Abstract
There is significant overlap in the genes and pathways that control liver development and those that regulate liver regeneration, hepatic progenitor cell expansion, response to injury and cancer. Additionally, defects in liver development may underlie some congenital and perinatal liver diseases. Thus, studying hepatogenesis is important for understanding not only how the liver forms, but also how it functions. Elegant work in mice has uncovered a host of transcription factors and signaling molecules that govern the early steps of hepatic specification, however the inherent difficulty of studying embryogenesis in utero has driven developmental biologists to seek new systems. The rapidly developing vertebrate zebrafish is a favorite model for embryology. The power of forward genetic screens combined with live real-time imaging of development in transparent zebrafish embryos has highlighted conserved processes essential for hepatogenesis and has uncovered some exciting new players. This review will present the advantages of zebrafish for studying liver development, underscoring how studies in zebrafish and mice complement each other. In addition to their value for studying development, zebrafish models of hepatic and biliary diseases are expanding, and using these small, inexpensive embryos for drug screening has become de rigueur. Zebrafish provide a shared platform for developmental biology and translational research, offering innovative methods for studying liver development and disease.
The story of hepatogenesis has something for everyone. It involves transcriptional regulation, cell-cell interaction, signaling pathways, control of cell proliferation and apoptosis plus morphogenic processes that sculpt vasculature, parenchymal cells and mesenchyme to form the multi-faceted liver. Decades of research on liver development in mice and other vertebrates offer valuable lessons in how the multi-potent endoderm is programmed to form a functional liver. Of equal importance are insights that have illuminated the mechanisms by which hepatic progenitors are activated in a damaged liver, how the adult liver regenerates and, possibly, the basis for engineering liver cells in vitro for cell transplantation to sustain patients with liver failure. Moreover, processes that are key to liver development are often co-opted during pathogenesis. Therefore, reviewing hepatogenesis is informative for both basic and translational researchers.
In this review, we bring to light the many advantages offered by the tropical freshwater vertebrate zebrafish (Danio rerio) in studying hepatogenesis. By comparing zebrafish and mice, we highlight how work in each system complements the other and emphasize novel paradigms that have been uncovered using zebrafish. Finally, we highlight exciting efforts using zebrafish to model hepatobiliary diseases.
Developmental Genetics in Zebrafish
Zebrafish are the darling of developmental biologists because they produce hundreds of offspring with every mating, and their transparent embryos develop outside of the mother, allowing constant visualization (see Fig. 1) and easy manipulation. Additionally, since they are sustained by nutrients in the yolk, the extra-embryonic structures that are essential for supporting the mammalian embryo do not form in zebrafish.
Figure 1. Overview of the genes involved in zebrafish liver development.
A. Each gene in this figure has been shown to be either expressed in the liver (listed in grey) or directly regulate liver development (listed in black). B. Live images of embryos during development. Outgrowth is imaged by detection of the red fluorescent transgene expressed in hepatocytes in Tg(fabp10:dsred) embryos.
Only a standard microscope is needed to observe developmental events such as a beating heart, twitching muscle and a growing liver. Organogenesis is underway in nearly all systems by 24 hours post fertilization (hpf) and the embryo is swimming by 2 days post fertilization (dpf). By 5 dpf, all major organ systems are established (Fig. 2) and larvae are ready to feed.
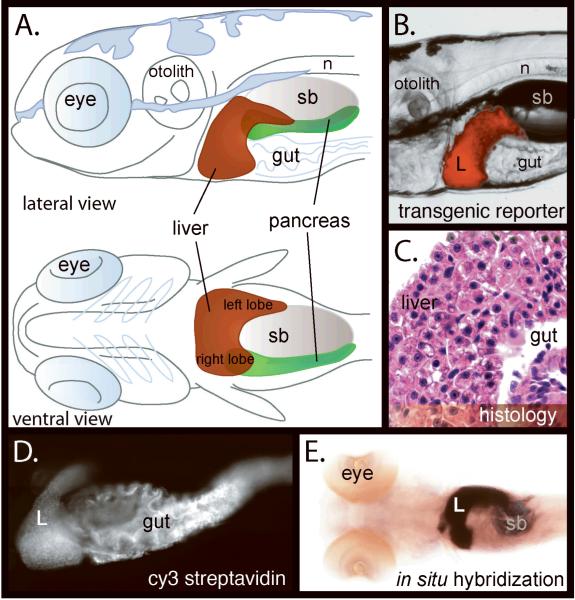
Figure 2. The liver in 5 dpf zebrafish is mature.
