Abstract
The targeted removal of damaged proteins by proteolysis is crucial for cell survival. We have shown previously that the Lon protease selectively degrades oxidized mitochondrial proteins, thus preventing their aggregation and cross-linking. We now show that the Lon protease is a stress-responsive protein that is induced by multiple stressors, including heat shock, serum starvation, and oxidative stress. Lon induction, by pre-treatment with low-level stress, protects against oxidative protein damage, diminished mitochondrial function, and loss of cell proliferation, induced by toxic levels of hydrogen peroxide. Blocking Lon induction, with Lon siRNA, also blocks this induced protection. We propose that Lon is a generalized stress-protective enzyme whose decline may contribute to the increased levels of protein damage and mitochondrial dysfunction observed in ageing and age-related diseases.
Keywords: Heat Shock, Mitochondria, Oxidative Stress, Proteolysis, Serum Starvation
INTRODUCTION
Protein oxidation may be one of the most important forms of oxidative damage. Some 30%-40% of proteins exhibit oxidative modification as a part of normal ageing[1, 2]. The buildup of oxidized proteins in cells can lead to failure of protein maintenance, loss of protein/enzyme function and a variety of deleterious alterations such as protein fragmentation and aggregation, that cause cellular toxicity[3-6]. Proteolytic pathways such as the 20S proteasome and lysosomes are critical in minimizing the levels of oxidized proteins and appear sufficient in young, healthy cells, to cope with increases in protein oxidation, thereby suppressing oxidative stress[7-9]. Conversely, inhibition of these pathways is sufficient to cause the accumulation of oxidized proteins, thus exacerbating oxidative stress[4, 8, 10]. The 20S Proteasome is the major proteolytic enzyme for the removal of oxidatively damaged proteins in the cytosol, nucleus and the endoplasmic reticulum[11-13], backed up by lysosomal autophagy[8].
Mitochondria are a major source of reactive oxygen species, but do not contain proteasome. Of the approximately 20 mitochondrial proteases and peptidases, the Lon protease appears to be mostly responsible for selectively degrading oxidized mitochondrial proteins[14]. Previously we reported that the Lon protease preferentially degrades oxidized mitochondrial aconitase, an enzyme that is highly susceptible to oxidative modification and is a marker of oxidative stress and ageing[14]. We also described the effects of chronic Lon down-regulation in human fibroblasts, where mitochondrial respiration is impaired, mitochondrial mass decreases, and massive cell death ensues[15]. Clearly, sufficient Lon is required to cope with oxidized proteins under normal conditions and prevent their accumulation, aggregation, and cross-linking. Whether mitochondria require ‘extra’ Lon to conduct the rapid elimination of aberrant, oxidized proteins, during oxidative stress is not known.
Metabolic stresses such as serum starvation, heat, and H2O2 can induce the production of damaged and oxidatively modified proteins[16, 17]. In addition, heat shock genes have a role in cellular resistance against oxidative stress and are increasingly expressed during oxidant exposure[18, 19]. In a study with glucose starved Escherichia coli cells, where ectopically high levels of heat shock transcription factor σ32 were induced, a reduction in protein carbonylation was observed; carbonylation was enhanced, however, when Lon was deleted from these cells[20]. We have previously shown that Lon protease conducts the degradation of oxidized proteins in mitochondria[14],[15],[21]. We now wanted to know if the transcription and/or translation of Lon would respond to stress conditions.
In the present report, we investigated the effects of oxidative stress, heat shock, and serum starvation on the Lon protease. During stress, which increases protein damage, we wondered if expression of the Lon protease might be induced, thus providing transient protection (an hormetic response) against the accumulation of damaged proteins. In other words, we hypothesized that Lon may be a stress protein.
METHODS
Cell Culture
Rhabdomyosarcoma (RD) cells were purchased from ATCC (Manassas, VA). RD Cells were maintained in a 37°C / 5% CO2 / 19.9% O2 incubator in DMEM supplemented with 2% horse serum unless otherwise stated.
Heat Stress, Serum Starvation, and Peroxide Pre-treatments to Induce Lon
RD cells were seeded at a density of 3 × 106 cells per 75cm2 flask, 24 hours prior to treatment. For heat stress, RD cells were incubated in a 45°C / 5%CO2 / 19.9% O2 incubator for one hour and then immediately returned to 37°C / 5%CO2 / 19.9% O2 for recovery. Serum Starvation involved incubating cells for one hour in serum free DMEM, washing twice with warm PBS, and then adding 2% horse serum supplemented DMEM for cell recovery. Hydrogen peroxide (H2O2) treatments involved incubating cells with various low concentrations (0-800μM) of H2O2 in DMEM for 1 hour. The cells were washed twice with warm PBS, and DMEM supplemented with 2% horse serum was then added back to the cells for recovery. For preconditioning experiments, RD cells were incubated in All-Star negative control (non-silencing) siRNA or Lon siRNA for 3 days, and then pre-treated for 1 hour in DMEM with 0-800μM H2O2. The cells were allowed to recover for 3 hours in DMEM supplemented with 2% DMEM and then challenged with a toxic dose of 2mM H2O2 in DMEM for 1 hour.
siRNA Incubations
RD cells were seeded at a concentration of 937,500 cells per 75cm2 flask. One day after seeding, cells were treated with 0.04% oligofectamine and either 0.04% All-Star negative control siRNA (Qiagen) or 0.04% Lon siRNA (target sequence: AAGCACGTCATGGATGTTGTG) in DMEM for 4 hours. All siRNAs were purchased from Qiagen (Valencia, CA). Cells were then incubated in DMEM supplemented with 2% horse serum without siRNA for 2 days, before seeding in preparation for H2O2 treatments. Cells were then trypsinized and re-seeded at a density of 3 × 106 cells per 75cm2 flask for 24 hours prior to H2O2 treatment.
