Abstract
Dicer is a central enzyme in microRNA (miRNA) processing. We identified a Dicer-independent miRNA biogenesis pathway that uses Argonaute2 (Ago2) slicer catalytic activity. In contrast to other miRNAs, miR-451 levels were refractory to dicer loss of function but were reduced in MZago2 (maternal-zygotic) mutants. We found that pre-miR-451 processing requires Ago2 catalytic activity in vivo. MZago2 mutants showed delayed erythropoiesis that could be rescued by wild-type Ago2 or miR-451-duplex but not by catalytically dead Ago2. Changing the secondary structure of Dicer-dependent miRNAs to mimic that of pre-miR-451 restored miRNA function and rescued developmental defects in MZdicer mutants, indicating that the pre-miRNA secondary structure determines the processing pathway in vivo. We propose that Ago2-mediated cleavage of pre-miRNAs, followed by uridylation and trimming, generates functional miRNAs independently of Dicer.
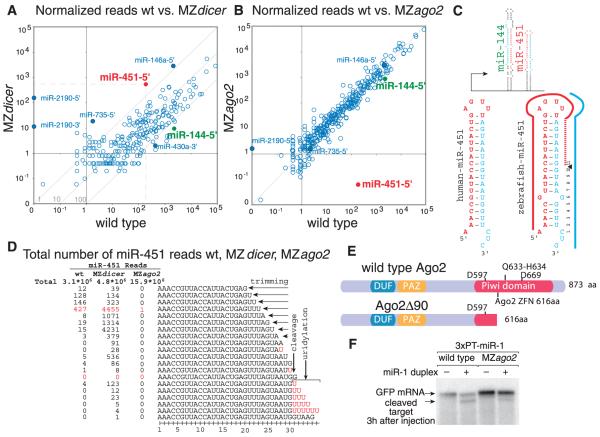
MicroRNAs (miRNAs) are ~22-nucleotide (nt) small RNAs that regulate dead-enylation, translation, and decay of their target mRNAs (1, 2). In animals, most miRNAs are processed from a primary transcript (termed pri-miRNA) by two ribonuclease III (RNase III) enzymes, Drosha and Dicer. Recent studies have identified several miRNA classes that bypass Drosha-mediated processing, namely miRtrons, tRNAZ, and small nucleolar RNA (snoRNA) (2–6). In contrast to Drosha, Dicer has been viewed as a central processing enzyme in the maturation of small RNAs (2). But are there functional miRNAs that bypass Dicer? To identify pathways that might process miRNAs in a Dicer-independent manner, we sequenced small RNAs (19 to 36 nt) in wild-type and maternal-zygotic dicer mutants (MZdicer)(7). We analyzed 48-hour-old embryos in two wild-type replicates and two dicer mutant alleles (8), dicerhu715 and dicerhu896 (fig. S1). Of the ~2 million reads per sample, 69 to 82% mapped to known 5′- or 3′-derived miRNAs in the wild type, whereas 4 to 9% mapped to miRNAs in the MZdicer mutants (fig. S2). Several miRNAs appeared refractory to dicer loss of function, notably miR-451-5′, miR-2190-5′, miR-2190-3′, and miR-735-5′ (Fig. 1A and figs. S3 and S4). On the basis of read frequency, reproducibility, and evolutionary conservation, we focused subsequent analysis on miR-451. miR-451 differs from other “canonical” miRNAs for several reasons: (i) It is encoded in a conserved 42-nt hairpin (fig. S5) with a 17-nt stem, whereas Dicer requires a >19-nt stem for efficient processing (9); (ii) miR-451 has a defined 5′ end but a variable 3′ end that extends over the loop region and ranges between 20 and 30 nt (Fig. 1, C and D); and (iii) reads stopped at nucleotide 30, and longer reads carried one to five nontemplated uridines, with nucleotide 31 mostly being a non-templated U (Fig. 1D). The final templated base pairs with nucleotide 10 of the mature miRNA (Fig. 1C and fig. S1), a site where slicer activity cleaves the passenger strand in siRNAs (10–12). These observations lead us to hypothesize that Ago2 slicer activity could participate in miRNA maturation (fig. S1).
