Not all patients respond to drug therapy in a uniform and beneficial fashion. The goal of this review is to describe the contribution of genetic variation to drug response, with a focus on drugs used in cardiovascular therapy. Genetic approaches used to analyze rare and common adverse effects as well as variability in efficacy are first presented. The challenges and potential solutions to incorporating this body of knowledge into contemporary medical practice are then discussed.
History of pharmacogenetics
The notion that genetic variants might modulate variability in drug actions was first proposed by the English physiologist Garrod.1 He suggested that enzymatic defects lead not only to accumulation of endogenous substrates in “inborn errors of metabolism” (a term that he coined), but also to accumulation of exogenously administered substrates such as drugs, foodstuffs, and toxins, with clinical consequences. Initial examples of genetically-determined variable drug actions were the identification of pseudocholinesterase deficiency in prolonged paralysis after administration of the muscle relaxant succinylcholine,2 deficient N-acetylation of isoniazid,3 and a high incidence of hemolytic anemia among African-Americans with G6PD deficiency receiving antimalarial drugs in the South Pacific during WWII.4 The latter observation highlights the principle in modern genomics and pharmacogenomics that ancestry may play a key role in modulating clinically-important phenotypes.
The term “pharmacogenetics” was coined in 19595 and the first textbook was published in 1962,6 well before methods for studying DNA sequence variation were available. The term “pharmacogenomics” has been more recently used to transmit the idea that variable drug response may reflect sets of variants within an individual or across a population. DNA variants can modulate protein function, and hence drug response, through multiple mechanisms. Much of the initial focus in the field was on non-synonymous DNA variants, i.e. those that alter protein function by changing the encoded amino acids. Non-coding variants that modulate gene expression represent another common candidate mechanism for variable drug responses. Contemporary genomics has uncovered multiple other mechanisms regulating gene function and expression, such as epigenetic changes and small mRNAs, and a role for these in determining drug response seems likely.7
Identifying genetic contributors to variable drug actions
Heritability and drug responses
Studies in families can define the extent to which common human disease phenotypes like myocardial infarction or sudden cardiac death include a heritable component. However, it is usually not possible to accumulate well-defined drug response phenotypes across multiple related patients with the same disease; as a result, the heritable component of variability in drug action may not be well-defined. An in vitro approach that has been useful to estimate heritability of cytotoxicity due to anticancer agents is exposure of lymphoblastoid cell lines from related subjects to the drug.8,9 Using this method, the heritability of cytotoxicity has been estimated at 0.25–0.65; one study went on to use linkage analysis to identify a potential locus mediating this toxicity.9
One approach when heritability is not well-understood is to quantify drug responses in multiple healthy members of a family. For example, very early studies in twins demonstrated far more variability in the urinary excretion of isoniazid within dizygotic than monozygotic twins,10 thus establishing that this trait – now known to reflect genetically-determined variable N-acetylation – is heritable. Similarly, digoxin clearance was much better correlated within monozygotic than dizygotic twins; the heritability of the area under the curve was >79%.11 ADP-stimulated platelet aggregation was studied before and after clopidogrel in the Amish, a founder population with extensive genealogical records: the investigators reported that heritability was 0.33 at baseline and 0.73 during drug treatment, indicating a strong genetic component in the drug response.12
Experimental approaches in pharmacogenetics
Defining mechanisms underlying variable drug concentrations and effects provides a starting point for identifying candidate genes for further pharmacogenetic study. As a result, many important examples in pharmacogenetics relate to variable drug uptake, metabolism, or elimination. Other contributors to variable drug responses identified by this physiologically-based candidate approach include variation in drug target molecules or in disease pathways. In some cases, variants in multiple genes have been implicated, as discussed below (“combinatorial pharmacogenetics”). More recently, technologies to search for previously unanticipated relationships between phenotypes and hundreds of thousands of common polymorphic sites across the genome (an unbiased approach)13 have been applied to the problem of variable drug actions; these genome-wide association studies (GWAS) have been conducted both in human cohorts as well as in cellular or model organ systems. Table 1 lists the experimental approaches that have been used in the field, along with their potential advantages and disadvantages.
Table 1.
Approaches to identifying and validating genetic influences on drug response
| Approach | Advantages | Disadvantages | Examples |
|---|---|---|---|
| Biological candidates | |||
| Candidate gene based on variable pharmacokinetics | Variability in these processes logically determine variable drug effects Candidate genes often identified |
Identification and replication of associations between variant genotypes and drug responses may require large populations, depending on the size of the genetic effect and the frequency of the variant allele | Warfarin/CYP2C914 Simvastatin/SLCO1B115 Clopidogrel/CYP2C1916–18 Metoprolol/CYP2D619 Atorvastatin/CYP3A520 |
| Candidate gene based on variable pharmacodynamics | Bucindolol/ADRB121 Beta-blockers in heart failure /ACE22 Antiarrhythmics in atrial fibrillation/ACE23 Warfarin/VKORC124,25 |
||
| Candidate pathway analysis | Possibly less biased than single gene approaches | Requires interrogation of large numbers of SNPs; replication may be difficult | HMG-CoA reductase haplotype as a predictor of statin response26 |
| Unbiased approaches | |||
| Candidate gene selected from GWAS or other unbiased approach | Unbiased | GWAS result must be available Replication may be difficult | NOS1AP as a predictor of mortality during calcium channel blocker therapy27 |
| Genome-wide association study | Sets of cases and controls generally need to be large; replication may be difficult | Simvastatin/SLCO1B128 Warfarin/VKORC1, CYP2C9, CYP4F229,30 Clopidogrel/CYP2C9/19 locus12 |
|
| Drug response in model organisms with manipulated genetic background | Assay may be difficult to establish Translation from model organism to human may be imperfect |
QT prolongation/GINS3 locus31 | |
Replication of genotype-phenotype relations can be a major issue in modern genomics, both when the effects of single candidate variants are examined32–34 as well as with genome-wide approaches.35 Pharmacogenetic studies may be especially difficult to replicate because large numbers of subjects with well-curated drug response phenotypes are often not available. Other challenges, notably for the use of GWAS, include choice of appropriate control groups matched for factors such as underlying and ancestry, contributions by DNA variants not captured by current platforms (e.g. rare variants or copy number variation), and analysis of gene-gene and gene-environment interactions in determining phenotype.
Variable drug actions and single gene variants
Large effect variants in drug-metabolizing enzymes
In the 1950s, McKusick and Price-Evans described variable N-acetylation,3 an important contributor to variable isoniazid hepatotoxicity and the lupus syndrome during treatment with procainamide and hydralazine. In the 1970s, two groups, studying different drugs (debrisoquine, an antihypertensive,36 and sparteine, being assessed as an antiarrhythmic37,38), reported a set of 5–10% of subjects with adverse effects due to apparent absence of a key enzyme mediating drug bioinactivation. The enzymes were initially termed debrisoquine 4-hydroxylase and sparteine N-oxidase, but subsequently it became clear that this was the same defect,39 now recognized to represent homozygosity for loss of function of a specific member of the cytochrome P450 (CYP) superfamily of drug metabolizing enzymes, CYP2D6.40 Dozens of variants have now been reported to reduce or eliminate CYP2D6 function (http://www.cypalleles.ki.se/cyp2d6.htm).