Western Analysis
Cells were harvested from 75cm2 flasks by trypsinization. Cells were washed twice with PBS to remove trypsin, and then lysed in RIPA buffer supplemented with protease inhibitors. Protein was quantified with the BCA Protein Assay Kit from Pierce (Rockford, IL) according to manufacturers instructions. For western analysis, 40μg of protein was run on SDS-PAGE and transferred to a PVDF membrane for antibody analysis. Custom Lon antibody was synthesized and utilized as described previously[14]. Porin antibody was purchased from Abcam (Cambridge, MA , #ab15895) and mitochondrial heat shock protein 70 (mtHSP-70) antibody was purchased from Affinity BioReagents (Golden, CO, #MA3028). Blots were blocked and probed in Startingblock™ buffer from Pierce (Rockford, IL). An enhanced chemiluminescence detection kit, purchased from Pierce (Rockford, IL), was used for chemiluminescence and membranes were developed onto Kodak biomax films (VWR, West Chester, PA) using the Kodak GBX developing system (VWR, West Chester, PA).
RNA Isolation and Quantitative Real Time PCR
TRIzol® (Invitrogen, Carlsbad, CA) was added directly into 75cm2 flasks immediately after treatment and then isolated following manufacturers instructions. RNA pellets were resuspended in 100μl of water and then treated with DNA-free™ kit (Ambion, Austin, TX) following the manufacturer’s instructions. RNA was quantified with the nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE). Amplification for Lon was carried out with the primer sequences: 5′-ATGGAGGACGTCAAGAAACG-3′ and 5′-GATCTCAGCCACGTCAGTCA-3′ and for actin: 5′-TTGTTACAGGAAGTCCCTTGCC-3′ and ATGCTATCACCTCCCCTGTGTG-3′. qRT-PCR reactions were performed in triplicate in a MJ Research Detection System (MJ Research, Waltham, MA) using SuperScript™ III Platinum® SYBR® Green One-Step qRT-PCR Kit (Invitrogen, Carlsbad, CA) as recommended by the manufacturer. The sensitivity of reactions and eventual amplification of contaminant products, such as primer–dimers, indiscriminately detected by the SYBR green chemistry, was evaluated by amplifying serial dilutions of cDNA and by performing a blank without cDNA. The amplification of a single-size product was verified by gel electrophoresis (data not shown). Induction Ratio was calculated, using the relative expression method where results with a confidence value greater than 17% were rejected. Actin mRNA was used as the internal control.
Detection of Carbonylated Proteins
Protein Oxidation Detection Kit, OxyBlot™ was obtained from Millipore (Billerica, MA) and was used to perform immunoblot detection of oxidatively modified proteins by the generation of carbonyl groups. 15 μg of protein was used for each reaction. Carbonyl groups in samples were derivatized to 2,4-dinitrophenylhydrazone (DNP-hydrazone) by reaction with 2,4-dinitrophenylhydrazine (DNPH). Samples were stored at 4°C until ready to load onto 12.5% polyacrylamide gels for western blot analysis as described above. Detection of carbonylated proteins was performed as suggested by the manufacturer, using an anti-DNP antibody.
Quantification of Surviving Cells
Cells were collected and cell numbers directly quantified using a Z1 series Beckman Coulter Particle Counter. 100,000 cells were seeded on 6-well plates, in triplicate, and cultured in in DMEM supplemented with 10% horse serum in a 37°C / 5%CO2 / 19.9% O2 incubator for 3 days, after which cells were counted again.
Another assay for mitochondrial function and cell viability is through the reduction of tetrazolium salt, known as the MTT [3- (4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assay. Reduction of the tetrazolium compound MTT is dependent upon the presence of nicotinamide adenine dinucleotide and nicotinamide adenine dinucleotide phosphate, as well as intact mitochondrial electron transport and, therefore, reflects mitochondrial function and metabolic activity [22]. MTT analysis was performed with the celltiter 96 nonradioactive cell proliferation assay® by Promega (Madison, WI). After hydrogen peroxide treatments, 40,000 cells were seeded in quadruplicate on a 96-well plate and incubated for 3 days in DMEM supplemented with 10% horse serum in a 37°C / 5%CO2 / 19.9% O2 incubator. Cellular reduction of the tetrazolium salt [3- (4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] into a formazan product was then quantified according to the manufacturer’s instructions.
RESULTS
Lon is a stress responsive protein
Metabolic stress can result in a rapid increase in damaged mitochondrial proteins that need to be rapidly removed, before they accumulate as insoluble aggregates, and disrupt mitochondrial function and cellular homeostasis. We hypothesized that Lon might be a general human stress responsive protein to combat homeostatic disruptions in mitochondria by degrading damaged proteins that accumulate under adverse conditions. To test this possibility, we designed studies in which human rhabdomyosarcoma cells (RD)* were exposed to three different types of stresses: oxidative stress, heat stress, and serum starvation. Primary cultures of muscle cells are difficult to attain and would be difficult to propagate to the large numbers needed for these exploratory experiments. Thus, although RD cells are substantially transformed, they are derived from human muscles, they are a robust culture line, and they divide at a rate that made it possible to perform the large-scale dose- and time-response studies presented in this paper. RD cells were exposed to each stress for 1 hour, then left to recover under normal conditions for a further 3, 6, or 24 hours and, finally, collected for analysis.
