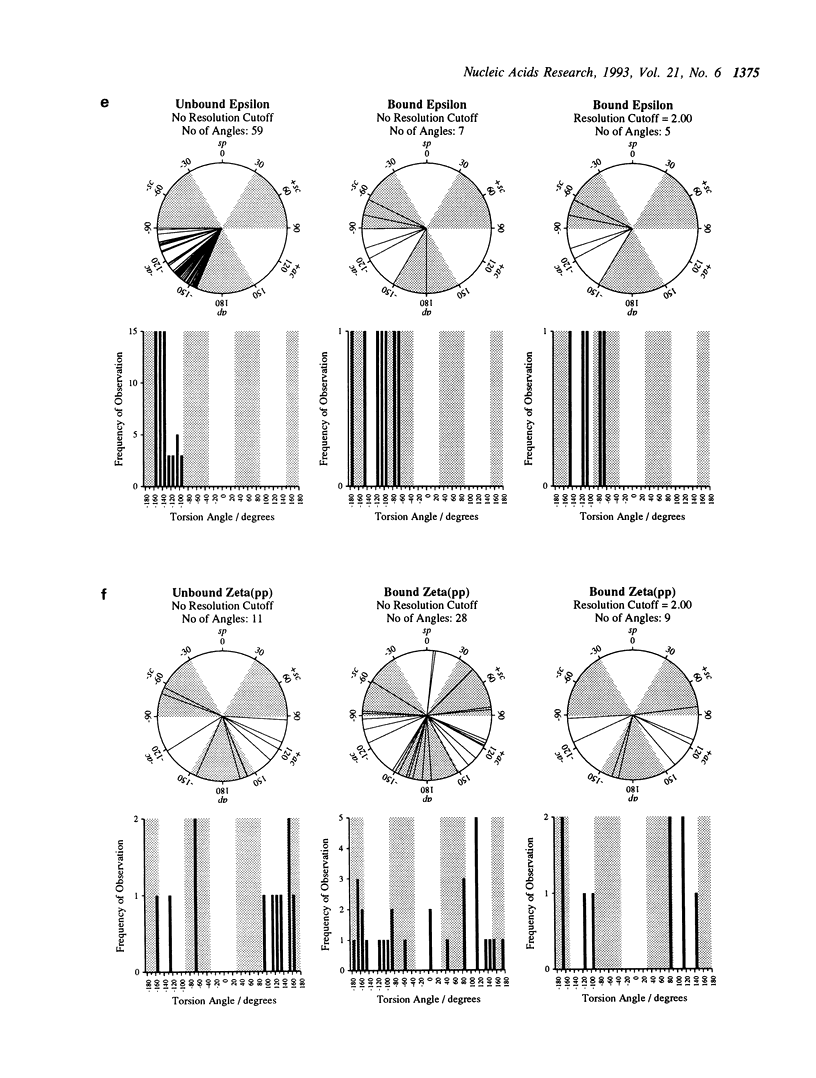
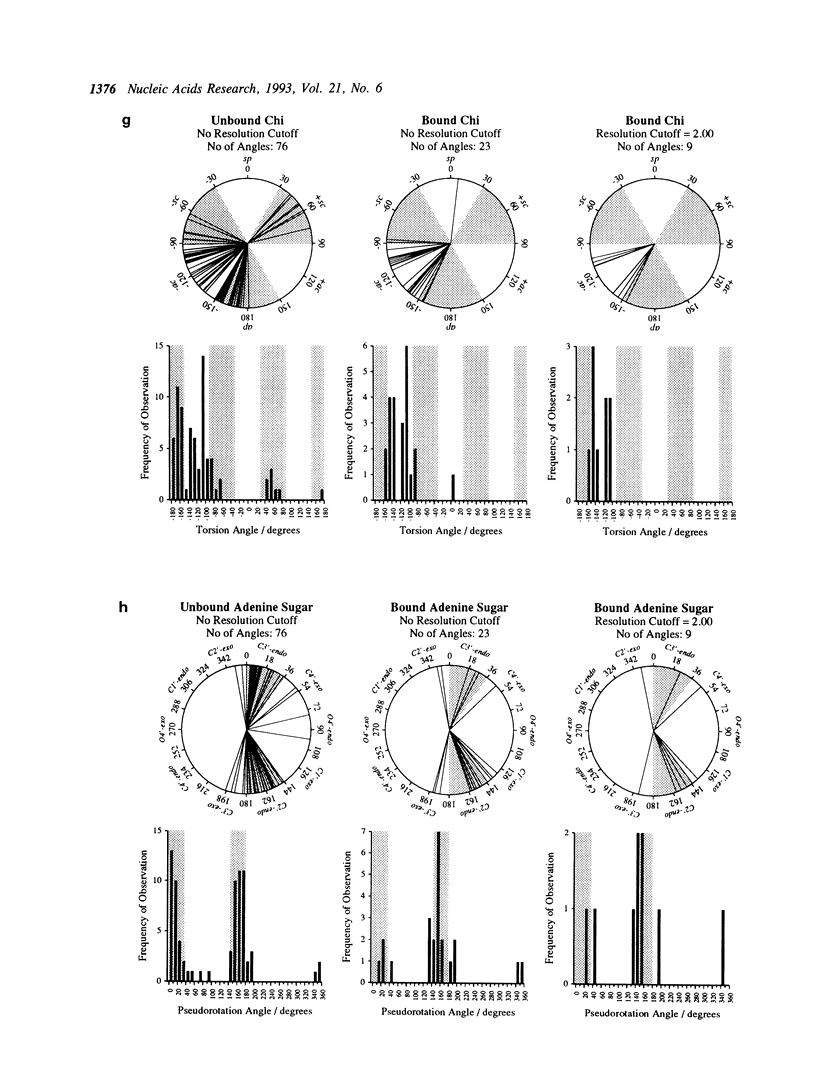
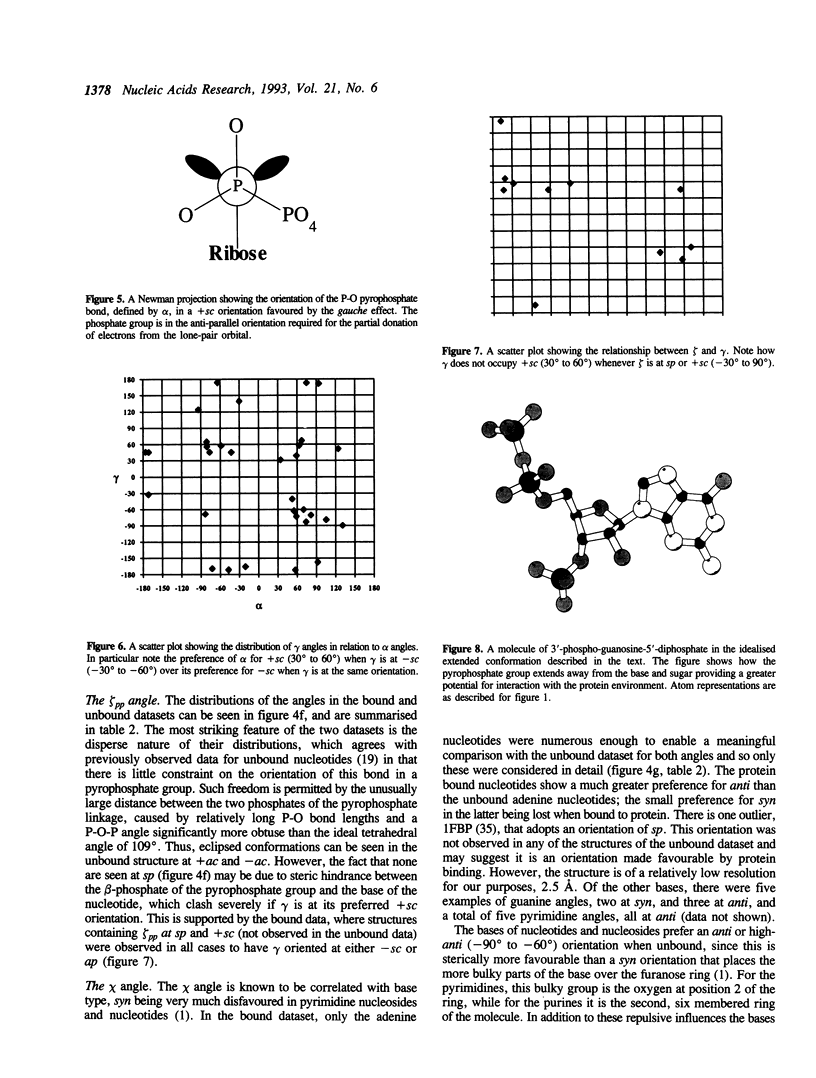
Abstract
In this study, we examine the effects of binding to protein upon nucleotide conformation, by the comparison of X-ray crystal structures of free and protein-bound nucleotides. A dataset of structurally non-homologous protein-nucleotide complexes was derived from the Brookhaven Protein Data Bank by a novel protocol of dual sequential and structural alignments, and a dataset of native nucleotide structures was obtained from the Cambridge Structural Database. The nucleotide torsion angles and sugar puckers, which describe nucleotide conformation, were analysed in both datasets and compared. Differences between them are described and discussed. Overall, the nucleotides were found to bind in low energy conformations, not significantly different from their 'free' conformations except that they adopted an extended conformation in preference to the 'closed' structure predominantly observed by free nucleotide. The archetypal conformation of a protein-bound nucleotide is derived from these observations.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abad-Zapatero C., Griffith J. P., Sussman J. L., Rossmann M. G. Refined crystal structure of dogfish M4 apo-lactate dehydrogenase. J Mol Biol. 1987 Dec 5;198(3):445–467. doi: 10.1016/0022-2836(87)90293-2. [DOI] [PubMed] [Google Scholar]
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Baudys M., Foundling S., Pavlík M., Blundell T., Kostka V. Protein chemical characterization of Mucor pusillus aspartic proteinase. Amino acid sequence homology with the other aspartic proteinases, disulfide bond arrangement and site of carbohydrate attachment. FEBS Lett. 1988 Aug 1;235(1-2):271–274. doi: 10.1016/0014-5793(88)81277-8. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Rhodes G., Banaszak L. J. Refined crystal structure of cytoplasmic malate dehydrogenase at 2.5-A resolution. Biochemistry. 1989 Jul 11;28(14):6065–6081. doi: 10.1021/bi00440a051. [DOI] [PubMed] [Google Scholar]