The liver (L) is analyzed through multiple techniques in the 5 dpf zebrafish. A. The bi-lobed zebrafish liver lies ventrally and anterior to the swim bladder (SB) and notochord (N). It can be visualized using a transgenic reporter expressing red fluorescent protein under a hepatocyte specific promoter (Tg(fabp10-dsred) (B) hematoxylin and eosin staining (C) by deconvoluted whole mount cy3-streptavidin staining (D) and whole mount in situ hybridization using the fabp10 probe (E).
The array of genetic tools and large number of progeny make zebrafish ideal for developmental genetic studies. Assembly of the zebrafish genome is nearly complete, and despite the genome duplication that occurred during teleost evolution, the high genetic conservation among vertebrates makes it straightforward to identify zebrafish orthologs of human genes. Moreover, each embryo is provided with mRNA and proteins from the mother to sustain early development. These maternal stores allow mutant embryos that lack a cell-essential gene to survive longer than their mammalian counterparts.
The ability to carry out forward genetic screens in zebrafish is among their most powerful attributes. Forward screening allows the investigator to take an unbiased approach to identify genes essential for a process of interest by identifying embryonic mutants that are defective in that process. For instance, our interest in hepatic outgrowth led us to screen for mutants that fail to expand their liver bud (1). Liver mutants have been identified in several large-scale forward screens which used either chemical or retroviral mutagens (Table 1). Their analysis has illuminated aspects of hepatogenesis that are conserved between mice and zebrafish and led to the discovery of new players.
Table 1.
Forward genetic screens identify liver mutants
| Laboratory | Location | Mutant | Gene | Reference |
|---|---|---|---|---|
| Nusslein-Volhard | Max-Planck Institute, Tubingen | lumpazi | unknown | (55) |
| gammler | unknown | |||
| tramp | unknown | |||
| tippelbruder | unknown | |||
|
| ||||
| Fishman | Harvard Medical School | beefeater | unknown | (56) |
| nil per os | rbm19 | (46) | ||
|
| ||||
| Hopkins | Massachusetts Institute of Technology | foie gras | fgr | (52) |
| neurofibromatosis 2 | nf2 | (52) | ||
| vps18 | vps18 | (52) | ||
| uhrf1 | uhrf1 | (1) | ||
| digestive-organ expansion | def | (45) | ||
| factor | (57) | |||
| pescadillo | pes | |||
|
| ||||
| Stainier | University of California, San Francisco | prometheus | wnt2bb | (20) |
| hdac1 | hdac1 | (48) | ||
| fgf10 | fgf10 | (11) | ||
Reverse genetics approaches, whereby a specific gene is targeted and the resulting phenotype is analyzed, are feasible in zebrafish in a limited fashion. Targeted gene mutation is being developed, but is not yet standardized. Instead, antisense morpholino oligonucleotides are routinely used for transient gene knock-down in embryos. Morpholinos injected into the yolk of the newly fertilized embryo diffuse quickly and uniformly into the cells, which are in syncytia with the yolk. As development progresses, the morpholinos are diluted with each cell division so that the degree of knock-down decreases as the embryo ages (2). Thus, morpholino knock-down is effective for examining early embryonic events, such as endodermal patterning, but less useful for affecting events later in development. The future of the field awaits improvement and expansion on the initial promising results provided by technologies such as TILLING (3) and zinc finger nucleases (4, 5) to carry out targeted gene mutation in zebrafish.
Creating transgenic zebrafish is easy, rapid and inexpensive. Fluorescent transgenes controlled by ubiquitous, inducible or tissue-specific promoters can be visualized in real-time as they proceed through development, enabling detailed fate mapping and spatio-temporal analysis of gene expression. For instance, the fabp10 promoter drives high transgene expression in hepatocytes (6) (Fig. 1) and the Tg(gutGFP)s854 line expresses GFP in endodermally-derived organs (7), allowing exquisite analysis of hepatogenesis. Additionally, crossing transgenic lines generates multi-labeled organs. For instance, fish expressing RFP in the liver and pancreatic islet cells, and GFP in the exocrine pancreas (Tg(fabp10:dsred;elas:GFP;ins:dsred)) allow evaluation of multiple organs at a glance (8).