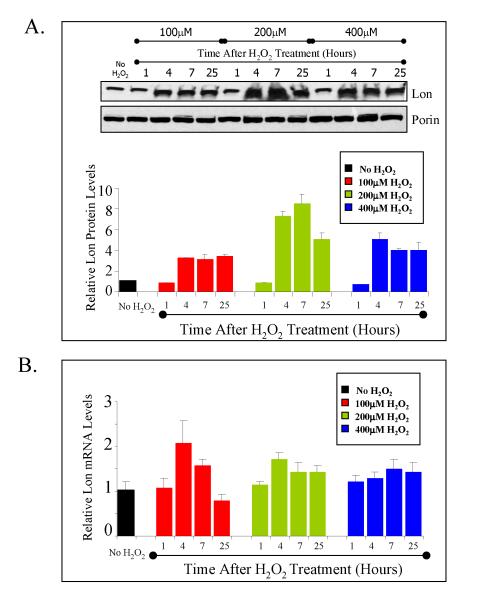
We first tested whether oxidative stress caused by hydrogen peroxide could induce Lon. We incubated RD cells in complete medium with either 0, 100, 200, or 400 μM of H2O2 for 1 hour and let the cells recover under normal conditions with fresh medium for a further 3, 6, or 24 hours. Induction of the Lon protease was observed at 4, 7, and 25 hours after treatment, with the highest protein induction, approximately 8 fold, observed 4 to 7 hours after treatment with 200μM peroxide (figure 1A). There was a modest induction of mRNA after H2O2 treatment (figure 1B), analyzed through quantitative real time PCR (qRT-PCR*), with the highest inductions of 2.3 fold (at 4 hrs) and 1.7-fold (at 7 hrs) observed when the cells were treated with 100 μM H2O2, and 1.9 fold (at 4hrs) and 1.6-fold (at 7 hrs) after 200μM H2O2 treatment.
Figure 1. Lon is induced with hydrogen peroxide treatment.
A, Lon protein analysis of RD cells treated with 0, 100, 200, or 400μM H2O2 for 1 hour and then left to recover for up to 25 hours after H2O2 treatment. ‘No H2O2’ represents samples that were incubated with vehicle instead of hydrogen peroxide. Blots were probed with anti-Lon antibody or anti-porin antibody as a mitochondrial loading control. The bar graph represents data normalized against porin. B, mRNA analysis by qRT-PCR of the same samples examined in A. Samples were amplified with Lon and GAPDH primers and calculation of Lon mRNA levels were normalized against GAPDH as an internal loading control. In both panels A and B, all data points shown are the means ± standard errors of three independent determinations.
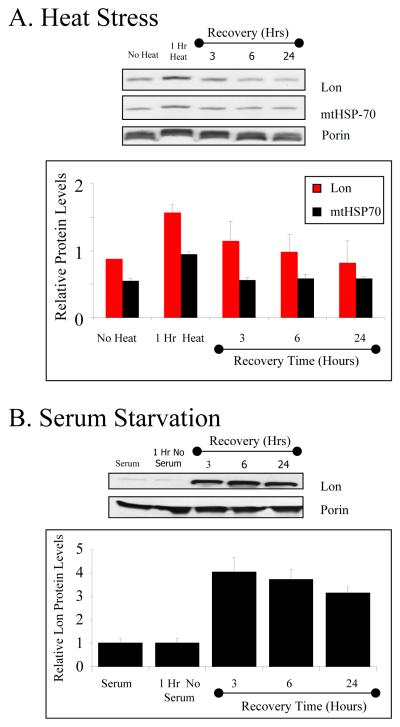
Our optimization studies for heat stress indicated that Lon induction was significant between 42 to 45°C degrees (a common range for heat experiments). Within this range, treatment at 45°C generated a significantly larger increase in Lon levels than did 42°C treatment (data not shown). To test for response to heat, RD cells were incubated at 45°C for 1 hour and then allowed to recover in a 37°C incubator for an additional 3, 6, or 24 hours. In figure 2A, cells exposed to a 45°C heat shock exhibited an 80% induction of Lon protein after 1 hour of shock, and 31% induction after 3 hours recovery, gradually returning towards basal levels at 6 and 24 hours (figure 2A). In our studies, Lon up-regulation was accompanied by a co-induction of mitochondrial heat shock protein 70 (mtHSP-70*), which exhibited a 70% increase after 1 hour of heat shock and declined to basal levels thereafter (figure 2A).
Figure 2. Lon is induced with heat stress and serum starvation.
A, Lon and mtHSP-70 western analysis of RD cells treated with a 45°C heat shock for one hour and then allowed to recover in 37°C for 3-24 hours. B, Western analysis of RD cells incubated in serum-free medium and then allowed to recover in complete medium with serum for 3-24 hours. Samples were probed with anti-Lon antibody or anti-porin antibody as a mitochondrial loading control. The bar graphs in both A and B represents Lon levels normalized against porin and all data points shown are the means ± standard errors of three independent determinations.
To study serum starvation stress, RD cells were incubated in medium without serum for 1 hour and then recovered with medium supplemented with 2% horse serum for 3, 6, or 24 hours. As seen in figure 2B, after the 1-hour serum starvation, there was no increase in Lon levels. After recovery of 3 hours in serum-supplemented medium, Lon protein levels exhibited a 4-fold increase (figure 2B). Lon protein levels after 6 and 24 hours of serum starvation stress recovery remained relatively high with protein inductions of about 3.8 and 3.1 fold, respectively (figure 2B).
Lon Induction protects cells against an increase in protein oxidative damage
Since hydrogen peroxide increases mitochondrial protein oxidative damage in cells[23], and Lon degrades oxidized proteins[14], we hypothesized that Lon induction should contribute to cellular protection against oxidized proteins. Cells treated with low levels of hydrogen peroxide showed strong induction of Lon protein levels 4 hours after treatment (see figure 1A). We took advantage of this knowledge and hypothesized that cells in which Lon was induced by pretreatment with low levels of H2O2, should exhibit greater ability to cope with oxidized proteins generated during a subsequent challenge (higher) dose of H2O2. In order to test for such inducible protective effects, we measured protein carbonyl content in these cells. Recognizing that H2O2 can induce a number of stress responsive genes however [24-28], we wanted to carefully test whether Lon induction specifically contributes to protection against an increase in the carbonyl content. We approached this by designing an experiment to test whether Lon can contribute to increased (adaptive) protection, sometimes called hormesis, by incubating cells with either control non-silencing siRNA (figure 3A) or Lon silencing siRNA (figure 3B), beginning three days prior to all H2O2 treatments.