Coding region variants in other members of this superfamily, such as CYP2C9 and CYP2C19, generate populations of “poor metabolizers” for substrates of each of these enzymes. Interestingly, CYP3A4, the enzyme most commonly implicated in the metabolism of clinically-used drugs,41 does not include common coding region polymorphisms that alter function; nevertheless, CYP3A4 activity varies widely across individuals, and at least some of this variability likely arises from genetic variation in the regulation of CYP3A4 gene expression.42 Another contributor to variability in CYP3A activity is a common intronic single nucleotide polymorphism (SNP) in a closely-related gene, CYP3A5;20,43 the variant CYP3A5*3 allele alters mRNA by creating a new splice site.
The incidence of functionally-important CYP alleles can vary strikingly by ancestry. For example, poor metabolizers with absent CYP2D6 function are found in 5–10% of European and African populations, but are less common in Asian subjects. By contrast, CYP2C19 poor metabolizers are commoner in Asian subjects compared to the other two major ancestry groups, and the frequency of the CYP3A5*3 variant is much higher in Caucasians (0.85) compared to African Americans (0.55), which correlates with higher hepatic CYP3A5 expression in African American subjects.43
High-risk pharmacokinetics
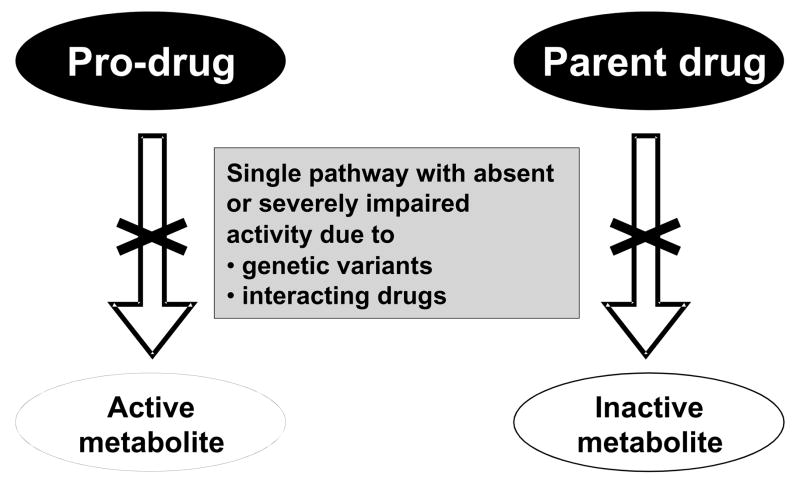
When drugs are eliminated by multiple pathways, absence of one of these (because of genetic variation or because of the presence of interacting inhibiting drugs) is unlikely to produce major variation in drug concentrations at the target site and thus in drug effect. However, the potential for highly variable drug concentrations increases dramatically when a drug is metabolized by a single pathway, a situation we have termed “high risk pharmacokinetics”.44 There are two scenarios in which this may occur (Figure 1). The first is the situation in which a prodrug must be metabolized, or bioactivated, to generate pharmacologic effects. In situations in which this bioactivation is accomplished by an enzyme with known loss-of-function variants, poor metabolizers will, predictably, display decreased drug action; clopidogrel and losartan are examples of cardiovascular drugs with this attribute (see Table 2), and codeine59,60 and tamoxifen61,62 are other prominent examples. Co-administration of commonly-used drugs that inhibit the bioactivating enzyme can result in a “phenocopy” of the poor metabolizer trait: that is, individuals who are genetically extensive metabolizers may display the same pharmacologic outcome as poor metabolizers if administered an interacting drug.
Figure 1.
High-risk pharmacokinetics. Drugs that are eliminated by a single pathway can generate aberrant responses if that pathway is absent on a genetic basis, or because of co-administration of inhibiting drugs. This figure illustrates the two scenarios underlying such “high-risk” pharmacokinetic situations. One (left) is the administration of a drug that is itself not active but requires drug metabolism to generate an active metabolite; the absence of the pathway can lead to failure to generate the desired drug effect. This is thought to underlie variability in response to clopidogrel, tamoxifen, losartan, and codeine, as described in the text. The second scenario (right) is the administration of a drug eliminated by single pathway. Absence of this pathway will result in accumulation of the parent drug and thus drug-related toxicity. Adapted, by permission.44
Table 2.
Genetic variants influencing cardiovascular drug therapy: examples
| Gene | Drug | Clinical effects |
|---|---|---|
| Drug metabolism | ||
| CYP2C9 | Losartan | Decreased bioactivation and effects (PMs)45 |
| Warfarin | Decreased dose requirements; possible increased bleeding risk (PMs)14,46 | |
| CYP2C19 | Clopidogrel | Decreased bioactivation and effect in PMs16–18 |
| CYP2D6 | metoprolol carvedilol timolol propafenone |
Increased beta-blockade in PMs19,47,48 |
| CYP3A5 | atorvastatin simvastatin lovastatin |
Increased lipid lowering efficacy49 increased severity of myotoxicity50 |
| NAT2 | hydralazine, procainamide |
Increased risk of toxicity in PMs51 |
| Drug transport | ||
| SLCO1B1 | simvastatin | Variant non-synonymous single nucleotide polymorphism alters efficacy and increases myopathy risk15,28,52 |
| ABCG2 | many statins | Altered pharmacokinetics52 |
| Drug targets | ||
| HMG-CoA reductase | pravastatin | Haplotype-dependent LDL lowering26 |
| VKORC1 | warfarin | Decreased dose requirements with variant promoter haplotype24 |
| ADRB1, ADRB2 | many beta- blockers | Altered vascular and heart rate effects53–55 |
| ACE | ACE inhibitors | No effect on drug response34 |
| Other genes | ||
| ACE | Beta-blockers in heart failure Antiarrhythmics in atrial fibrillation | Decreased response in subjects with DD genotype22,23 |
| G-protein β3 subunit, kininogen, other loci | Thiazide diuretics | Greater reduction in diastolic and systolic blood pressure56–58 |
As discussed in the text, there is variability in the size of the genetic effects and in the extent to which these findings have been replicated.
Further data at the Pharmacogenetics Research Network/Knowledge base: http://www.pharmgkb.org
The second “high risk pharmacokinetic” scenario is seen when a substrate drug undergoes bioinactivation via a single metabolic pathway. In the absence of this pathway, much higher concentrations of active parent drug will accumulate. For compounds with a wide therapeutic margin, such accumulation may be without clinical implications; conversely for other drugs such accumulation predictably results in serious toxicity. An example is the active S-enantiomer of warfarin which undergoes CYP2C9-mediated metabolism to inactive forms (Figure 2). As discussed further below, patients with common reduction-of-function alleles have higher S-warfarin concentrations and thus lower dose requirements to achieve steady state anticoagulation.14,46 However, there are rare patients with near-complete loss of CYP2C9 function (homozygotes for the *3 variant, arising from 1075A>C encoding I359L), and these patients may be very difficult to manage clinically because of low, and often unstable, warfarin dose requirements.64
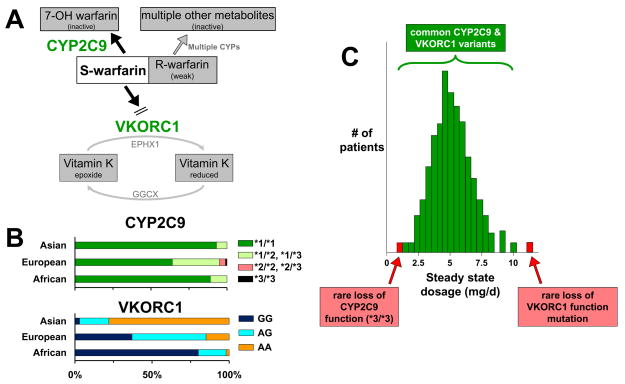
Figure 2.