Fig. 1.
MicroRNA analysis in wild type (wt) and in MZdicer and MZago2 mutants. (A and B) Normalized reads from wild type versus MZdicer (A) or MZago2 (B) libraries for all annotated zebrafish miRNAs. Some miRNAs are shown as a reference for enhanced and reduced miRNAs (solid circles); miR-144-5′ (green) and miR-451-5′ (red) are expressed in the same pri-miRNA. (C) Scheme of miR-144/miR-451 genomic loci and predicted secondary structure of both human and zebrafish pre-miR-451 (mature miRNA in red). (D) Total number of reads that matchmiR-451inwildtype, MZdicer, and MZago2. Nontemplated uridines are shown in red. (E)Domain organization of Ago2. The 90-nt deletion (Δ90) results in a predicted truncated protein lacking two of three catalytic residues. Amino acid positions are based on the mammalian Ago2. (F) Northern blot of embryos to detect slicer cleavage of an injected GFP target mRNA with three complementary targets to miR-1 (3×PT-miR-1) in the presence (+) or absence (−) of miR-1 (7). Slicer activity is indicated by the higher-mobility product (fig. S1) (18).
To determine whether Ago2 participates in miRNA maturation, we generated a deletion in the Piwi domain of the ago2 gene (ago2Δ90) with the use of zinc finger nucleases (13–15) (Fig. 1E and fig. S1). Because argonaute genes are maternally expressed (fig. S6), we generated maternal-zygotic ago2 mutants (MZago2). Indeed, slicer cleavage of an mRNA with perfectly complementary targets to miR-1 was severely reduced in MZago2 but not Zago2 relative to wild-type embryos (Fig. 1F and fig. S1).
To investigate the role of Ago2 in miRNA processing, we sequenced small RNAs (19 to 36 nt) from 48-hour-old MZago2 mutant embryos. Comparing the normalized read frequency for each 5′- and 3′-mature miRNA between wild-type and MZago2 mutants revealed a reduction in the number of reads that mapped to miR-451 (Fig. 1, B and D). In contrast, other miRNAs remained largely unchanged. miR-451 and miR-144 are coexpressed from a common primary transcript in the erythroid lineage (16, 17) (Fig. 1C). Whereas miR-451 accumulated in the absence of Dicer (factor of ~3 increase), miR-144 reads were reduced by a factor of >200 in MZdicer mutants (18) (Fig. 1A). Conversely, ago2 loss of function did not affect the read frequency of miR-144 (Fig. 1B) but did reduce miR-451 levels by a factor of >8000. Taken together, these results indicate that Ago2 regulates miR-451 levels posttranscriptionally by affecting either its processing or stability.
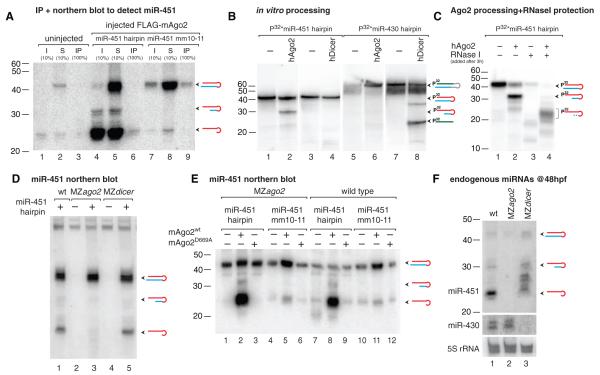