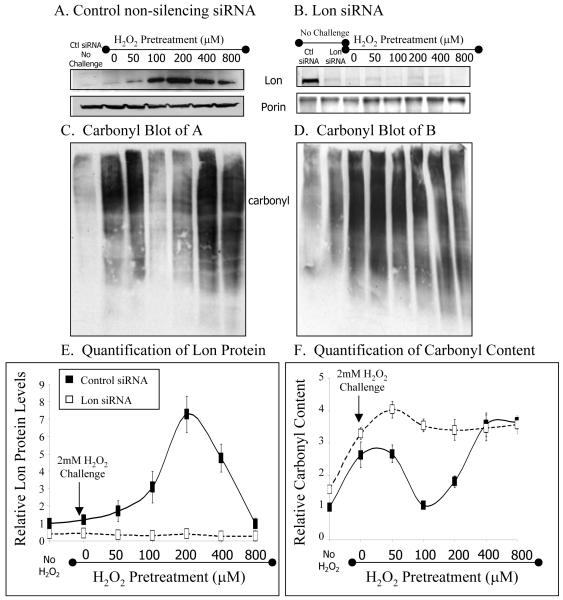
Figure 3. Preconditioned cells are better protected against oxidative damage.
Western Analysis of RD cells. Cells were incubated with A, control non-silencing siRNA or B, Lon siRNA. Three days later these samples were pretreated with between 0-800μM H2O2 for 1 hour and then allowed to recover for a further 3 hours in medium without H2O2. ‘No challenge’ samples represent cells incubated with siRNA but not with H2O2. The blot in B was overexposed to show silenced Lon bands. The same cells were subsequently challenged 4 hours after pretreatment with 2mM hydrogen peroxide for 1 hour, after which carbonyl content was analyzed in C, control non-silencing siRNA, or D, Lon siRNA. Oxidatively modified proteins were detected upon treatment of cell extracts with 2,4-dinitrophenylhydrazine to derivitize protein carbonyl groups, followed by Western blot analysis using a polyclonal antibody to the 2,4 dinitrophenylhydrazine moeity. E,F, Quantification of immununoblots from A-D, all data points shown are the means ± standard errors of three independent determinations.
We pretreated our cells with 0, 50, 100, 200, 400, or 800μM hydrogen peroxide; from pilot experiments (figure1), less than 100μM H2O2 was expected to induce only small increases in Lon expression, 100, 200 and 400μM H2O2 were expected to induce strong Lon expression, and H2O2 pretreatments of above 400μM were again expected to induce smaller Lon increases. After 1 hour of H2O2 pre-treatment, cells were allowed to recover in fresh medium, without peroxide, for an additional 3 hours to allow Lon protein induction. The data presented in figure 3 panels A-B are an extension of the experiments from figure 1, with data collected after 4 hours of peroxide treatment (which was the most effective time point according to fig. 1). In fig. 3, we extended the concentrations to 50 and 800uM to determine a more dosage effect, since 100uM in fig. 1 already showed a rather strong inducing effect on Lon, and since 400um (in fig. 1) appeared to suggest the beginnings of a loss of induction or cellular toxicity. Please note that Lon induction levels in fig. 3 are entirely comparable (at the same peroxide doses) to those shown in figure 1 after four hours treatment. It is also worth reiterating that the cells from figure 3 panels A-B, were not challenged with 2mM peroxide, however, the carbonyl data from panels C-F were obtained from both non-challenged cells, and cells challenged with 2mM hydrogen peroxide.
The pre-treated cells were then challenged with a high, toxic, dose of 2mM H2O2 for one hour. The cells were then harvested and western blots were analyzed. As expected (see figure 1), Western analysis of cells incubated with control non-silencing siRNA revealed that the 100, 200, and 400μM H2O2 pretreatments induced the highest Lon expression, whereas higher and lower H2O2 pretreatments were not so effective (figure 3A, E). In contrast, incubation with Lon siRNA prevented increases in Lon expression at all H2O2 pretreatment concentrations (figure 3B,E).
Exposure of control, non-silencing siRNA incubated cells to the 2mM challenge dose of H2O2 (without H2O2 pretreatment to induce Lon) caused oxidative protein damage, as evidenced by significantly increased levels of protein carbonyls (figure 3C, left 2 lanes, and panel F). Pretreatment of cells with 50μM H2O2, which was not effective in inducing Lon, did not show protection against carbonyls generated by 2mM H2O2. Similarly, pretreatment with 800 μM H2O2, which also caused no increase in Lon, again failed to block carbonyl production caused by the 2mM H2O2 challenge (fig. 3C,E). In contrast, pretreatment with 100 or 200μM H2O2 (both of which strongly increased Lon expression) blocked carbonyl production (protein oxidation) by as much as 95% (figure 3C,E,F). This suggests that pre-treatment of cells with low levels of H2O2, which induce Lon, protects cells against carbonyl production when challenged with a subsequent high dose of H2O2. This protection may only be effective up to a certain pretreatment concentration threshold, however, at which point the pretreatment itself becomes toxic enough to cause significant carbonyl production. We propose that this explains the results of 400μM H2O2 pretreatment, which induced Lon but failed to protect against carbonyl production (Fig. 3C,E,F).
Incubation of cells with Lon siRNA firmly confirmed the protective effect of Lon against protein oxidation. As shown in figure 3B and E, Lon siRNA effectively blocked Lon induction at all H2O2 pretreatment concentrations, and even knocked-down Lon to about one-third of basal levels. In keeping with our hypothesis that Lon is a stress-protective enzyme, carbonyl production generated by the challenge dose of 2mM H2O2 in these Lon siRNA incubated cells was greater (about four-fold) than under any other conditions (figure 3F).