A framework for analyzing contributions of multiple genes to a clinical phenotype. The example of warfarin maintenance dose requirement is shown here. A. A simple pathway analysis of the key molecular determinants of warfarin response. The drug is administered as a racemate, and most anticoagulant action is mediated by the more potent S-enantiomer. S-warfarin is bioinactivated primarily by CYP2C9. The pharmacologic target for the drug is encoded by VKORC1, important for maintaining active Vitamin K. The role for other drug metabolizing pathways and for other enzymes that influence Vitamin K metabolism (EPHX1, GGCX) are shown in gray. B. Distribution of CYP2C9 and VKORC1 variants as a function of ancestry.63 For CYP2C9 (top panel), the *1 allele has the highest activity, *2 is a reduction of function variant, and *3 is a near loss of function variant. For the VKORC1 promoter variant shown, the G allele results in greater liver expression than does the A allele.24 These distributions of genotypes largely explain ancestry-dependent variability in warfarin dosing.63 C. Contribution of common and rare variants to warfarin dose requirements in a population. The normally-distributed dose requirements predominantly reflect the common variants shown in panel (B). However, individuals with rare VKORC1 coding region variants and individuals with the rare CYP2C9*3/*3 genotype may display unusually high or low dose requirements.
Large effect variants in other genes
Single variants in genes not involved in drug metabolism can also confer high risk for variable drug responses. These may involve variants in genes encoding the target molecules or pathways with which drugs interact, or those encoding genes unrelated to the therapeutic effect. In the latter group, one well-studied example is variants in the HLA system. Individuals with a single HLA B*5701 variant are at high risk for potentially fatal skin reactions during treatment with the antiretroviral drug abacavir,65,66 and similarly B*1502 (an allele seen primarily among Asians) has been linked to severe skin reactions during treatment with carbamazepine.67 As discussed further below, the V174A variant in SLCO1B1, that encodes a transport molecule responsible for uptake of simvastatin in liver, has been associated with a markedly increased risk for myopathy.28
An example of a large effect of a variant in a drug target molecule is the reported association of the R389G variant in the beta1-adrenergic receptor gene with outcomes during treatment of heart failure with the adrenergic receptor blocker bucindolol.21 This variant is known to strongly modulate β1-mediated pharmacologic responses in vivo and in vitro, and outcomes in individuals with the G variant were very close to those treated with placebo. This finding suggests that pre-prescription genotyping could be used to target therapy to those predicted to derive benefit. However, the association is not replicated and other studies have implicated variants in other genes as potential contributors to outcomes of drug therapy in heart failure: examples are ACE,22 CYP2D6,47,68 GRK5,69 and alpha2C receptors.70,71 The relationship between warfarin dose and variants in VKORC1, encoding the warfarin target, are discussed below and other examples are listed in Table 2.
Combinatorial pharmacogenetics
Another approach to analyzing variability in complex traits like heart failure and its response to drugs is to study not single genetic variants, but combinations across multiple genes.72
Two recent examples, warfarin and clopidogrel, illustrate how the interrogation of very large clinical datasets for candidate polymorphisms in multiple candidate genes in combination can help establish the role of these variants in determining a drug’s action. As discussed further below, the understanding that relatively large effects of single genetic variants modulate the effects of these drugs has prompted the US Food and Drug Administration to include genetic information in the labels for these and other agents, triggering a debate about how this new knowledge can be incorporated into practice.
An extension of this idea is interrogation of hundreds of SNPs in multiple genes in a candidate “pathway” to identify loci modulating a drug response.26 Drug-induced prolongation of the QT interval has also been analyzed in this fashion. Almost all drugs that prolong QT do so by blocking a specific cardiac potassium current, IKr. However, studies examining variation in the QT interval itself73,74 or its response to drug challenge,75,76 have implicated multiple other ion channel and other genes. This supports a view in which control of the QT interval relies on diverse mechanisms (almost all unrelated to IKr), and variation in these mechanisms then leads to variability in the extent to which IKr blockers prolong QT. This idea, termed “repolarization reserve,”77,78 is a specific example of the more general concept that variability in physiologic and drug response phenotypes reflects the interplay of multiple biologic pathways.
The warfarin example (Figure 2)
In 2004, the disease gene for a very rare pharmacogenetic syndrome, warfarin resistance (in which patients displayed little change in INR upon challenge with extremely high doses of warfarin) was identified.79 The gene, VKORC1, encodes the component of the vitamin K receptor complex that is the warfarin target, thus explaining the rare genetic trait. Identification of VKORC1 as the warfarin target led rapidly to identification of common variant promoter haplotypes which correlated well with variable liver expression of the protein,24 and are associated with decreased steady state dose requirement and shorter time to therapeutic anticoagulation.24,25 One study in 539 Caucasian patients receiving steady state warfarin therapy reported that 25% of the variability in warfarin dose could be accounted for by common VKORC1 promoter SNPs, and 9% by variants in CYP2C9; these are very large genetic effects.24 Warfarin dose requirements vary strikingly by ancestry (highest in African-Americans, lowest in Asians), and much of the difference can be attributed to the frequency of common VKORC1 promoter variants (Figure 2B).80 In addition, rare VKORC1 coding region variants have been described in some subjects with unusually high warfarin dosages; for example, in one study, 4.3% of Ashkenazi subjects were found to have a variant resulting in D36Y, associated with high dose requirements (>10mg/day).81
The International Warfarin Pharmacogenetics Consortium studied the relationship between genotypes and steady state warfarin dose in >5,000 patients of diverse ancestries.63 There was clear ancestry-dependent variation in dose requirement (highest in subjects of African descent, lowest in subjects of Asian descent), and the differences could be attributed to variation in VKORC1 and CYP2C9 (Figure 2B). As discussed below, trials are now underway to compare outcomes in patients using genetically- versus clinically-guided therapy.
The clopidogrel example
Several studies in early 2009 reported that reduction-of-function variants in CYP2C19 (the enzyme responsible for the bioactivation of clopidogrel) increase the risk of cardiovascular events after stent placement.16–18 One of these18 also examined the potential contribution of multiple other candidate genes to variable clopidogrel effects, including those encoding other CYPs, the P2Y12 receptor (the drug target), other molecules known to interact with the receptor, and the drug efflux transporter P-glycoprotein. The latter is encoded by ABCB1, and P-glycoprotein is known to modulate absorption and elimination of many other drugs. The result of that study was that, in addition to the CYP2C19 effect, individuals homozygous for a variant ABCB1 coding region allele were more likely to display failure of efficacy during clopidogrel therapy. Although the role of CYP2C19 is now described in the clopidogrel label, the way in which clinicians should respond to this information remains uncertain.82,83
Unbiased approaches to identifying genes modulating drug actions
The Human Genome Project and subsequent increasingly detailed maps of human genetic variation are providing tools to interrogate the relationship between genetic variation across the human genome and important human physiology and disease traits in a relatively unbiased fashion. A conventional GWAS design generates a set of SNPs associated with the trait under study, and then seeks to replicate these associations in other (often larger) clinical datasets.13 The contribution of variants identified by GWAS to susceptibility to common diseases is usually modest: it is likely that high risk alleles do not accumulate in populations because of the evolutionary disadvantage such accumulation confers.