Recent studies suggest that Ago2 binds pre-miRNAs and miRNA:miRNA* duplexes (19–22), where miRNA* denotes the complementary strand. Ago2 interacted with radiolabeled synthetic pre-miR-451 in vitro (fig. S7). Coexpression of Flag-mouse-Ago2 (mAgo2) with pre-miR-451 or a mutant pre-miR-451mm10-11 (with two mismatches in the predicted slicer cleavage site) followed by Ago2 immunoprecipitation showed that Ago2 bound to both mature miR-451 and pre-miR-451mm10-11 (Fig. 2A). Incubation of human Ago2 (hAgo2) with pre-miR-451 but not pre-miR-430 resulted in a sharp 30-nt band corresponding with the predicted slicer cleavage product of miR-451 (Fig. 2B). Conversely, recombinant Dicer bound both pre-miRNAs (fig. S7) but could only process pre-miR-430 (Fig. 2B). To investigate whether Ago2 processes miR-451, we injected pre-miRNAs into one-cell-stage embryos. Synthetic and endogenous pre-miR-451 hairpins were processed into ~30-nt intermediates and a ~22- to 26-nt mature miR-451 in wild-type and MZdicer mutant but not in MZago2 mutant embryos (Fig. 2, D and F). In contrast, a canonical mature miR-430 was processed in both wild-type and MZago2 mutant embryos but not in MZdicer (Fig. 2F). On the basis of the sequencing results, we hypothesized that Ago2-processed hairpin might undergo nucleolytic trimming at the 3′ end (Fig. 1D). We observed that Ago2 protected the ~30-nt slicer-cleaved intermediate from RNase I in vitro, resulting in a ~20- to 26-nt 3′-end trimmed product (Fig. 2C), similar to the mature miRNAs observed in vivo (Fig. 2, D to F). Ago2 slicer activity depends on its catalytic triad (DDH) and the pairing between the guide and the target mRNA (23–25). Expressing wild-type but not catalytically dead (D669A) mAgo2 in MZago2 mutants rescued pre-miR-451 processing in vivo (Fig. 2E). Furthermore, a hairpin with mismatches that disrupt pairing in the predicted slicer cleavage was bound by Ago2 (fig. S7) but was inefficiently processed into mature miR-451 (Fig. 2E). These results indicate that Ago2 binds and cleaves pre-miR-451 in a process that requires the slicer catalytic activity and is independent of Dicer.
Fig. 2.
Ago2 binds and processes pre-miR-451. (A) Immunoprecipitation of FLAG-mAgo2 in wild-type and mutant embryos injected with pre-miR-451 followed by Northern blot analysis to detect bound miR-451. Input (I), supernatant (S), and immunoprecipitate (IP) are indicated. (B and C) In vitro cleavage assay using hAgo2 or hDicer protein and 5′-radiolabeled pre-miR-430 or pre-miR-451. (C) Ago2 processing reactions were treated with (+) or without (−) RNase I to assay protection of the processed hairpin by Ago2. (D to F) Northern blot analyses to detect mature miR-451 after injection with pre-miR-451 (+) [(D) and (E)] or endogenous miR-451 and miR-430 (F). Injection of wild-type mAgo2 but not a catalytically dead mAgo2D669A rescues pre-miR-451 processing in vivo (E). The processing of miR-451mm10-11 is strongly reduced. Endogenous pre-miR-451 at 48 hpf is processed in wild type and MZdicer but not in MZago2 mutants. Diagrams for predicted hairpins, cleavage intermediates, mature miR-451 (red), miR-430 (green), and miRNA* (blue) are shown. P32* indicates that injected hairpins were radiolabeled (18).