The zebrafish community is well organized and open access. The growing attendance at the biennial International Meeting on Zebrafish Development and Genetics reflects the expanding popularity of this model, both for biomedical scientists and educators. A comprehensive catalog of zebrafish mutants, morphants, transgenics as well as images of the developmental expression pattern of thousands of transcripts are maintained on the community website, zfin.org. Most published mutants and transgenics are maintained and distributed by the stock center (http://zebrafish.org/zirc/home/guide). The Trans-NIH Zebrafish Initiative supports grants and other resources for improving and expanding the use, mutagenesis and analysis of this organism.
Of Mice and Zebrafish
Decades of outstanding studies using reverse genetics, embryo explants and tissue culture have uncovered a network of transcription factors and signaling pathways required for forming the mammalian liver (9–11). Hepatogenesis in mice begins approximately 1/3 of the way through gestation and concludes near birth, whereas in zebrafish, the liver is fully formed in five days. In mice, members of the FoxA, Gata and Hnf families of transcription factors combine with signaling pathways initiated by members of the fibroblast growth factor (Fgf) and bone morphogenic protein (Bmp) families to activate genes important for driving hepatic lineage and to suppress pancreas formation (10, 11). The Hhex and Prox1 transcription factors are activated in hepatic progenitors and are required for hepatoblast specification. Many of the same genes and pathways are also essential for zebrafish hepatogenesis (8, 12–17).
Hepatic outgrowth begins in mice when hepatoblasts invade the surrounding septum transversum mesenchyme (STM) and continues through late gestation. Moreover, the hepatic vasculature co-develops with hepatoblasts and is required for liver development (18) and fetal hematopoiesis. Thus, vasculogenesis, hematopoiesis and hepatogenesis are intertwined in mice: dysfunction of one of these processes prevents the proper development of the others. In contrast, embryonic hematopoiesis does not take place in the zebrafish liver; therefore abnormalities in the embryonic liver will not lead to early lethality associated with hematopoietic dysfunction. Early liver development in zebrafish is independent of vasculogenesis – the embryo can develop for several days without circulation (7, 19, 20). An illustrative example is provided by our analysis of Klf6, a gene hypothesized to be important for liver growth and function. In previous work, loss of Klf6 in mice caused early embryonic lethality due to defective hematopoiesis (21), precluding analysis of the liver. However, our analysis of klf6 zebrafish morphants revealed a role in development of endodermally-derived organs, especially the liver, distinct from its role in hematopoiesis (Zhao, Sadler and Friedman, unpublished). This demonstrates how zebrafish can complement and extend findings in mice.
There are obvious differences between fish and mammals, yet evolution has conserved many players essential for hepatogenesis, tailoring the specifics to match the unique attributes of each species. For example, the hepatic vasculature and hematopoiesis are all essential for hepatogenesis in mammals, but not in zebrafish. Moreover, secretion hepatic specification depends on Fgfs secreted from the mammalian heart (22), which is nestled against the hepatic bud as the embryo curls into the fetal position. In contrast, the early zebrafish embryo stretches along the top of their yolk (Fig. 1), creating distance between the liver and the heart and making it unlikely that these organs communicate. Interestingly, however, it is clear that Fgfs play an essential role in hepatic development in zebrafish (8, 15) implying that the signal has been conserved, but the source can change. Another difference is the role of the STM, which provides important inductive signals to the mammalian hepatic bud. The existence of the STM in zebrafish is questionable; instead, the lateral plate mesoderm (LPM) appears to serve an analogous function in zebrafish (13, 23). Third, the cellular and anatomical architecture of the zebrafish liver differs from mammals: instead of a portal triad, plates of hepatocytes lined by sinusoids and biliary ductules (24–26) compose the teleost liver, while maintaining the same functions. This has been comprehensively reviewed for Medaka (27). Finally, as in mammals, the biliary system consists of extra and intrahepatic ducts and ductules. However, the preductular epithelial cell is an extra branch on the teleost biliary tree (25, 26), analogous to the Canal of Hering. These cells form junctions with canaliculi to collect the bile, which they transport to the ductules through intracellular lumen (25, 26, 27). These unusual cells may be the fish version of hepatic progenitors (28) and warrant further study.