Lon Induction Preserves Cell Viability During Stress
Since Lon induction appeared to protect cells from the accumulation of damaged protein, we wanted to test whether Lon induction would also improve cell growth and mitochondrial function after a strong oxidant challenge. For these studies, cells were taken from the experiments of figure 3, after incubating with control siRNA or Lon siRNA, pre-treating with 0-800μM H2O2, and challenging with 2mM H2O2. Cell growth was assessed by plating 100,000 cells in six-well plates, incubating them for three days, and then counting cell numbers in a particle counter (figure 4A). Mitochondrial functionality was also estimated by reduction of the tetrazolium compound, MTT (figure 4B).
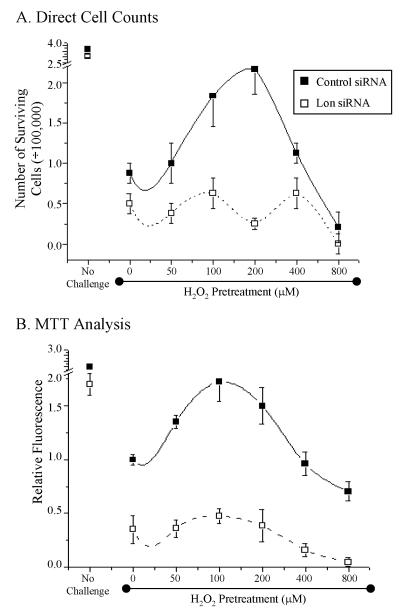
Figure 4. Preconditioned cells exhibit increased cell viability.
Cells from the experiments of figure 3 were collected and incubated for an additional 3 days to determine survival number and viability. In short, cells were incubated with control or Lon siRNA followed by H2O2 pretreatment (concentrations indicated) and challenged with 2mM hydrogen peroxide for 1 hour. ‘No Challenge’ samples are cells that were treated with siRNA but not treated with hydrogen peroxide. A, Number of cells that survived after hydrogen peroxide treatments determined by direct cell counting. B, Relative fluorescence of cells which represent cell viability and mitochondrial activity with the MTT assay. In both panels A and B, all data points shown are the means ± standard errors of four independent determinations.
Challenging cells with 2mM H2O2 caused a drastic decline in the number of viable cells (figure 4A) and in MTT reduction capacity (figure 4B) in both control siRNA and Lon siRNA incubated cells. In control siRNA incubated cells, however, Lon induction by pretreatment with 100μM or 200μM H2O2 (the pretreatments that produced greatest Lon induction and allowed least protein carbonyl accumulation – figure 3) limited the loss of cell number (figure 4A) and MTT reduction (figure 4B) to one-third or less of the decline seen without H2O2 pretreatment. In contrast, incubation with Lon siRNA, which blocked Lon induction completely (figure 3) also completely prevented improvements in both cell number (figure 4A) and MTT reduction results (figure 4B). Thus, Lon induction appears to be important for cell survival following an oxidative stress. It should be noted that the cells used in the studies of figure 4 were taken directly from the experiments of figure 3 – the cells were halved and used for both sets of tests – so that the Lon induction and carbonyl detection results of figure 3 may be directly applied to the cells used in figure 4.
DISCUSSION
Although the importance of Lon protease in normal mitochondrial function is clear, Lon transcription, translation, and activity seem to vary with age. The livers of old rats exhibit less Lon protease activity and higher amounts of oxidized and Nε-carboxymethyllysine protein content than do livers of young rats [29]. Similarly, the Lon content and activity in skeletal muscle mitochondria from old mice are significantly decreased from values found in young mice, whereas the levels of oxidized mitochondrial proteins are increased; furthermore, increased chronic oxidative stress, experienced by transgenic mice that are heterozygous for mitochondrial manganese-superoxide dismutase (50% loss of activity) exacerbates the effects of ageing on both diminished Lon and increased protein oxidation[21]. In contrast to these results in rodent liver and skeletal muscle, the activity of Lon remains unchanged in heart muscle from old rats, although, Lon protein and mRNA levels both increase with age: suggesting an accumulation of inactive Lon[30]. These data suggest that the ability of ageing mitochondria to respond to stress may be compromised, due, at least in part, to the loss of Lon activity. Mitochondrial dysfunction has been implicated in the ageing process[31] as well as in age-related diseases such as Alzheimer’s[32, 33], Parkinson’s[34, 35], and inclusion body myositis[36, 37].
In this study, we tested whether Lon is a stress responsive protein. Our analysis of multiple stressors on Lon reveals that Lon is induced under these conditions. Following oxidative stress, the induction of Lon protein seems to be much higher than the increases in steady-state Lon mRNA levels. Post-transcriptional regulation has been observed in a number of stress-responsive proteins as a means to rapidly increase protein content without the considerable time lag associated with the synthesis, processing, and export of de novo synthesized mRNA[38]. Oxidative stress may result in a rapid increase in damaged proteins that need immediate removal. Such a case would require rapid synthesis of Lon proteins or an increase in Lon activity. Our main focus in this paper was to determine whether Lon might respond to oxidative stress. We also performed mini studies on multiple stresses because we wanted to test our hypothesis that Lon may be a more general shock or stress protein, and because of published work indicating that heat and serum starvation can result in oxidative damage in cells. The induction of Lon with heat and serum starvation supports the hypothesis that Lon is protective under multiple stress conditions. It should be noted that Lon protein levels were significantly increased with only a minor elevation of lon transcripts after oxidative stress (fig. 1). Furthermore, Hori et al (39) reported no increase in lon mRNA levels in HeLa cells. We are currently working to test a model that Lon is translationally regulated during stress (although both transcriptional and translational regulation may be involved in different stresses), but such experiments are beyond the scope of the current paper.