Applying the GWAS paradigm to pharmacogenomics faces the obstacle that very large sets of patients with well-defined drug response phenotypes are unusual (Table 1). On the other hand, since there may be no selection pressure on genes encoding proteins mediating drug action, functionally-important variants with large effects may have accumulated in populations. Another distinctive feature of GWAS studies of drug response is the nature of the signals identified. GWAS approaches to disease have often identified new susceptibility loci. By contrast, many (but not all84–86) GWAS studies in pharmacogenomics have yielded signals in previously-studied pathways, presumably reflecting large effects readily derived from candidate gene approaches.
One notable success using the GWAS approach in cardiovascular pharmacogenomics was a study that examined simvastatin-associated myopathy.28 Among 6033 patients receiving 20 mg/day, there were only 8 possible cases, so the study focused on 98/6031 patients who developed myotoxicity during high dose (80 mg/day) therapy. A GWAS comparing 85 cases of definite or incipient myopathy to 90 controls identified a single SNP (rs4363657) in SLCO1B1 at genomewide significance. This SNP is in near perfect linkage disequilibrium with a previously-studied non-synonymous variant (V174A) in OATP1B1, the drug uptake transporter that SLCO1B1 encodes; studies prior to the GWAS showed that V174A impairs elimination of simvastatin acid, and thus implicated it as a potential modulator of drug efficacy and toxicity.15 Patients with the rare (2.1%) homozygous phenotype had an 18% 5-year incidence of myopathy, compared to 3% among heterozygotes, and 0.6% among those lacking the risk allele. The findings were replicated in a separate study of subjects a lower dose, 40 mg/day, with a smaller effect size (relative risk 2.6 per C allele).28 To date, one group has reported a similar finding across multiple statin drugs in a smaller trial.87
A recent GWAS examining predictors of therapeutic response to statin therapy in three randomized treatment trials has implicated multiple known lipid control loci, as well as identifying a novel association near calmin, a gene not previously known to influence lipid homeostasis.88 This analysis of thousands of patients exposed to atorvastatin, pravastatin or simvastatin used Bayesian statistical approaches to suggest that the relationship between genetic variability at the calmin locus and statin-related change in fasting lipid levels may represent a class effect.
GWAS results to aid study design
In 2003, the FDA proposed a prospective evaluation of the utility of CYP2C9 genotyping as a guide to warfarin therapy. This effort was suspended with the recognition that VKORC1 contributes importantly to variability in warfarin action. While interest in a prospective trial remained high, the new finding raised the question of how many other as-yet-unidentified genes might also contribute to warfarin actions. GWAS studies examining steady state warfarin dose demonstrated that VKORC1 variants were the most important contributors to this phenotype, and while a number of other genes were implicated, none except CYP2C9 (and in one study CYP4F2) survived replication.29,30 Thus, although variants in other genes may still contribute to variability in warfarin actions (CYP4F2,89 CYP2C18,90 EPHX1,91 GGCX92) these contributions seem to be small30 or affect only a small number of individuals and prospective trials are now ongoing in the US and Europe to examine the utility of genotyping to guide dosage of warfarin and other vitamin K antagonists. Similarly, a GWAS examining clopidogrel effects on ADP-mediated platelet aggregation ex vivo demonstrated that variability at a single locus, encompassing CYP2C19, was the major contributor to the phenotype.12 Interestingly, ~70% of the variability in clopidogrel dose was found to be heritable, and this locus contributed ~15% to that variability. This is a very large effect of a single common polymorphism, that also highlights the “missing heritability” that may reflect gene-gene interactions or multiple rare genetic variants.93
Model organisms and mechanisms
In addition to the cell-based approaches mentioned above (immortalized lymphoblastoid cell lines),8,9 unbiased studies in model organisms have also been used to identify contributions of loci across the genome to drug response phenotypes. Milan et al. demonstrated that QT prolonging drugs reproducibly produce bradycardia and atrioventricular block in zebrafish embryos.94 Challenging wild-type of mutagenized fish with dofetilide, a prototypical QT prolonging agent, identified multiple loci modulating this drug response phenotype.31 Interestingly, one of these, GINS3, also was implicated as a modulator of the normal QT duration in GWAS analyses of tens of thousands of subjects.73,74 An example of how unbiased discovery approaches can lead to new understanding new mechanisms is the finding that a SNP in the HMG-CoA reductase gene, initially identified as a modulator of response to inhibitor drugs, generates an alternatively spliced mRNA that encodes a protein with reduced drug sensitivity.95
How will pharmacogenetic information be incorporated into the fabric of healthcare?
One simple approach to the problem of drugs with genetically-determined variable outcomes is to replace them with other drugs that lack these genetic features. In fact, because of delineation of the role of common variants in CYP2D6, candidate drugs whose elimination occurs largely via this enzyme are generally not developed. For available drugs that display genetically-determined variable metabolism, dose adjustment or use of alternate drugs seems a logical strategy to reduce that variability. For example, the clinical effects of the P2Y12 inhibitor prasugrel appear to be independent of CYP2C19,96 and so this drug may be especially desirable in patients who would ordinarily receive clopidogrel but have variant enzyme. Conversely, it may be cheaper and just as effective to use the older (off-patent) drug in patients lacking the clinically important genetic variants.
Another approach to adopting pharmacogenomic data to practice is to design clinical trials that do not simply examine the effect of genotypes on outcomes post hoc, but use genotypes as entry or stratification criteria. This idea has only occasionally been attempted to date in cardiovascular medicine,97 although it is rapidly becoming an important part of cancer therapeutics, where mutations in the tumor genome are increasingly identified as drivers of disease and of response to therapy.98,99 The evidence that genomic variants can clearly influence the outcome of drug therapy has led the Food and Drug Administration to include this information in drug labels.100,101 However, it is fair to say that application of this information to the bedside care of patients has been slow to be adopted. Multiple reasons can be proposed for this apparent paradox:
It is burdensome for healthcare providers to incorporate genetic testing into prescribing. The results of genetic testing take at least hours (if not days or longer) to generate and once the result is obtained, the patient must be recontacted to inform them that the drug and dose selected are or are not appropriate. Further, consensus recommendations on dose adjustment may not be available, even when large effects of genetic variants on drug response have been identified.
The level of evidence required to adopt a specific genetic test as a guide to practice is not established. The gold standard for evaluating therapeutic approaches in populations is the randomized clinical trial (RCT); however, given the number of variants implicated as modulators of drug action, it is not possible to mount an RCT to evaluate each one. Moreover, variants that are relatively uncommon across populations may still exert large and clinically important effects in individual subjects, and there is ample precedent for incorporating these into practice in the absence of RCTs: the clinical practice of decreasing dose of renally-excreted drugs in patients with elevated creatinine is one simple example.