The challenges in using zebrafish for studying liver biology are primarily technical: the lack of an easy technology for knocking out genes and the relatively few useful antibodies. Moreover, as the field is still young, stellate, Kupffer and oval cells have not yet been identified in zebrafish. Since much of the power of the zebrafish model rests in the attributes of their embryos, little has been done to understand adult hepatic biology, although it is clear that liver regeneration (1) and hepatocarcinogenesis are similar to mammals (29).
Overview of Zebrafish Hepatogenesis
As in their terrestrial vertebrate relatives, the zebrafish liver arises from endoderm. Here, we focus on events in hepatogenesis that occur after endoderm is formed, as zebrafish endoderm development has been reviewed elsewhere (30). There are three main stages of hepatogenesis: (i) specification (as part of endoderm patterning), (ii) differentiation and (iii) hepatic outgrowth, accompanied by morphogenesis. The zebrafish liver develops from anterior endodermal progenitor cells (7), identifiable by 22 hpf through expression of hhex and prox1 (15). The liver primordium is clearly distinguishable by 48 hpf as a prominent bud extending left of the midline over the yolk. Expression of genes indicative of hepatocyte function, such as ceruloplasmin, are first detectable by 32 hpf (31) and other markers of differentiation follow (Fig. 1). As hepatocytes differentiate, proliferation accelerates. Hepatic outgrowth begins between 60–72 hpf and continues until the liver achieves its appropriate proportional size. By the end of outgrowth on 5 dpf, the bi-lobed, boomerang-shaped liver consists of a larger left lobe which crosses the midline under the anterior gut and the smaller right lobe which extends ventrally towards the head of the pancreas (Fig. 2). Few studies thus far have focused on genes required for outgrowth and little is known about morphogenesis.
Transcriptional Control
The same transcriptional networks control hepatogenesis in mice and in zebrafish (Fig. 1). The earliest transcription factors expressed in the zebrafish hepatic bud are prox1 and hhex (12, 15, 30). prox1 is first detected at 22 hpf and persists well through differentiation (15, 23). hhex is expressed in the hepatic bud from 22–50 hpf (32–34) and hhex morphants have a small liver (12). Hepatic nuclear factors (hnf1, 3, and 4) have been extensively studied for their roles in mammalian hepatogenesis (11). hnf1b (also called vhnf1) is essential for several key developmental decisions (35), including hepatic specification in zebrafish (32). A genome-wide scan in zebrafish identified a long list of genes specifically expressed in the liver, nearly 50 of which are targets of an Hnf family member (17).
Some transcription factors required for endoderm formation may also function later in liver development. For instance, gata family members (15, 16, 36, 37), sox17 (7), foxa2 and foxa3 (7, 15, 30) are expressed in the early endoderm and their loss impairs proper endoderm development. These same genes are expressed again during hepatic budding (30), where they also might play a role. The difficulty in addressing this lies in the ability to distinguish which phenotypes are due to the loss of a particular gene versus those secondary effects due to the global, early abnormalities observed in some mutants and morphants, including those with defects in endoderm formation. We addressed this by devising a scoring system that evaluated overall health, size and achievement of developmental milestones so that a finding of disrupted events late in hepatogenesis is less likely to be attributed as a secondary consequence of severe global defects or developmental delay.
Conserved Signaling Pathways Drive Hepatic Specification
Fgfs, Bmps, and the Wnt/β-catenin pathway are the primary signaling pathways required for mammalian hepatogenesis (22, 38–40). These three pathways also control early hepatic development in zebrafish, albeit with some differences.
Fgf signaling is essential for hepatic specification in mice and zebrafish, even though the heart does not seem to be the likely source of Fgfs in zebrafish. Over-expressing dominant negative Fgf-receptor in zebrafish embryos between 18 and 26 hpf, decreases the later expression of hhex, prox1, gata4, gata6 and ceruloplasmin (15). While it is unclear which Fgf family member is most important for specification, fgf10 released from the mesenchyme prevents cells fated to be pancreas, hepatopancreatic duct or intestine from becoming hepatocytes (8). It is not yet clear whether the mesenchyme is the only source of Fgfs required for zebrafish hepatic specification.