In HeLa cells, it has been shown that Lon expression is enhanced with endoplasmic reticulum stress[39]. Heat shock induction of Lon has been shown in E.coli [40, 41], however lon mRNA induction was not observed in human Hela cells [39]. Our heat shock experiments were performed at 45°C for 1 hour, which may explain the apparent discrepancy between our results and those of a previous study with HeLa cells, which were tested at 43°C. It should also be noted that protein induction was not analyzed in the previous study by Hori et al.[39]. We speculate that the induction of Lon observed 1- 3 hours after stress may be sufficient to help refold or assemble damaged protein products generated during the stress, or to degrade those damaged proteins that are non-repairable. Since these cells survived a one-hour heat shock with no indication of extensive cellular damage or death, we speculate that after 3 hours of recovery, elevated Lon levels were no longer required. The mechanism(s) by which excess Lon protein is removed following a successful stress response is an intriguing question that we are now trying to address. The co-induction of mtHSP-70 with Lon supports the (previously proposed) concept that Lon may be working in conjunction of mtHSP-70 as both a chaperone and a protease [39, 42]. Further experiments are in progress to better define and understand the mechanism in which Lon is protective under heat shock.
We demonstrate that Lon induction provides important cellular protection against the accumulation of oxidized proteins during metabolic stress: a true hormetic response, up to a threshold. Through the use of siRNA, we were able to show that the level of Lon induction is dependent upon the level of oxidative damage, in a dose-responsive manner. Oxidative damage caused by low to intermediate levels of peroxide, such as 50 to 400 μM, is effective at inducing Lon, while high levels of oxidative damage caused by a high dose of 800μM peroxide is no longer effective. We speculate that mitochondrial damage occurring after pretreatment with a high dose of 800μM may have sensitized these cells, and is the approximate upper threshold for the Lon stress response, after which a different mechanism may take effect, such as apoptosis, which was also encountered in our previous work (Bota et al. 2004). The present results (figs. 3 and 4) make it clear that Lon induction significantly contributes to increased protection against the accumulation of carbonylated proteins, and also increases cellular viability during oxidative stress. The increased cell viability may be due, at least in part, to Lon’s ability to maintain mitochondrial function.
In the present manuscript, we find that the induction of Lon is protective against oxidative protein damage in whole cells, and that the oxidative (carbonyl) damage is not limited to mitochondrial proteins. Since mitochondria are a major source of free radical generation in cells, and the loss of Lon results in damaged mitochondria, this might contribute to a vicious cycle of ROS generation within the cell. It is also entirely possible that some proteins (e.g. aconitase) are selectively or preferentially damaged, whereas others may undergo more random oxidation. A categorization of the nature of the proteins oxidized, as well as those that are degraded or protected by Lon, would certainly be interesting, but is beyond the scope of this manuscript. Although mitochondrial functionality after hydrogen peroxide treatment during Lon downregulation has not been studied in detail, MTT analysis suggests that mitochondrial function is seriously impaired, and this is further supported by data from our previous work (Bota et al., 2004).
An emerging body of evidence suggests that the Lon protease is important during cellular stress. In addition to increased Lon activity and expression during heat shock[40, 41] and ER stress[39], hypoxia induces Lon mRNA through the hypoxia-inducible factor 1 (HIF-1), a nuclear transcription factor that can regulate Lon expression[43]. In addition, ischemia/reperfusion in mouse heart results in marked increases in Lon activity, while preventing the accumulation of carbonylated mitochondrial proteins[44]. This report indicates that Lon protein is induced with oxidative stress, heat stress, and serum starvation, and protects against the production of protein oxidation in cells. Our data suggests that the majority of the regulation may occur post-transcriptionally. This may permit a more rapid adaptive response that can facilitate cellular proliferation by minimizing mitochondrial damage during a toxic stress.
The induction of Lon during stress may be of even greater importance due to its potential role in mtDNA binding. Lon degrades the Kreb’s cycle enzyme aconitase, which has recently been shown to also have a role in mtDNA packaging[45]. In addition, there is evidence that Lon binds to the mtDNA genome[46], including the mtDNA light strand promoter overlapping the mtTFA binding site[47, 48], and such mtDNA proteins as mtSSB, polymerase γ, twinkle helicase, and PDIP38[49, 50]. Involvement of Lon in proteolysis, chaperoning, and mtDNA binding, may contribute to the impairment of mitochondrial function and cell death observed in our previous study[15]. With its multiple features, Lon might be poised as a major regulator of mitochondrial homeostasis that can respond to transient changes in the mitochondrial environment. During ageing, accumulation of oxidized mitochondrial proteins, and impaired mitochondrial function may be due, at least in part, to the decline of Lon and its diminished ability to cope with stress.