The numbers of variants and their potential effects during treatment with specific drugs is growing extraordinarily rapidly. It is not possible for individual practitioners to keep track of these without assistance from information technology systems. Medical school and post-graduate education is only now beginning to incorporate new genomics into curricula.
In the face of these obstacles, a positive and potentially enabling development is that the costs of genotyping at target loci or across large sections of the genome are plummeting: the cost for the first full human genome sequence, completed a decade ago, was ~$3,000,000,000, and is expected to fall to $1000 within a year (a cost that begins to compare favorably to that for individual genotyping tests). A number of companies are now offering direct to consumer wide-scale genotyping, and this includes some of the common CYP and other variants described above.
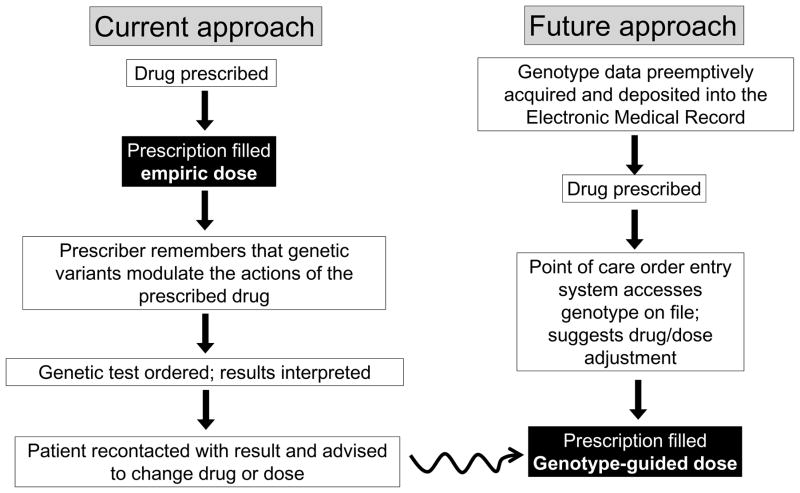
These developments set the stage for a potential paradigm shift in the use genomic, and in particular pharmacogenomic, information in clinical medicine. Currently (Figure 3; left), genetic information is used on an ad hoc and as needed basis: the prescriber must remember or be reminded to order a specific genetic test, and once the result is delivered, recontact the patient. Figure 3 (right) illustrates the new paradigm: genomic data is deposited preemptively into individual subjects’ electronic medical records (EMRs), to be accessed at the point of care, as needed. The incorporation of genetic information in this fashion into EMRs could also enable a future vision in which drug outcomes can be not only queried retrospectively but also followed prospectively, to generate new genotype-drug response relationships. Francis Collins enunciated this vision after being appointed NIH director, when he said: “The limiting factor right now is thatoften times, if you are ready to write a prescription, you donot want to wait a week to find out the genotype before you decide whether you've got the right dose and the right drug.But if everybody's DNA sequence is already in their medicalrecord and it is simply a click of the mouse to find out allthe information you need, then there is going to be a much lower barrier to beginning to incorporate that information into drug prescribing. If you have the evidence, it will be hard, I think, to say that this is not a good thing. And once you've got the sequence, it's not going to be terribly expensive. And it should improve outcomes and reduce adverse events.”102
Figure 3.
Contrasting approaches to incorporating genomic information into prescribing. The pathway on the left illustrates current practice, genetic testing on an as needed basis. The pathway on the right illustrates how preemptive deposit of genotypic data into a genome-enabled electronic medical record would result rapid and efficient genotype-guided therapy.
The barriers to executing this idea are considerable. Whole genome sequencing is likely to be very inexpensive, but the ethical issues inherent in use of large scale sequencing in clinical practice are daunting, especially at the dawn of the era of individualized genomes, when multiple rare variants of unknown significance will be routinely discovered in each of us.103 One solution might be to acquire whole genome sequence data but to mask all but the “actionable” portions of the genome, such as high-impact pharmacogenetic variants. The sequence data will need to meet standards for use in clinical settings, and extensive annotation and curation will be required to ensure that traits like “poor metabolizer” are correctly inferred. As whole genomes become more widely available, methods to ensure that costs do not spiral in pursuit of incidental findings must be developed.104 It is clear that this vision of preemptive genotyping cannot be executed without electronic systems to manage the data. Levels of evidence that justify acting on a particular genotype or set of genotypes and smart informatics systems to deliver prescribing advice in a simple but comprehensive format will need to be developed. New algorithms to mine outcomes in EMRs and to link these to embedded genomic markers would be highly desirable. . The costs of preemptive genotyping and the extent to which costs associated with ineffective or dangerous drug therapy can be avoided will need to be assessed. Overcoming all these barriers will enable a new paradigm of practice that moves away from one size fits all medicine towards a more personalized approach to therapy.
Acknowledgments
Grant Support
Supported in part by grants from the US National Institutes of Health (U01 HL065962, U01 HL069757, R01 DK080007) and the Deutsche Forschungsgemeinschaft (SFB TR19)
Footnotes
Disclosures
Dr. Roden reports receiving royalties from Clinical Data Inc., for a patent relating to genetic testing for arrhythmia susceptibility. There are no other disclosures relevant to the content of this manuscript.