Several studies highlight the utility of zebrafish to undertake detailed analysis of developmental processes on a fine-scale time course and the ability to identify new genes regulating these common pathways. For example, Bmps are required to induce hepatic differentiation in mammalian embryo explants (22, 38, 41) and hepatic progenitors in vitro (42). The STM is the likely source of Bmp4 and potentially other Bmps in mammals. Although zebrafish lack the STM, strong data demonstrates that Bmps are essential for zebrafish hepatic specification: lost-a-fin mutants lack a bmp/activin receptor and display reduced expression of many hepatic specification genes, as do embryos that over-express a dominant negative Bmp receptor (15). Interestingly, the requirement for Bmps decreases as the embryo ages, with the susceptible window between 18–26 hpf, overlapping with the time frame in which Fgfs are required (15) (Fig. 1). A forward genetic screen revealed that proper positioning and morphology of the LPM and subsequent hepatic induction by Bmp2a both rely on an unforeseen player, myosin phosphatase targeting subunit 1 (14). bmp2b is also expressed in the LPM (13), yet it is not clear exactly which Bmps contribute to hepatic specification.
In the canonical Wnt signaling pathway, Wnt binds to its cell surface receptor, causing dissociation of β-catenin from the APC complex and preventing degradation (40). This pathway drives hepatic outgrowth in mice (43–46) and is also essential for hepatoblast maturation (46). This complex pathway also plays multiple roles in zebrafish liver development. Inducing wnt expression in early somitogenesis blocks liver specification, whereas inducing wnt just a few hours later creates an enlarged liver. Expanding the liver at the expense of pancreatic cells also occurs when β-catenin is stabilized due to loss of one copy of apc (47). wnt2bb is expressed in the LPM from 18–52 hpf and a forward genetic screen identified prometheus mutants which display delayed hepatic specification due to mutation of wnt2bb (23). These data combined with studies from other vertebrates suggest a model whereby Wnt signaling acts in several phases of hepatogenesis: (i) during early somitogenesis, Wnt signaling to the anterior endoderm suppresses hepatic fate (48) and (ii) just a short time later, Wnt secretion from the LPM promotes hepatic formation (23, 44, 47). (iii) Later, β-catenin activation drives hepatic differentiation and outgrowth and reigning in this pathway is essential to achieve correct liver size and morphogenesis (44, 46, 47). It will be of interest to investigate the interplay between Wnt, Bmp and Fgf signaling.
Much of the zebrafish work on signaling pathways controlling hepatogenesis conform to the framework provided by reverse genetic studies in other vertebrates, however forward genetic screens are often most powerful in uncovering novel paradigms. This is exemplified by the identification of mutants with a putative “developmental checkpoint” where endodermal organs fail to expand. For instance, mutation of the novel digestive-organ expansion factor (def) gene (49) and the RNA binding gene in nil per os (npo) (50), as well as embryos with a block in the TOR pathway (51), undergo proper specification of endodermal organs but then fail to mature and expand. def mutants have activation of a novel p53 isoform causing cell cycle arrest (49). A lack of apoptosis is a key distinction between these putative “checkpoint” mutants and other outgrowth mutants, such as uhrf1 (1), suggesting multiple mechanisms control liver outgrowth and maturation.
Fate Choice
The plasticity of the primitive endoderm is well established; these cells can form the liver, foregut or exocrine pancreas (10). Understanding the genetic decisions that drive this choice has implications beyond embryology. The ability to manipulate endodermal cells in culture to form differentiated functioning hepatocyte could lead to stem cell therapy for patients with liver failure.
Zebrafish studies prove that while cells of the anterior endoderm remain multipotent, the fate choices come at a price. For instance, histone deacetylase 1 (hdac1) mutants fail to fully form a liver or pancreas; instead they have an enlarged foregut (52). Similarly, enlarging the liver by over-expressing wnt8 during somitogenesis reduces the size of the pancreatic bud (47). Work by Chung et al. (2008) exemplifies how detailed analysis of liver development is feasible in zebrafish. Using embryos expressing tissue-specific transgenes combined with a photoactivatable dye for lineage tracing, they demonstrate that the precise timing of Bmp2b expression directs endoderm to become liver instead of pancreas. Conversely, loss-a-fin mutants are in Bmp signaling, and have a smaller liver bud and expanded expression of a pancreatic marker (13). It is interesting that studies with mouse endodermal cells driven to adopt a hepatic fate in vivo also require Bmp signaling (42), further verifying the commonalities between zebrafish and mice.