- Blundell T. L., Jenkins J. A., Sewell B. T., Pearl L. H., Cooper J. B., Tickle I. J., Veerapandian B., Wood S. P. X-ray analyses of aspartic proteinases. The three-dimensional structure at 2.1 A resolution of endothiapepsin. J Mol Biol. 1990 Feb 20;211(4):919–941. doi: 10.1016/0022-2836(90)90084-Y. [DOI] [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Cotton F. A., Hazen E. E., Jr, Legg M. J. Staphylococcal nuclease: proposed mechanism of action based on structure of enzyme-thymidine 3',5'-bisphosphate-calcium ion complex at 1.5-A resolution. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2551–2555. doi: 10.1073/pnas.76.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foundling S. I., Cooper J., Watson F. E., Cleasby A., Pearl L. H., Sibanda B. L., Hemmings A., Wood S. P., Blundell T. L., Valler M. J. High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes. 1987 May 28-Jun 3Nature. 327(6120):349–352. doi: 10.1038/327349a0. [DOI] [PubMed] [Google Scholar]
- Grau U. M., Trommer W. E., Rossmann M. G. Structure of the active ternary complex of pig heart lactate dehydrogenase with S-lac-NAD at 2.7 A resolution. J Mol Biol. 1981 Sep 15;151(2):289–307. doi: 10.1016/0022-2836(81)90516-7. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- James M. N., Sielecki A. R. Molecular structure of an aspartic proteinase zymogen, porcine pepsinogen, at 1.8 A resolution. Nature. 1986 Jan 2;319(6048):33–38. doi: 10.1038/319033a0. [DOI] [PubMed] [Google Scholar]
- James M. N., Sielecki A. R. Structure and refinement of penicillopepsin at 1.8 A resolution. J Mol Biol. 1983 Jan 15;163(2):299–361. doi: 10.1016/0022-2836(83)90008-6. [DOI] [PubMed] [Google Scholar]