References
- 1.Garrod AE. Inborn errors of metabolism. 2. London: Henry Frowde and Hodder Stroughton; 1923. [Google Scholar]
- 2.Kalow W. Familial incidence of low pseudocholinesterase level. Lancet. 1956;268:576–577. doi: 10.1016/s0140-6736(56)90869-8. [DOI] [PubMed] [Google Scholar]
- 3.Price-Evans DA, Manley FA, McKusick VA. Genetic control of isoniazid metabolism in man. Br Med J. 1960;2:485–491. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutler E, Dern RJ, Alving AS. The hemolytic effect of primaquine. VI. An in vitro test for sensitivity of erythrocytes to primaquine. Journal of Laboratory & Clinical Medicine. 1955;45:40–50. [PubMed] [Google Scholar]
- 5.Vogel F. Moderne Probleme der Humangenetik. Ergebn Inn Med Kinderheilkd. 1959;12:52–125. [Google Scholar]
- 6.Kalow W. Pharmacogenetics -- Heredity and responses to drugs. Philadelphia: W.B.Saunders; 1962. [Google Scholar]
- 7.Sadee W, Dai Z. Pharmacogenetics/genomics and personalized medicine. Hum Mol Genet. 2005;14(Spec No 2):R207–R214. doi: 10.1093/hmg/ddi261. [DOI] [PubMed] [Google Scholar]
- 8.Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc Natl Acad Sci U S A. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, Jr, Badner JA. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 10.Bönicke R, Lisboa BP. Uber die Erbbedingtheit der intraindividuellen Konstantz der Isoniazidausscheidung beim Menshen (Untersuchen an eineiggen und zweieiggen Zwilligen) Naturwissenschafften. 1957;44:314. [Google Scholar]
- 11.Birkenfeld AL, Jordan J, Hofmann U, Busjahn A, Franke G, Kruger N, Igel S, Murdter T, Drescher S, Shi S, Engeli S, Schwab M, Eichelbaum M, Luft FC, Fromm MF. Genetic influences on the pharmacokinetics of orally and intravenously administered digoxin as exhibited by monozygotic twins. Clin Pharmacol Ther. 2009;86:605–608. doi: 10.1038/clpt.2009.170. [DOI] [PubMed] [Google Scholar]
- 12.Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, Damcott CM, Pakyz R, Tantry US, Gibson Q, Pollin TI, Post W, Parsa A, Mitchell BD, Faraday N, Herzog W, Gurbel PA. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 15.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006;16:873–879. doi: 10.1097/01.fpc.0000230416.82349.90. [DOI] [PubMed] [Google Scholar]
- 16.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS. Cytochrome P-450 Polymorphisms and Response to Clopidogrel. N Engl J Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 17.Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, Payot L, Brugier D, Cayla G, Beygui F, Bensimon G, Funck-Brentano C, Montalescot G. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373:309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 18.Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, Steg PG, Ferrieres J, Danchin N, Becquemont L. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 19.Lennard MS, Silas JH, Freestone S, Ramsay LE, Tucker GT, Woods HF. Oxidation phenotype--a major determinant of metoprolol metabolism and response. N Engl J Med. 1982;307:1558–1560. doi: 10.1056/NEJM198212163072505. [DOI] [PubMed] [Google Scholar]
- 20.Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005;15:415–421. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, Nelson B, Morrison J, Domanski MJ, Wagoner LE, Abraham WT, Anderson JL, Carlquist JF, Krause-Steinrauf HJ, Lazzeroni LC, Port JD, Lavori PW, Bristow MR. A polymorphism within a conserved {beta}1-adrenergic receptor motif alters cardiac function and {beta}-blocker response in human heart failure. PNAS. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNamara DM, Holubkov R, Janosko K, Palmer A, Wang JJ, MacGowan GA, Murali S, Rosenblum WD, London B, Feldman AM. Pharmacogenetic Interactions Between beta-Blocker Therapy and the Angiotensin-Converting Enzyme Deletion Polymorphism in Patients With Congestive Heart Failure. Circulation. 2001;103:1644–1648. doi: 10.1161/01.cir.103.12.1644. [DOI] [PubMed] [Google Scholar]
- 23.Darbar D, Motsinger AA, Ritchie MD, Gainer JV, Roden DM. Polymorphism modulates symptomatic response to antiarrhythmic drug therapy in patients with lone atrial fibrillation. Heart Rhythm. 2007;4:743–749. doi: 10.1016/j.hrthm.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, Kim RB, Roden DM, Stein CM. Genetic Determinants of Response to Warfarin during Initial Anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291:2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 27.Becker ML, Visser LE, Newton-Cheh C, Hofman A, Uitterlinden AG, Witteman JC, Stricker BH. A common NOS1AP genetic polymorphism is associated with increased cardiovascular mortality in users of dihydropyridine calcium channel blockers. Br J Clin Pharmacol. 2009;67:61–67. doi: 10.1111/j.1365-2125.2008.03325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 29.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, Veenstra DL, Rettie AE, Rieder MJ. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, Arking DE, Amsterdam AH, Sabeh KM, Mably JD, Rosenbaum DS, Peterson RT, Chakravarti A, Kaab S, Roden DM, Macrae CA. A Drug-Sensitized Zebrafish Screen Identifies Multiple Genes, Including GINS3, as Regulators of Myocardial Repolarization. Circulation. 2009;120:553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genetics. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 33.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 34.Arnett DK, Davis BR, Ford CE, Boerwinkle E, Leiendecker-Foster C, Miller MB, Black H, Eckfeldt JH. Pharmacogenetic association of the angiotensin-converting enzyme insertion/deletion polymorphism on blood pressure and cardiovascular risk in relation to antihypertensive treatment: the Genetics of Hypertension-Associated Treatment (GenHAT) study. Circulation. 2005;111:3374–3383. doi: 10.1161/CIRCULATIONAHA.104.504639. [DOI] [PubMed] [Google Scholar]
- 35.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahgoub A, Idle RJ, Dring LG, Lancaster R, Smith RL. Polymorphic hydroxylation of debrisoquine in man. Lancet. 1977;2:584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- 37.Eichelbaum M, Spannbrucker N, Dengler HJ. Proceedings: N-oxidation of sparteine in man and its interindividual differences. Naunyn Schmiedebergs Arch Pharmacol. 1975;287(Suppl):R94. [PubMed] [Google Scholar]
- 38.Eichelbaum M, Spannbrucker N, Steincke B, Dengler HJ. Defective N-oxidation of sparteine in man: a new pharmacogenetic defect. Eur J Clin Pharmacol. 1979;16:183–187. doi: 10.1007/BF00562059. [DOI] [PubMed] [Google Scholar]
- 39.Bertilsson L, Dengler HJ, Eichelbaum M, Schulz HU. Pharmacogenetic covariation of defective N-oxidation of sparteine and 4-hydroxylation of debrisoquine. Eur J Clin Pharmacol. 1980;17:153–155. doi: 10.1007/BF00562624. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez FJ, Skoda RC, Kimura S, Umeno M, Zanger UM, Nebert DW, Gelboin HV, Hardwick JP, Meyer UA. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature. 1988;331:442–446. doi: 10.1038/331442a0. [DOI] [PubMed] [Google Scholar]
- 41.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 42.Lamba V, Panetta JC, Strom S, Schuetz EG. Genetic predictors of inter-individual variability in hepatic CYP3A4 expression. J Pharmacol Exp Ther. 2010;332:1088–1099. doi: 10.1124/jpet.109.160804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genetics. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 44.Roden DM, Stein CM. Clopidogrel and the concept of high risk pharmacokinetics. Circulation. 2009;119:2127–2130. doi: 10.1161/CIRCULATIONAHA.109.865907. [DOI] [PubMed] [Google Scholar]