Footnotes
RD - Rhabodmyosarcoma
qRT-PCR – Quantitative real time PCR
mtHSP-70 – Mitochondrial heat shock protein
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Davies KJ. Oxidative stress: the paradox of aerobic life. Biochem Soc Symp. 1995;61:1–31. doi: 10.1042/bss0610001. [DOI] [PubMed] [Google Scholar]
- 2.Sohal RS. Role of oxidative stress and protein oxidation in the aging process. Free Radic Biol Med. 2002;33:37–44. doi: 10.1016/s0891-5849(02)00856-0. [DOI] [PubMed] [Google Scholar]
- 3.Giulivi C, Davies KJ. Mechanism of the formation and proteolytic release of H2O2-induced dityrosine and tyrosine oxidation products in hemoglobin and red blood cells. J Biol Chem. 2001;276:24129–24136. doi: 10.1074/jbc.M010697200. [DOI] [PubMed] [Google Scholar]
- 4.Ding Q, Dimayuga E, Keller JN. Proteasome regulation of oxidative stress in aging and age-related diseases of the CNS. Antioxid Redox Signal. 2006;8:163–172. doi: 10.1089/ars.2006.8.163. [DOI] [PubMed] [Google Scholar]
- 5.Friguet B. Oxidized protein degradation and repair in ageing and oxidative stress. FEBS Lett. 2006;580:2910–2916. doi: 10.1016/j.febslet.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 6.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 7.Bulteau AL, Moreau M, Saunois A, Nizard C, Friguet B. Algae extract-mediated stimulation and protection of proteasome activity within human keratinocytes exposed to UVA and UVB irradiation. Antioxid Redox Signal. 2006;8:136–143. doi: 10.1089/ars.2006.8.136. [DOI] [PubMed] [Google Scholar]
- 8.Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- 9.Pettinari A, Amici M, Cuccioloni M, Angeletti M, Fioretti E, Eleuteri AM. Effect of polyphenolic compounds on the proteolytic activities of constitutive and immuno-proteasomes. Antioxid Redox Signal. 2006;8:121–129. doi: 10.1089/ars.2006.8.121. [DOI] [PubMed] [Google Scholar]
- 10.Squier TC. Redox modulation of cellular metabolism through targeted degradation of signaling proteins by the proteasome. Antioxid Redox Signal. 2006;8:217–228. doi: 10.1089/ars.2006.8.217. [DOI] [PubMed] [Google Scholar]
- 11.Grune T, Reinheckel T, Davies KJ. Degradation of oxidized proteins in mammalian cells. Faseb J. 1997;11:526–534. [PubMed] [Google Scholar]
- 12.Davies KJ. Protein damage and degradation by oxygen radicals. I. general aspects. J Biol Chem. 1987;262:9895–9901. [PubMed] [Google Scholar]
- 13.Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJ. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc Natl Acad Sci U S A. 1999;96:6223–6228. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 15.Bota DA, Ngo JK, Davies KJ. Downregulation of the human Lon protease impairs mitochondrial structure and function and causes cell death. Free Radic Biol Med. 2005;38:665–677. doi: 10.1016/j.freeradbiomed.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Verbeke P, Deries M, Clark BF, Rattan SI. Hormetic action of mild heat stress decreases the inducibility of protein oxidation and glycoxidation in human fibroblasts. Biogerontology. 2002;3:117–120. doi: 10.1023/a:1015284119308. [DOI] [PubMed] [Google Scholar]
- 17.Boraldi F, Annovi G, Paolinelli-Devincenzi C, Tiozzo R, Quaglino D. The effect of serum withdrawal on the protein profile of quiescent human dermal fibroblasts in primary cell culture. Proteomics. 2008;8:66–82. doi: 10.1002/pmic.200700833. [DOI] [PubMed] [Google Scholar]
- 18.Ueom J, Kwon S, Kim S, Chae Y, Lee K. Acquisition of heat shock tolerance by regulation of intracellular redox states. Biochim Biophys Acta. 2003;1642:9–16. doi: 10.1016/s0167-4889(03)00081-8. [DOI] [PubMed] [Google Scholar]
- 19.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 20.Fredriksson A, Ballesteros M, Dukan S, Nystrom T. Defense against protein carbonylation by DnaK/DnaJ and proteases of the heat shock regulon. J Bacteriol. 2005;187:4207–4213. doi: 10.1128/JB.187.12.4207-4213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bota DA, Van Remmen H, Davies KJ. Modulation of Lon protease activity and aconitase turnover during aging and oxidative stress. FEBS Lett. 2002;532:103–106. doi: 10.1016/s0014-5793(02)03638-4. [DOI] [PubMed] [Google Scholar]
- 22.Grazette LP, Boecker W, Matsui T, Semigran M, Force TL, Hajjar RJ, Rosenzweig A. Inhibition of ErbB2 causes mitochondrial dysfunction in cardiomyocytes: implications for herceptin-induced cardiomyopathy. J Am Coll Cardiol. 2004;44:2231–2238. doi: 10.1016/j.jacc.2004.08.066. [DOI] [PubMed] [Google Scholar]
- 23.Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 24.Crawford DR, Schools GP, Salmon SL, Davies KJ. Hydrogen peroxide induces the expression of adapt15, a novel RNA associated with polysomes in hamster HA-1 cells. Arch Biochem Biophys. 1996;325:256–264. doi: 10.1006/abbi.1996.0032. [DOI] [PubMed] [Google Scholar]
- 25.Crawford DR, Leahy KP, Wang Y, Schools GP, Kochheiser JC, Davies KJ. Oxidative stress induces the levels of a MafG homolog in hamster HA-1 cells. Free Radic Biol Med. 1996;21:521–525. doi: 10.1016/0891-5849(96)00160-8. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Crawford DR, Davies KJ. adapt33, a novel oxidant-inducible RNA from hamster HA-1 cells. Arch Biochem Biophys. 1996;332:255–260. doi: 10.1006/abbi.1996.0340. [DOI] [PubMed] [Google Scholar]