Epigenetic Control: A New Paradigm
In addition to the specific genes targeted by individual transcription factors, recent evidence from zebrafish points to the importance of epigenetic regulation of gene expression in liver development. Histone acetylation and DNA methylation are major means of epigenetic control of gene expression. Analyses of embryos that lack either Hdac or DNA methyltransferase (Dnmt) activities demonstrate that epigenetic controls both hepatic specification and outgrowth.
hdac1 mutants develop small livers attributable to hepatic patterning defects. Moreover, only a subset of hdac1 embryos express an hepatocyte-specific transgene (52), indicating that hdac1 is required for both hepatic specification and for differentiation. Treating embryos with an Hdac inhibitor prior to 24 hpf reduces hhex and prox1 expression, resulting in a small liver. Moreover, while hdac1 and hdac3 knock-down cause multiple defects in embryos, loss of hdac3 appears more specific for the liver. hdac3 morphants have both patterning and hepatic outgrowth defects attributed to the repression of the Tgfβ family member, gdf11 (19).
A direct role for epigenetics is provided by analysis of dnmt1 mutants. These embryos undergo normal hepatic patterning and differentiation, but fail to undergo outgrowth due to massive hepatocyte apoptosis (Anderson and Stainier, personal communication). This finding is reminiscent of our observations in uhrf1 mutants, which do not expand the liver bud due to both a block in cell proliferation and a marked increase in hepatocyte apoptosis (1). Uhrf1 is essential for DNA methylation by recruiting Dnmt1 to hemimethylated DNA (53, 54). Thus, loss of either dnmt1 or uhrf1 results in aberrant DNA methylation and hepatocyte apoptosis. Understanding epigenetic control of liver development is in its infancy and represents a fertile new direction.
Disease Models
While zebrafish are celebrated for their utility in studying development, recent studies to model hepatobiliary diseases are particularly exciting. Hepatocytes in fish and mammals carry out similar functions, and the histopathology of steatosis, cholestasis, and neoplasia are alike in both organisms. Histologically, hepatocytes are identifiable by 36 hpf, the extrahepatic biliary duct by 52 hpf, and endothelial-lined sinusoids at 3 dpf with blood flow easily observed in 4 dpf embryos. Bile production, serum protein secretion, glycogen storage, lipogenesis and xenobiotic metabolism are fully operational by 5 dpf and various analyses can be carried out on the complex larvae (Fig. 2).
The most straightforward disease modeling efforts are focused on understanding congenital disorders that result from defects in embryo morphogenesis. For instance, the genetic basis of bile duct paucity in Alagille Syndrome can be comprehensively investigated in zebrafish (55). We carried out a forward genetic screen for mutants with features of liver disease and identified a model for hepatic steatosis in foie gras mutants, choledochal cysts in nf2 mutants and cholestasis in vps18 mutants (56). Another interesting model of fatty liver and hepatic injury mediated by Tnfα was recently described in ducttrip mutants, which are defective in s-adenosylhomocystine hydrolase (57).
A distinct advantage of using fish to study liver disease is the opportunity to test effects of drug and toxin exposure by their simple addition to the water. We used this approach to develop a model of acute alcoholic liver disease. With continuous exposure to 2% ethanol, zebrafish larvae develop hepatomegaly, steatosis, and changes in hepatic gene expression, all indicating that alcohol is metabolized by the zebrafish and provokes oxidative stress (58). In addition to alcohol, the zebrafish system facilitates testing of the liver's response to a plethora of toxins, enabling high-throughput toxicity detection and drug screening and discovery.
Future Directions
In the nearly 30 years since researchers began using zebrafish as a developmental genetic model, work from many model organisms has significantly advanced our understanding of liver development. We now understand that transcriptional and signaling networks guide cellular decision-making and drive hepatic progenitors to create the machinery needed to perform the vast array functions carried out by the liver, to proliferate and to organize multiple cell types into the mature liver. There are, however, a many areas that remain under explored: how do different cell types interact during development? what is the embryological origins of stellate and Kupffer cells? do the pathways regulating cell proliferation in the embryo also control regeneration in adults? Zebrafish are a well established model to address each of these questions. Of particular importance is the link between genes that govern development and those that contribute to pathology: how are they deregulated during liver disease, and can they be manipulated as a means of treatment? Zebrafish provides a powerful system capable of detailed developmental genetic analysis and is especially suited for the study of hepatogenesis. This combined with the growing number of liver disease models provides an exciting framework for understanding development, disease, and potential new therapies.