- Jaskólski M., Miller M., Rao J. K., Leis J., Wlodawer A. Structure of the aspartic protease from Rous sarcoma retrovirus refined at 2-A resolution. Biochemistry. 1990 Jun 26;29(25):5889–5898. doi: 10.1021/bi00477a002. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Daniels M. J., Herriott J. R. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science. 1991 Jan 4;251(4989):60–66. [PubMed] [Google Scholar]
- Karpusas M., Holland D., Remington S. J. 1.9-A structures of ternary complexes of citrate synthase with D- and L-malate: mechanistic implications. Biochemistry. 1991 Jun 18;30(24):6024–6031. doi: 10.1021/bi00238a028. [DOI] [PubMed] [Google Scholar]
- Ke H. M., Zhang Y. P., Lipscomb W. N. Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 6-phosphate, AMP, and magnesium. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5243–5247. doi: 10.1073/pnas.87.14.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepke J., Maslowska M., Heinemann U., Saenger W. Three-dimensional structure of ribonuclease T1 complexed with guanylyl-2',5'-guanosine at 1.8 A resolution. J Mol Biol. 1989 Apr 5;206(3):475–488. doi: 10.1016/0022-2836(89)90495-6. [DOI] [PubMed] [Google Scholar]
- Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. Stereochemical quality of protein structure coordinates. Proteins. 1992 Apr;12(4):345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- Nachman J., Miller M., Gilliland G. L., Carty R., Pincus M., Wlodawer A. Crystal structure of two covalent nucleoside derivatives of ribonuclease A. Biochemistry. 1990 Jan 30;29(4):928–937. doi: 10.1021/bi00456a012. [DOI] [PubMed] [Google Scholar]
- Pearl L., Blundell T. The active site of aspartic proteinases. FEBS Lett. 1984 Aug 20;174(1):96–101. doi: 10.1016/0014-5793(84)81085-6. [DOI] [PubMed] [Google Scholar]
- Pullman B., Perahia D., Saran A. Molecular orbital calculations on the conformation of nucleic acids and their constituents. 3. Backbone structure of di- and polynucleotides. Biochim Biophys Acta. 1972 Apr 26;269(1):1–14. doi: 10.1016/0005-2787(72)90068-8. [DOI] [PubMed] [Google Scholar]