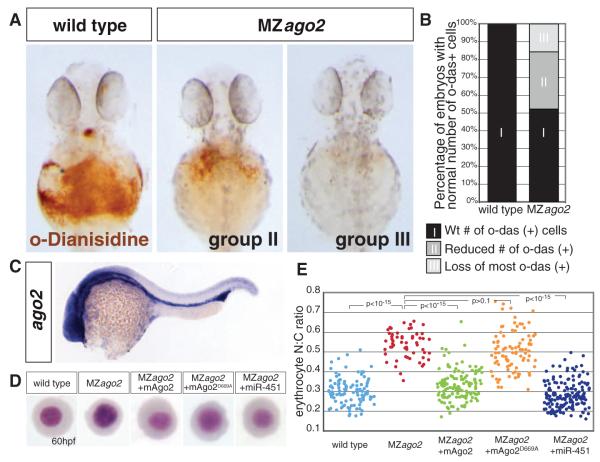
MZago2 mutant embryos displayed normal morphogenesis during gastrulation, brain development, and heart development (fig. S8). Ago2 is maternally expressed, and later in development it acquires tissue-specific expression in the brain and intermediate cell mass (ICM) (Fig. 3C and fig. S6). The ICM corresponds to the hematopoietic precursors and overlaps with the expression domain of miR-451 (16), which plays an important role in erythrocyte maturation in zebrafish (16, 17). Consistent with the Ago2-dependent processing of miR-451, MZago2 but not MZdicer mutants showed a reduction in the number of hemoglobinized erythrocytes (Fig. 3, A and B, and fig. S8). In zebrafish, erythrocyte maturation can be monitored by changes in erythrocyte morphology and reduced nuclear/cytoplasmic (N:C) ratio (17, 26, 27). Erythrocyte maturation was delayed in MZago2 mutants, as manifested by a significant increase in N:C ratio at 60 hours post-fertilization (hpf) (P < 10−15) (Fig. 3, D and E). Providing back wild-type mAgo2 or mature miR-451-duplex but not catalytically dead mAgo2D669A rescued erythrocyte maturation in MZago2 mutants (Fig. 3, D and E). Thus, Ago2 catalytic function plays an important role during erythrocyte maturation.
Fig. 3.
MZago2 mutants show reduced erythropoiesis. (A) Expression of hemoglobin (brown) visualized by the oxidation of o-dianisidine (o-das) at 48 hpf in the ducts of Cuvier. Hemoglobinized cells accumulate in wild type but are reduced in MZago2 mutants [group II (mild) and group III (severe) reduction of o-das–positive cells]. (B) Percentage of embryos with hemoglobinized cells in MZago2 mutants (n = 61) compared to wild-type embryos (n = 200), showing strongly reduced (group III; light gray) and partially reduced (group II; gray) numbers of o-das(+) cells(χ2 test, P < 0.001). (C) Whole-mount in situ hybridization of ago2 expression at 24 hpf. (D) May-Grünwald/Giemsa stain of erythrocytes from wild-type, MZago2 mutants, and MZago2 injected at one-cell stage with various RNAs as shown (+). Erythrocytes are representative of the mean for each group. (E)Scatterplotof the nuclear cytoplasmic ratio (N:C) for each genotype in (D) as a readout of erythrocyte maturation (17). Distributions of the N:C ratios in wild-type compared to MZago2 differed significantly (Wilcoxon rank-sum test after Bonferroni correction, P < 10−15). Erythrocyte maturation is rescued by miR-451-duplex (MZago2 and MZago2+ miR-451, P < 10−15) and wild-type mAgo2 (MZago2 and MZago2+mAgo2, P < 10−15) but not catalytically dead mAgo2D669A (MZago2 and MZago2+mAgo2D669A, P > 0.1).