Acknowledgements
We are indebted to Chris Monson for expert fish care and preparation of the figures. Our gratitude extends to Elke Ober, Scott Friedman, and Chinwe Ukomadu for their critical reading of this manuscript and to Didier Stainier for sharing unpublished data. Funding was provided by the March of Dimes, Breast Cancer Alliance, AGA and NIH.
References
- 1.Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci U S A. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hogan BM, Verkade H, Lieschke GJ, Heath JK. Manipulation of gene expression during zebrafish embryonic development using transient approaches. Methods Mol Biol. 2008;469:273–300. doi: 10.1007/978-1-60327-469-2_19. [DOI] [PubMed] [Google Scholar]
- 3.Moens CB, Donn TM, Wolf-Saxon ER, Ma TP. Reverse genetics in zebrafish by TILLING. Brief Funct Genomic Proteomic. 2008;7:454–459. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Her GM, Chiang CC, Chen WY, Wu JL. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio) FEBS Lett. 2003;538:125–133. doi: 10.1016/s0014-5793(03)00157-1. [DOI] [PubMed] [Google Scholar]
- 7.Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 8.Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, et al. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- 9.Zhao R, Duncan SA. Embryonic development of the liver. Hepatology. 2005;41:956–967. doi: 10.1002/hep.20691. [DOI] [PubMed] [Google Scholar]
- 10.Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 12.Wallace KN, Yusuff S, Sonntag JM, Chin AJ, Pack M. Zebrafish hhex regulates liver development and digestive organ chirality. Genesis. 2001;30:141–143. doi: 10.1002/gene.1050. [DOI] [PubMed] [Google Scholar]
- 13.Chung WS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H, Ruan H, Aw MY, Hussain A, Guo L, Gao C, Qian F, et al. Mypt1-mediated spatial positioning of Bmp2-producing cells is essential for liver organogenesis. Development. 2008;135:3209–3218. doi: 10.1242/dev.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, et al. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- 16.Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- 17.Cheng W, Guo L, Zhang Z, Soo HM, Wen C, Wu W, Peng J. HNF factors form a network to regulate liver-enriched genes in zebrafish. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver Organogenesis Promoted by Endothelial Cells Prior to Vascular Function. Science. 2001;294:559–563. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- 19.Farooq M, Sulochana KN, Pan X, To J, Sheng D, Gong Z, Ge R. Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 20.Korzh S, Pan X, Garcia-Lecea M, Winata CL, Wohland T, Korzh V, Gong Z. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol. 2008;8:84. doi: 10.1186/1471-213X-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumoto N, Kubo A, Liu H, Akita K, Laub F, Ramirez F, Keller G, et al. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood. 2006;107:1357–1365. doi: 10.1182/blood-2005-05-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- 23.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006 doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 24.Matthews RP, Lorent K, Russo P, Pack M. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–259. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 25.Hampton JA, Lantz RC, Goldblatt PJ, Lauren DJ, Hinton DE. Functional units in rainbow trout (Salmo gairdneri, Richardson) liver: II. The biliary system. Anat Rec. 1988;221:619–634. doi: 10.1002/ar.1092210208. [DOI] [PubMed] [Google Scholar]
- 26.Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardman RC, Volz DC, Kullman SW, Hinton DE. An in vivo look at vertebrate liver architecture: three-dimensional reconstructions from medaka (Oryzias latipes) Anat Rec (Hoboken) 2007;290:770–782. doi: 10.1002/ar.20524. [DOI] [PubMed] [Google Scholar]
- 28.Okihiro MS, Hinton DE. Partial hepatectomy and bile duct ligation in rainbow trout (Oncorhynchus mykiss): histologic, immunohistochemical and enzyme histochemical characterization of hepatic regeneration and biliary hyperplasia. Toxicol Pathol. 2000;28:342–356. doi: 10.1177/019262330002800215. [DOI] [PubMed] [Google Scholar]
- 29.Lam SH, Wu YL, Vega VB, Miller LD, Spitsbergen J, Tong Y, Zhan H, et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat Biotechnol. 2006;24:73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- 30.Ober EA, Field HA, Stainier DY. From endoderm formation to liver and pancreas development in zebrafish. Mech Dev. 2003;120:5–18. doi: 10.1016/s0925-4773(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 31.Korzh S, Emelyanov A, Korzh V. Developmental analysis of ceruloplasmin gene and liver formation in zebrafish. Mech Dev. 2001;103:137–139. doi: 10.1016/s0925-4773(01)00330-6. [DOI] [PubMed] [Google Scholar]