- Pullman B., Saran A. Quantum-mechanical studies on the conformation of nucleic acids and their constituents. Prog Nucleic Acid Res Mol Biol. 1976;18:215–322. doi: 10.1016/s0079-6603(08)60589-9. [DOI] [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Saenger W., Reddy B. S., Mühlegger K., Weimann G. X-ray study of the lithium complex of NAD. Nature. 1977 May 19;267(5608):225–229. doi: 10.1038/267225a0. [DOI] [PubMed] [Google Scholar]
- Sali A., Veerapandian B., Cooper J. B., Moss D. S., Hofmann T., Blundell T. L. Domain flexibility in aspartic proteinases. Proteins. 1992 Feb;12(2):158–170. doi: 10.1002/prot.340120209. [DOI] [PubMed] [Google Scholar]
- Saran A., Govil G. Quantum chemical studies on the conformational structure of nucleic acids. 3. Calculation of backbone structure by extended Hückel theory. J Theor Biol. 1971 Nov;33(2):407–418. doi: 10.1016/0022-5193(71)90074-9. [DOI] [PubMed] [Google Scholar]
- Sevcik J., Dodson E. J., Dodson G. G. Determination and restrained least-squares refinement of the structures of ribonuclease Sa and its complex with 3'-guanylic acid at 1.8 A resolution. Acta Crystallogr B. 1991 Apr 1;47(Pt 2):240–253. [PubMed] [Google Scholar]
- Stevens R. C., Gouaux J. E., Lipscomb W. N. Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution. Biochemistry. 1990 Aug 21;29(33):7691–7701. doi: 10.1021/bi00485a019. [DOI] [PubMed] [Google Scholar]
- Suguna K., Bott R. R., Padlan E. A., Subramanian E., Sheriff S., Cohen G. H., Davies D. R. Structure and refinement at 1.8 A resolution of the aspartic proteinase from Rhizopus chinensis. J Mol Biol. 1987 Aug 20;196(4):877–900. doi: 10.1016/0022-2836(87)90411-6. [DOI] [PubMed] [Google Scholar]
- Weber D. J., Mullen G. P., Mildvan A. S. Conformation of an enzyme-bound substrate of staphylococcal nuclease as determined by NMR. Biochemistry. 1991 Jul 30;30(30):7425–7437. doi: 10.1021/bi00244a009. [DOI] [PubMed] [Google Scholar]