- 27.Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D. Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell. 2004;15:2361–2374. doi: 10.1091/mbc.E03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandenbroucke K, Robbens S, Vandepoele K, Inze D, Van de Peer Y, Van Breusegem F. Hydrogen Peroxide-Induced Gene Expression across Kingdoms: A Comparative Analysis. Mol Biol Evol. 2008 doi: 10.1093/molbev/msm276. [DOI] [PubMed] [Google Scholar]
- 29.Bakala H, Delaval E, Hamelin M, Bismuth J, Borot-Laloi C, Corman B, Friguet B. Changes in rat liver mitochondria with aging. Lon protease-like reactivity and N(epsilon)-carboxymethyllysine accumulation in the matrix. Eur J Biochem. 2003;270:2295–2302. doi: 10.1046/j.1432-1033.2003.03598.x. [DOI] [PubMed] [Google Scholar]
- 30.Delaval E, Perichon M, Friguet B. Age-related impairment of mitochondrial matrix aconitase and ATP-stimulated protease in rat liver and heart. Eur J Biochem. 2004;271:4559–4564. doi: 10.1111/j.1432-1033.2004.04422.x. [DOI] [PubMed] [Google Scholar]
- 31.Bulteau AL, Szweda LI, Friguet B. Mitochondrial protein oxidation and degradation in response to oxidative stress and aging. Exp Gerontol. 2006;41:653–657. doi: 10.1016/j.exger.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 32.Castellani R, Hirai K, Aliev G, Drew KL, Nunomura A, Takeda A, Cash AD, Obrenovich ME, Perry G, Smith MA. Role of mitochondrial dysfunction in Alzheimer’s disease. J Neurosci Res. 2002;70:357–360. doi: 10.1002/jnr.10389. [DOI] [PubMed] [Google Scholar]
- 33.Gibson GE, Sheu KF, Blass JP. Abnormalities of mitochondrial enzymes in Alzheimer disease. J Neural Transm. 1998;105:855–870. doi: 10.1007/s007020050099. [DOI] [PubMed] [Google Scholar]
- 34.Mandemakers W, Morais VA, De Strooper B. A cell biological perspective on mitochondrial dysfunction in Parkinson disease and other neurodegenerative diseases. J Cell Sci. 2007;120:1707–1716. doi: 10.1242/jcs.03443. [DOI] [PubMed] [Google Scholar]
- 35.Fukae J, Mizuno Y, Hattori N. Mitochondrial dysfunction in Parkinson’s disease. Mitochondrion. 2007;7:58–62. doi: 10.1016/j.mito.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Oldfors A, Moslemi AR, Jonasson L, Ohlsson M, Kollberg G, Lindberg C. Mitochondrial abnormalities in inclusion-body myositis. Neurology. 2006;66:S49–55. doi: 10.1212/01.wnl.0000192127.63013.8d. [DOI] [PubMed] [Google Scholar]
- 37.Askanas V, McFerrin J, Baque S, Alvarez RB, Sarkozi E, Engel WK. Transfer of beta-amyloid precursor protein gene using adenovirus vector causes mitochondrial abnormalities in cultured normal human muscle. Proc Natl Acad Sci U S A. 1996;93:1314–1319. doi: 10.1073/pnas.93.3.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 39.Hori O, Ichinoda F, Tamatani T, Yamaguchi A, Sato N, Ozawa K, Kitao Y, Miyazaki M, Harding HP, Ron D, Tohyama M, D MS, Ogawa S. Transmission of cell stress from endoplasmic reticulum to mitochondria: enhanced expression of Lon protease. J Cell Biol. 2002;157:1151–1160. doi: 10.1083/jcb.200108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips TA, VanBogelen RA, Neidhardt FC. lon gene product of Escherichia coli is a heat-shock protein. J Bacteriol. 1984;159:283–287. doi: 10.1128/jb.159.1.283-287.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goff SA, Casson LP, Goldberg AL. Heat shock regulatory gene htpR influences rates of protein degradation and expression of the lon gene in Escherichia coli. Proc Natl Acad Sci U S A. 1984;81:6647–6651. doi: 10.1073/pnas.81.21.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savel’ev AS, Novikova LA, Kovaleva IE, Luzikov VN, Neupert W, Langer T. ATP-dependent proteolysis in mitochondria. m-AAA protease and PIM1 protease exert overlapping substrate specificities and cooperate with the mtHsp70 system. J Biol Chem. 1998;273:20596–20602. doi: 10.1074/jbc.273.32.20596. [DOI] [PubMed] [Google Scholar]
- 43.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 44.Bulteau AL, Lundberg KC, Ikeda-Saito M, Isaya G, Szweda LI. Reversible redox-dependent modulation of mitochondrial aconitase and proteolytic activity during in vivo cardiac ischemia/reperfusion. Proc Natl Acad Sci U S A. 2005;102:5987–5991. doi: 10.1073/pnas.0501519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen XJ, Wang X, Kaufman BA, Butow RA. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- 46.Lu B, Yadav S, Shah PG, Liu T, Tian B, Pukszta S, Villaluna N, Kutejova E, Newlon CS, Santos JH, Suzuki CK. Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. J Biol Chem. 2007;282:17363–17374. doi: 10.1074/jbc.M611540200. [DOI] [PubMed] [Google Scholar]
- 47.Lu B, Liu T, Crosby JA, Thomas-Wohlever J, Lee I, Suzuki CK. The ATP-dependent Lon protease of Mus musculus is a DNA-binding protein that is functionally conserved between yeast and mammals. Gene. 2003;306:45–55. doi: 10.1016/s0378-1119(03)00403-7. [DOI] [PubMed] [Google Scholar]
- 48.Fu GK, Markovitz DM. The human LON protease binds to mitochondrial promoters in a single-stranded, site-specific, strand-specific manner. Biochemistry. 1998;37:1905–1909. doi: 10.1021/bi970928c. [DOI] [PubMed] [Google Scholar]
- 49.Cheng X, Kanki T, Fukuoh A, Ohgaki K, Takeya R, Aoki Y, Hamasaki N, Kang D. PDIP38 Associates with Proteins Constituting the Mitochondrial DNA Nucleoid. J Biochem (Tokyo) 2005;138:673–678. doi: 10.1093/jb/mvi169. [DOI] [PubMed] [Google Scholar]
- 50.Liu T, Lu B, Lee I, Ondrovicova G, Kutejova E, Suzuki CK. DNA and RNA binding by the mitochondrial lon protease is regulated by nucleotide and protein substrate. J Biol Chem. 2004;279:13902–13910. doi: 10.1074/jbc.M309642200. [DOI] [PubMed] [Google Scholar]