- 32.Lokmane L, Haumaitre C, Garcia-Villalba P, Anselme I, Schneider-Maunoury S, Cereghini S. Crucial role of vHNF1 in vertebrate hepatic specification. Development. 2008;135:2777–2786. doi: 10.1242/dev.023010. [DOI] [PubMed] [Google Scholar]
- 33.Bogue CW, Ganea GR, Sturm E, Ianucci R, Jacobs HC. Hex expression suggests a role in the development and function of organs derived from foregut endoderm. Dev Dyn. 2000;219:84–89. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1028>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 34.Liao W, Ho CY, Yan YL, Postlethwait J, Stainier DY. Hhex and scl function in parallel to regulate early endothelial and blood differentiation in zebrafish. Development. 2000;127:4303–4313. doi: 10.1242/dev.127.20.4303. [DOI] [PubMed] [Google Scholar]
- 35.Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DY. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiter JF, Kikuchi Y, Stainier DY. Multiple roles for Gata5 in zebrafish endoderm formation. Development. 2001;128:125–135. doi: 10.1242/dev.128.1.125. [DOI] [PubMed] [Google Scholar]
- 38.Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calmont A, Wandzioch E, Tremblay KD, Minowada G, Kaestner KH, Martin GR, Zaret KS. An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev Cell. 2006;11:339–348. doi: 10.1016/j.devcel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 40.Thompson MD, Monga SP. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- 41.Deutsch G, Jung J, Zheng M, Lora J, Zaret KS. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- 42.Gouon-Evans V, Boussemart L, Gadue P, Nierhoff D, Koehler CI, Kubo A, Shafritz DA, et al. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol. 2006;24:1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- 43.Apte U, Zeng G, Thompson MD, Muller P, Micsenyi A, Cieply B, Kaestner KH, et al. beta-Catenin is critical for early postnatal liver growth. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1578–1585. doi: 10.1152/ajpgi.00359.2006. [DOI] [PubMed] [Google Scholar]
- 44.Decaens T, Godard C, de Reynies A, Rickman DS, Tronche F, Couty JP, Perret C, et al. Stabilization of beta-catenin affects mouse embryonic liver growth and hepatoblast fate. Hepatology. 2008;47:247–258. doi: 10.1002/hep.21952. [DOI] [PubMed] [Google Scholar]
- 45.Tan X, Behari J, Cieply B, Michalopoulos GK, Monga SP. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 46.Tan X, Yuan Y, Zeng G, Apte U, Thompson MD, Cieply B, Stolz DB, et al. Beta-catenin deletion in hepatoblasts disrupts hepatic morphogenesis and survival during mouse development. Hepatology. 2008;47:1667–1679. doi: 10.1002/hep.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goessling W, North TE, Lord AM, Ceol C, Lee S, Weidinger G, Bourque C, et al. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 48.McLin VA, Rankin SA, Zorn AM. Repression of Wnt/{beta}-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development. 2007;134:2207–2217. doi: 10.1242/dev.001230. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W, Zhang Z, et al. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19:2900–2911. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayer AN, Fishman MC. Nil per os encodes a conserved RNA recognition motif protein required for morphogenesis and cytodifferentiation of digestive organs in zebrafish. Development. 2003;130:3917–3928. doi: 10.1242/dev.00600. [DOI] [PubMed] [Google Scholar]
- 51.Makky K, Tekiela J, Mayer AN. Target of rapamycin (TOR) signaling controls epithelial morphogenesis in the vertebrate intestine. Dev Biol. 2007;303:501–513. doi: 10.1016/j.ydbio.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noel ES, Casal-Sueiro A, Busch-Nentwich E, Verkade H, Dong PD, Stemple DL, Ober EA. Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev Biol. 2008;322:237–250. doi: 10.1016/j.ydbio.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 54.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 55.Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development. 2004;131:5753–5766. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- 56.Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- 57.Matthews RP, Lorent K, Manoral-Mobias R, Huang Y, Gong W, Murray IV, Blair IA, et al. TNF{alpha}-dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish s-adenosylhomocysteine hydrolase. Development. 2009;136:865–875. doi: 10.1242/dev.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Passeri MJ, Cinaroglu A, Gao C, Sadler KC. Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology. 2008;49:443–452. doi: 10.1002/hep.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]