- 45.Yasar U, Tybring G, Hidestrand M, Oscarson M, Ingelman-Sundberg M, Dahl ML, Eliasson E. Role of cyp2c9 polymorphism in losartan oxidation. Drug Metab Dispos. 2001;29:1051–1056. [PubMed] [Google Scholar]
- 46.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 47.Zhou HH, Wood AJ. Stereoselective disposition of carvedilol is determined by CYP2D6. Clin Pharmacol Ther. 1995;57:518–524. doi: 10.1016/0009-9236(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 48.Siddoway LA, Thompson KAMKT, McAllister CB, Wang T, Wilkinson GR, Roden DM, Woosley RL. Polymorphism of propafenone metabolism and disposition in man: clinical and pharmacokinetic consequences. Circulation. 1987;75:785–791. doi: 10.1161/01.cir.75.4.785. [DOI] [PubMed] [Google Scholar]
- 49.Kivisto KT, Niemi M, Schaeffeler E, Pitkala K, Tilvis R, Fromm MF, Schwab M, Eichelbaum M, Strandberg T. Lipid-lowering response to statins is affected by CYP3A5 polymorphism. Pharmacogenetics. 2004;14:523–525. doi: 10.1097/01.fpc.0000114762.78957.a5. [DOI] [PubMed] [Google Scholar]
- 50.Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005;15:415–421. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 51.Grant DM, Blum M, Meyer UA. Polymorphisms of N-acetyltransferase genes. Xenobiotica. 1992;22:1073–1081. doi: 10.3109/00498259209051861. [DOI] [PubMed] [Google Scholar]
- 52.Niemi M. Transporter pharmacogenetics and statin toxicity. Clin Pharmacol Ther. 2010;87:130–133. doi: 10.1038/clpt.2009.197. [DOI] [PubMed] [Google Scholar]
- 53.Johnson JA, Zineh I, Puckett BJ, McGorray SP, Yarandi HN, Pauly DF. Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin Pharmacol Ther. 2003;74:44–52. doi: 10.1016/S0009-9236(03)00068-7. [DOI] [PubMed] [Google Scholar]
- 54.Dishy V, Sofowora GG, Xie HG, Kim RB, Byrne DW, Stein CM, Wood AJ. The effect of common polymorphisms of the beta2-adrenergic receptor on agonist-mediated vascular desensitization. N Engl J Med. 2001;345:1030–1035. doi: 10.1056/NEJMoa010819. [DOI] [PubMed] [Google Scholar]
- 55.Sofowora GG, Dishy V, Muszkat M, Xie HG, Kim RB, Harris PA, Prasad HC, Byrne DW, Nair UB, Wood AJ, Stein CM. A common beta1-adrenergic receptor polymorphism (Arg389Gly) affects blood pressure response to beta-blockade. Clin Pharmacol Ther. 2003;73:366–371. doi: 10.1016/s0009-9236(02)17734-4. [DOI] [PubMed] [Google Scholar]
- 56.Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. C825T polymorphism of the G protein beta(3)-subunit and antihypertensive response to a thiazide diuretic. Hypertension. 2001;37:739–743. doi: 10.1161/01.hyp.37.2.739. [DOI] [PubMed] [Google Scholar]
- 57.Turner ST, Bailey KR, Fridley BL, Chapman AB, Schwartz GL, Chai HS, Sicotte H, Kocher JP, Rodin AS, Boerwinkle E. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension. 2008;52:359–365. doi: 10.1161/HYPERTENSIONAHA.107.104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, Gums J, Curry RW, Gong Y, Beitelshees AL, Schwartz G, Turner ST. Pharmacogenomics of antihypertensive drugs: rationale and design of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study. Am Heart J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caraco Y, Sheller JR, Wood AJJ. Polymorphic drug metabolism and codeine pharmacodynamics. Clin Res. 1993;41:204A. [Google Scholar]
- 60.Tirkkonen T, Laine K. Drug interactions with the potential to prevent prodrug activation as a common source of irrational prescribing in hospital inpatients. Clin Pharmacol Ther. 2004;76:639–647. doi: 10.1016/j.clpt.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 61.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, Hayes DF, Desta Z, Flockhart DA. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 62.Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, Fritz P, Simon W, Suman VJ, Ames MM, Safgren SL, Kuffel MJ, Ulmer HU, Bolander J, Strick R, Beckmann MW, Koelbl H, Weinshilboum RM, Ingle JN, Eichelbaum M, Schwab M, Brauch H. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.International Warfarin Pharmacogenetics Consortium. Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, Wagner MJ, Caldwell MD, Johnson JA. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ablin J, Cabili S, Eldor A, Lagziel A, Peretz H. Warfarin therapy is feasible in CYP2C9*3 homozygous patients. Eur J Intern Med. 2004;15:22–27. doi: 10.1016/j.ejim.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 65.Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C, Sayer D, Castley A, Mamotte C, Maxwell D, James I, Christiansen FT. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 66.Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, Jagel-Guedes E, Rugina S, Kozyrev O, Cid JF, Hay P, Nolan D, Hughes S, Hughes A, Ryan S, Fitch N, Thorborn D, Benbow A the PRED. HLA-B*5701 Screening for Hypersensitivity to Abacavir. N Engl J Med. 2008;358:568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 67.Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, Wu JY, Chen YT. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 68.Lennard MS, Silas JH, Freestone S, Trevethick J. Defective metabolism of metoprolol in poor hydroxylators of debrisoquine. Br J Clin Pharmacol. 1982;14:301–303. doi: 10.1111/j.1365-2125.1982.tb01982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liggett SB, Cresci S, Kelly RJ, Syed FM, Matkovich SJ, Hahn HS, Diwan A, Martini JS, Sparks L, Parekh RR, Spertus JA, Koch WJ, Kardia SL, Dorn GW. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sehnert AJ, Daniels SE, Elashoff M, Wingrove JA, Burrow CR, Horne B, Muhlestein JB, Donahue M, Liggett SB, Anderson JL, Kraus WE. Lack of association between adrenergic receptor genotypes and survival in heart failure patients treated with carvedilol or metoprolol. J Am Coll Cardiol. 2008;52:644–651. doi: 10.1016/j.jacc.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 71.Bristow MR, Murphy GA, Krause-Steinrauf H, Anderson JL, Carlquist JF, Thaneemit-Chen S, Krishnan V, Abraham WT, Lowes BD, Port JD, Davis GW, Lazzeroni LC, Robertson AD, Lavori PW, Liggett SB. An alpha2C-adrenergic receptor polymorphism alters the norepinephrine-lowering effects and therapeutic response of the beta-blocker bucindolol in chronic heart failure. Circ Heart Fail. 2010;3:21–28. doi: 10.1161/CIRCHEARTFAILURE.109.885962. [DOI] [PubMed] [Google Scholar]
- 72.Wilke RA, Reif DM, Moore JH. Combinatorial pharmacogenetics. Nat Rev Drug Discov. 2005;4:911–918. doi: 10.1038/nrd1874. [DOI] [PubMed] [Google Scholar]
- 73.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, Ehret GB, Orru M, Pattaro C, Kottgen A, Perz S, Usala G, Barbalic M, Li M, Putz B, Scuteri A, Prineas RJ, Sinner MF, Gieger C, Najjar SS, Kao WHL, Muhleisen TW, Dei M, Happle C, Mohlenkamp S, Crisponi L, Erbel R, Jockel KH, Naitza S, Steinbeck G, Marroni F, Hicks AA, Lakatta E, Muller-Myhsok B, Pramstaller PP, Wichmann HE, Schlessinger D, Boerwinkle E, Meitinger T, Uda M, Coresh J, Kaab S, Abecasis GR, Chakravarti A. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PIW, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, Sotoodehnia N, Smith NL, Rotter JI, Kors JA, Witteman JCM, Hofman A, Heckbert SR, O'Donnell CJ, Uitterlinden AG, Psaty BM, Lumley T, Larson MG, Ch Stricker BH. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaab S, Sinner MF, George AL, Jr, Wilde AA, Bezzina CR, Schulze-Bahr E, Guicheney P, Bishopric NH, Myerburg RJ, Schott J, Pfeufer A, Hinterseer M, Beckmann BM, Steinbeck G, Perz S, Meitinger T, Wichmann H, Ingram C, Carter S, Norris K, Crawford DC, Ritchie M, Roden D. Abstract 4412: High Density tagSNP Candidate Gene Analysis Identifies IKs as a Major Modulator of Genetic Susceptibility to Drug Induced Long QT Syndrome. Circulation. 2008;118:S_884. [Google Scholar]