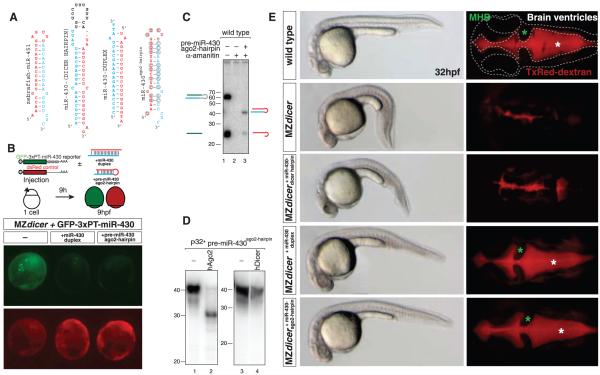
Whereas miR-451 is a 42-nt miRNA hairpin, canonical vertebrate miRNAs are ~60 nt, and unlike most miRNAs, mature miR-451 extends into the loop of the hairpin where it overlaps with the miRNA* (Fig. 4A and fig. S5). We hypothesized that selection of the processing pathway may be determined by structural differences or by specific sequence motifs. To distinguish between these two scenarios, we modified the sequence of pre-miR-451 to encode a Dicer-dependent miRNA (miR-430c or miR-1) mimicking pre-miR-451 structure and length (pre-miRNAago2-hairpin)(Fig. 4A and fig. S10). miR-430c is a member of a zygotically expressed miRNA family that regulates maternal mRNA clearance, gastrulation, and brain morphogenesis (7, 28). These processes are disrupted in MZdicer mutants but can be rescued by injection of a Dicer-independent miR-430-duplex (7, 28). Three lines of evidence indicate that pre-miR-430ago2-hairpin is processed and functional independently of Dicer: (i) Synthetic pre-miRNAago2-hairpin was processed into a ~23-nt mature miRNA in vivo (Fig. 4C and fig. S10) and processed by recombinant hAgo2 but not hDicer in vitro (Fig. 4D); (ii) injection of miR-430cago2-hairpin into MZdicer embryos repressed translation of a green fluorescent protein miR-430 reporter (GFP-miR-430) relative to a dsRed control (Fig. 4B); and (iii) injection of pre-miR-430cago2-hairpin into MZdicer mutants rescued the gastrulation and brain morphogenesis defects similarly to a miR-430-duplex (Fig. 4E). In contrast, equimolar levels of the annotated Dicer-dependent pre-miR-430 did not rescue the MZdicer phenotype (Fig. 4E). A second engineered miRNA (miR-1ago2-hairpin) was also processed independently of Dicer and down-regulated a GFP-miR-1 reporter in vivo (fig. S10). These results support a model in which the secondary structure of the hairpin determines whether a pre-miRNA is processed by Ago2 to form a physiologically functional Dicer-independent miRNA.
Fig. 4.
A Dicer-independent miRNA. (A) Zebrafish pre-miRNAs and duplexes as indicated. pre-miR-430ago2-hairpin is a miR-430c hairpin that has been mutated and shortened to form a 42-nt hairpin mimicking pre-miR-451 (ago2-hairpin). (B) GFP-reporter mRNA (green) was co-injected at the one-cell stage with control dsRed mRNA (red). The GFP reporter contains three complementary target sites to miR-430 in its 3′-untranslated region. (C) Northern blot to detect miR-430 in wild-type embryos injected with hairpins as indicated. α-Amanitin was co-injected to inhibit transcription of endogenous pri-miR-430. (D) Northern blot to detect 5′-radiolabeled pre-miR-430ago2-hairpin after in vitro processing by recombinant hAgo2 and hDicer. (E) In vivo assay to rescue miR-430 function in MZdicer mutants. Bright-field and fluorescent images of the dorsal view of the brain after injection of TxRed dextran in the ventricles (right) in 32-hpf embryos. Brain outline (dashed line), mid-hindbrain boundary (green asterisk), and ventricles (red, white asterisk) are shown. Morphogenesis defects are rescued by injection of a Dicer-independent pre-miR-430ago2-hairpin or a miR-430-duplex but not a Dicer-dependent pre-miR-430.
Our study defines a Dicer-independent pathway for miRNA processing that is dependent on Ago2 catalytic activity. We propose a model whereby Ago2 binds the pre-miRNA and cleaves the paired miRNA* passenger strand 10 nucleotides away from the 5′ end of the Ago2-bound miRNA guide strand (18). On the basis of our small RNA sequencing, this intermediate would undergo polyuridylation and nuclease-mediated removal of uridines and templated nucleotides not protected by Ago2 to generate the mature miRNA (fig. S11). Previous studies have shown that the terminal uridylyl transferase (TUT4) is recruited by lin-28 to uridylate pre-let7 (29), which blocks miRNA maturation and accelerates its degradation. Although we cannot exclude the possibility that miR-451–uridylated intermediates are targeted for complete degradation, our model favors a scenario where uridylated Ago2-cleaved pre-miRNAs are trimmed by a cellular nuclease to generate mature miRNA sequences protected by Ago2.
