEPES: N-(2-hydroxyethyl)-piperzine-N′-2-ethanesulfonic acid;TCEP: (tris(2-carboxyethyl)phosphine); PMSF: phenylmethylsulfonyl fluoride; LB: Luria broth; IPTG: isopropyl-β-D-1-thiogalactopyranoside; EDTA: ethylenediaminetetraacetic acid; DTT: DL-dithiothreitol; PBS: phosphate buffered saline; BS3: bis(sulfosuccinimidyl) suberate; PVDF: polyvinylidene fluoride; SDS: sodium dodecyl sulfate; PAGE: polyacrylamide gel electrophoresis; PEG: polyethylene glycol; ESI: electrospray ionization; ML: mid-loop; EH: end-helix
Supporting Information Available. Two figures showing an analysis of the anisotropic diffraction and electron density maps are included as supplemental information. This material is available free of charge on the internet: http://pubs.acs.org.
References
- 1.Schoch S, Gundelfinger ED. Molecular organization of the presynaptic active zone. Cell Tissue Res. 2006;326:379–391. doi: 10.1007/s00441-006-0244-y. [DOI] [PubMed] [Google Scholar]
- 2.Serra-Pagès C, Medley QG, Tang M, Hart A, Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem. 1998;273:15611–15620. doi: 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- 3.Olsen O, Moore KA, Fukata M, Kazuta T, Trinidad JC, Kauer FW, Streuli M, Misawa H, Burlingame AL, Nicoll RA, Bredt DS. Neurotransmitter release regulated by a MALS-liprin-alpha presynaptic complex. J Cell Biol. 2005;170:1127–1134. doi: 10.1083/jcb.200503011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko J, Na M, Kim S, Lee J, Kim E. Interaction of the ERC family of RIM-binding proteins with the liprin-alpha family of multidomain proteins. J Biol Chem. 2003;278:42377–42385. doi: 10.1074/jbc.M307561200. [DOI] [PubMed] [Google Scholar]
- 5.Ko J, Kim S, Valtschanoff JG, Shin H, Lee J, Sheng M, Premont RT, Weinberg RJ, Kim E. Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting. J Neurosci. 2003;23:1667–1677. doi: 10.1523/JNEUROSCI.23-05-01667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin H, Wyszynski M, Huh K, Valtschanoff JG, Lee J, Ko J, Streuli M, Weinberg RJ, Sheng M, Kim E. Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. J Biol Chem. 2003;278:11393–11401. doi: 10.1074/jbc.M211874200. [DOI] [PubMed] [Google Scholar]
- 7.Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, Südhof TC. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- 8.Patel MR, Shen K. RSY-1 is a local inhibitor of presynaptic assembly in C. elegans. Science. 2009;323:1500–1503. doi: 10.1126/science.1169025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhen M, Jin Y. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature. 1999;401:371–375. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
- 10.Hofmeyer K, Maurel-Zaffran C, Sink H, Treisman JE. Liprin-alpha has LAR-independent functions in R7 photoreceptor axon targeting. Proc Natl Acad Sci U S A. 2006;103:11595–11600. doi: 10.1073/pnas.0604766103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choe K, Prakash S, Bright A, Clandinin TR. Liprin-alpha is required for photoreceptor target selection in Drosophila. Proc Natl Acad Sci U S A. 2006;103:11601–11606. doi: 10.1073/pnas.0601185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ackley BD, Harrington RJ, Hudson ML, Williams L, Kenyon CJ, Chisholm AD, Jin Y. The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J Neurosci. 2005;25:7517–7528. doi: 10.1523/JNEUROSCI.2010-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunah AW, Hueske E, Wyszynski M, Hoogenraad CC, Jaworski J, Pak DT, Simonetta A, Liu G, Sheng M. LAR receptor protein tyrosine phosphatases in the development and maintenance of excitatory synapses. Nat Neurosci. 2005;8:458–467. doi: 10.1038/nn1416. [DOI] [PubMed] [Google Scholar]
- 14.Nachat R, Cipolat S, Sevilla LM, Chhatriwala M, Groot KR, Watt FM. KazrinE is a desmosome-associated liprin that colocalises with acetylated microtubules. J Cell Sci. 2009;122:4035–4041. doi: 10.1242/jcs.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Astigarraga S, Hofmeyer K, Farajian R, Treisman JE. Three Drosophila liprins interact to control synapse formation. J Neurosci. 2010;30:15358–15368. doi: 10.1523/JNEUROSCI.1862-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 17.Wolf E, Kim PS, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 19.Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 20.Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadav MK, Leman LJ, Price DJ, Brooks CL, Stout CD, Ghadiri MR. Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution. Biochemistry. 2006;45:4463–4473. doi: 10.1021/bi060092q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apgar JR, Gutwin KN, Keating AE. Predicting helix orientation for coiled-coil dimers. Proteins. 2008;72:1048–1065. doi: 10.1002/prot.22118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiao F, Bowie JU. The many faces of SAM. Sci STKE 2005. 2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 24.Meruelo AD, Bowie JU. Identifying polymer-forming SAM domains. Proteins. 2009;74:1–5. doi: 10.1002/prot.22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohn EJ, Edsall JT. Proteins, amino acids and peptides as ions and dipolar ions. Reinhold; New York: 1943. pp. 370–381. [Google Scholar]
- 27.Laue TM, Shah BD, Ridgeway SL, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. The Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- 28.Minor W, Otwinowski Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 34.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walshaw J, Woolfson DN. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J Mol Biol. 2001;307:1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 36.Strelkov SV, Burkhard P. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J Struct Biol. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- 37.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 38.Spangler SA, Hoogenraad CC. Liprin-alpha proteins: scaffold molecules for synapse maturation. Biochem Soc Trans. 2007;35:1278–1282. doi: 10.1042/BST0351278. [DOI] [PubMed] [Google Scholar]
- 39.Kurabi A, Brener S, Mobli M, Kwan JJ, Donaldson LW. A nuclear localization signal at the SAM-SAM domain interface of AIDA-1 suggests a requirement for domain uncoupling prior to nuclear import. J Mol Biol. 2009;392:1168–1177. doi: 10.1016/j.jmb.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Kim CA, Sawaya MR, Cascio D, Kim W, Bowie JU. Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer. J Biol Chem. 2005;280:27769–27775. doi: 10.1074/jbc.M503055200. [DOI] [PubMed] [Google Scholar]
- 41.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Offer G, Hicks MR, Woolfson DN. Generalized Crick equations for modeling noncanonical coiled coils. J Struct Biol. 2002;137:41–53. doi: 10.1006/jsbi.2002.4448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.