- 76.Kaab S, Ritchie MD, Crawford DC, Sinner M, Kannankeril PJ, Wilde AA, Bezzina C, Schulze-Bahr E, Guicheney P, Bishopric N, Schott JJ, Pfeufer A, Nakamura Y, Tanaka T, Ingram CR, Carter S, Bradford Y, George AL, Jr, Roden DM. Abstract 1971: Genome-wide Association Study Identifies Novel Genomic Regions Associated With Drug-induced Long Qt Syndrome. Circulation. 2009;120:S580. [Google Scholar]
- 77.Roden DM. Taking the idio out of idiosyncratic - predicting torsades de pointes. PACE. 1998;21:1029–1034. doi: 10.1111/j.1540-8159.1998.tb00148.x. [DOI] [PubMed] [Google Scholar]
- 78.Roden DM. Repolarization Reserve: A Moving Target. Circulation. 2008;118:981–982. doi: 10.1161/CIRCULATIONAHA.108.798918. [DOI] [PubMed] [Google Scholar]
- 79.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K, Pelz HJ, Lappegard K, Seifried E, Scharrer I, Tuddenham EG, Muller CR, Strom TM, Oldenburg J. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 80.The International Warfarin Pharmacogenetics Consortium. Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin Pharmacogenetics: CYP2C9 and VKORC1 Genotypes Predict Different Sensitivity and Resistance Frequencies in the Ashkenazi and Sephardi Jewish Populations. Am J Hum Genet. 2008;82:495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holmes DR, Jr, Dehmer GJ, Kaul S, Leifer D, O'Gara PT, Stein CM. ACCF/AHA Clopidogrel Clinical Alert: Approaches to the FDA "Boxed Warning". A Report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association. Circulation. 2010;122:537–557. doi: 10.1161/CIR.0b013e3181ee08ed. [DOI] [PubMed] [Google Scholar]
- 83.Roden DM, Shuldiner AR. Responding to the Clopidogrel Warning by the US Food and Drug Administration. Real Life Is Complicated. Circulation. 2010;122:445–448. doi: 10.1161/CIRCULATIONAHA.110.973362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang JJ, Cheng C, Yang W, Pei D, Cao X, Fan Y, Pounds SB, Neale G, Trevino LR, French D, Campana D, Downing JR, Evans WE, Pui CH, Devidas M, Bowman WP, Camitta BM, Willman CL, Davies SM, Borowitz MJ, Carroll WL, Hunger SP, Relling MV. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aberg K, Adkins DE, Bukszar J, Webb BT, Caroff SN, Miller dD, Sebat J, Stroup S, Fanous AH, Vladimirov VI, McClay JL, Lieberman JA, Sullivan PF, van den Oord EJ. Genomewide association study of movement-related adverse antipsychotic effects. Biol Psychiatry. 2010;67:279–282. doi: 10.1016/j.biopsych.2009.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 87.Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, Ginsburg GS. The SLCO1B1*5 Genetic Variant Is Associated With Statin-Induced Side Effects. J Am Coll Cardiol. 2009;54:1609–1616. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, Li X, Wilke RA, Rieder MJ, Williams PT, Ridker PM, Chatterjee A, Rotter JI, Nickerson DA, Stephens M, Krauss RM. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS ONE. 2010;5:e9763. doi: 10.1371/journal.pone.0009763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C, King CR, Brower A, Schmelzer JR, Glurich I, Vidaillet HJ, Yale SH, Qi ZK, Berg RL, Burmester JK. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teichert M, Eijgelsheim M, Rivadeneira F, Uitterlinden AG, van Schaik RH, Hofman A, De Smet PA, van GT, Visser LE, Stricker BH. A genome-wide association study of acenocoumarol maintenance dosage. Hum Mol Genet. 2009;18:3758–3768. doi: 10.1093/hmg/ddp309. [DOI] [PubMed] [Google Scholar]
- 91.Pautas E, Moreau C, Gouin-Thibault I, Golmard JL, Mahe I, Legendre C, Taillandier-Heriche E, Durand-Gasselin B, Houllier AM, Verrier P, Beaune P, Loriot MA, Siguret V. Genetic factors (VKORC1, CYP2C9, EPHX1, and CYP4F2) are predictor variables for warfarin response in very elderly, frail inpatients. Clin Pharmacol Ther. 2010;87:57–64. doi: 10.1038/clpt.2009.178. [DOI] [PubMed] [Google Scholar]
- 92.Cha PC, Mushiroda T, Takahashi A, Saito S, Shimomura H, Suzuki T, Kamatani N, Nakamura Y. High-resolution SNP and haplotype maps of the human gamma-glutamyl carboxylase gene (GGCX) and association study between polymorphisms in GGCX and the warfarin maintenance dose requirement of the Japanese population. J Hum Genet. 2007;52:856–864. doi: 10.1007/s10038-007-0183-9. [DOI] [PubMed] [Google Scholar]
- 93.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Milan DJ, Peterson TA, Ruskin JN, Peterson RT, Macrae CA. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation. 2003;107:1355–8. doi: 10.1161/01.cir.0000061912.88753.87. [DOI] [PubMed] [Google Scholar]
- 95.Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118:355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias WL, Braunwald E, Sabatine MS. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation. 2009;119:2553–2560. doi: 10.1161/CIRCULATIONAHA.109.851949. [DOI] [PubMed] [Google Scholar]
- 97.Hakonarson H, Thorvaldsson S, Helgadottir A, Gudbjartsson D, Zink F, Andresdottir M, Manolescu A, Arnar DO, Andersen K, Sigurdsson A, Thorgeirsson G, Jonsson A, Agnarsson U, Bjornsdottir H, Gottskalksson G, Einarsson A, Gudmundsdottir H, Adalsteinsdottir AE, Gudmundsson K, Kristjansson K, Hardarson T, Kristinsson A, Topol EJ, Gulcher J, Kong A, Gurney M, Thorgeirsson G, Stefansson K. Effects of a 5-lipoxygenase-activating protein inhibitor on biomarkers associated with risk of myocardial infarction: a randomized trial. JAMA. 2005;293:2245–2256. doi: 10.1001/jama.293.18.2245. [DOI] [PubMed] [Google Scholar]
- 98.Kantarjian H, Sawyers C, Hochhaus A, Guilhot F, Schiffer C, Gambacorti-Passerini C, Niederwieser D, Resta D, Capdeville R, Zoellner U, Talpaz M, Druker B the International STI. Hematologic and Cytogenetic Responses to Imatinib Mesylate in Chronic Myelogenous Leukemia. N Engl J Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 99.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras Mutations and Benefit from Cetuximab in Advanced Colorectal Cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 100.Lesko LJ, Woodcock J. Translation of pharmacogenomics and pharmacogenetics: a regulatory perspective. Nat Rev Drug Discov. 2004;3:763–769. doi: 10.1038/nrd1499. [DOI] [PubMed] [Google Scholar]
- 101.Woodcock J, Lesko LJ. Pharmacogenetics--tailoring treatment for the outliers. N Engl J Med. 2009;360:811–813. doi: 10.1056/NEJMe0810630. [DOI] [PubMed] [Google Scholar]
- 102.Collins F. Opportunities and challenges for the NIH--an interview with Francis Collins. Interview by Robert Steinbrook. N Engl J Med. 2009;361:1321–1323. doi: 10.1056/NEJMp0905046. [DOI] [PubMed] [Google Scholar]
- 103.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kohane IS, Masys DR, Altman RB. The incidentalome: a threat to genomic medicine. JAMA. 2006;296:212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]