Ago2 has been reported to cleave siRNAs and pre-miRNAs (21). Ago2-cleaved precursors (ac-pre-miRNAs) can serve as Dicer substrates, but their physiological functions remain unclear (21). Here, we show that Ago2 cleavage is necessary for the generation of a functional miRNA (Figs. 1, 2, and 4).The identification of a miRNA-processing pathway that bypasses Dicer function might have wide implications for the processing of canonical miRNAs. Our study provides a biological context in which Ago2 slicer activity is needed to process a blood-specific miRNA, miR-451 (30). Although it is likely that Ago2 has additional roles in the cell by cleaving perfectly complementary targets (1), the strong conservation of the sequence and secondary structure of miR-451 across vertebrates suggests that constraints are in place to maintain this Ago2-mediated miRNA processing pathway through evolution (18).
Supplementary Material
Acknowledgments
31. We thank J. Doudna, D. O’Carroll, L. Zon, S. Lacadie, G. Lieschke, D. Krausse, and S. Halene for reagents and protocols; A. Enright, C. Abreu-Goodger, and J. Brennecke for initial small RNA analysis; and B. Schachter, C. Takacs, V. Greco, and D. Cazalla for discussions and manuscript editing. Supported by Fundación Ramón Areces (D.C.), a Human Frontier Science Program fellowship (H.X.), NIH grants R01GM081602-03/03S1 (A.J.G.) and R01HL093766 (N.D.L. and S.A.W.), the Yale Scholar program, and the Pew Scholars Program in the Biomedical Sciences (A.J.G.). Contributions: D.C. and A.J.G. designed and performed experiments; H.X. performed computational analysis; D.C. and D.W.T. performed in vitro assays; H.P. performed in situ hybridizations; Y.M., G.J.H., and S.C. helped with initial small RNA library sequencing and discussion; E.M. provided recombinant Ago2; S.M. provided small RNA sequencing; S.A.W. and N.L. designed the zinc finger nucleases; and A.J.G. wrote the manuscript. Sequencing data are deposited in Gene Expression Omnibus (accession number GSE21503).
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/science.1190809/DC1 Materials and Methods Figs. S1 to S11 References
References and Notes
- 1.Bartel DP. Cell. 2009;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Cell. 2009;136:642. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Genes Dev. 2008;22:2773. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mol. Cell. 2007;28:328. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. Cell. 2007;130:89. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruby JG, Jan CH, Bartel DP. Nature. 2007;448:83. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giraldez AJ, et al. Science. 2005;308:833. [Google Scholar]
- 8.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. Nat. Genet. 2003;35:217. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 9.Siolas D, et al. Nat. Biotechnol. 2004;23:227. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 10.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Cell. 2005;123:607. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 11.Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Cell. 2002;110:563. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 12.Czech B, et al. Mol. Cell. 2009;36:445. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyon Y, et al. Nat. Biotechnol. 2008;26:702. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeder ML, et al. Mol. Cell. 2008;31:294. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Nat. Biotechnol. 2008;26:695. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dore LC, et al. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3333. [Google Scholar]
- 17.Pase L, et al. Blood. 2009;113:1794. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.See supporting material on Science Online.
- 19.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Science. 2001;293:1146. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, et al. Nat. Struct. Mol. Biol. 2009;16:1259. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- 21.Diederichs S, Haber DA. Cell. 2007;131:1097. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 22.Tan GS, et al. Nucleic Acids Res. 2009;37:7533. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, et al. Science. 2004;305:1437. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 24.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Science. 2004;305:1434. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 25.Tolia NH, Joshua-Tor L. Nat. Chem. Biol. 2007;3:36. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 26.Weinstein BM, et al. Development. 1996;123:303. doi: 10.1242/dev.123.1.303. [DOI] [PubMed] [Google Scholar]
- 27.Qian F, et al. PLoS Biol. 2007;5:e132. doi: 10.1371/journal.pbio.0050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giraldez AJ, et al. Science. 2006;312:75. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 29.Kai ZS, Pasquinelli AE. Nat. Struct. Mol. Biol. 2010;17:5. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. Nature. 2010 doi: 10.1038/nature09092. 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.