The signal that directs newly synthesized mitochondrial β-barrel proteins from the cytosol to the organelle is poorly defined. The findings of this study demonstrate that, rather than a linear sequence, the structural information in four β-strands is sufficient for the mitochondria to recognize and assemble β-barrel protein.
Abstract
β-barrel proteins are found in the outer membranes of eukaryotic organelles of endosymbiotic origin as well as in the outer membrane of Gram-negative bacteria. Precursors of mitochondrial β-barrel proteins are synthesized in the cytosol and have to be targeted to the organelle. Currently, the signal that assures their specific targeting to mitochondria is poorly defined. To characterize the structural features needed for specific mitochondrial targeting and to test whether a full β-barrel structure is required, we expressed in yeast cells the β-barrel domain of the trimeric autotransporter Yersinia adhesin A (YadA). Trimeric autotransporters are found only in prokaryotes, where they are anchored to the outer membrane by a single 12-stranded β-barrel structure to which each monomer is contributing four β-strands. Importantly, we found that YadA is solely localized to the mitochondrial outer membrane, where it exists in a native trimeric conformation. These findings demonstrate that, rather than a linear sequence or a complete β-barrel structure, four β-strands are sufficient for the mitochondria to recognize and assemble a β-barrel protein. Remarkably, the evolutionary origin of mitochondria from bacteria enables them to import and assemble even proteins belonging to a class that is absent in eukaryotes.
INTRODUCTION
Membrane-embedded β-barrel proteins transverse the membrane in the form of a cylindrically shaped structure built by interconnected β-strands (Wimley, 2003). These proteins are found in both prokaryotic and eukaryotic organisms. In prokaryotes, β-barrel proteins are found in the outer membrane of Gram-negative bacteria, whereas in eukaryotes they reside exclusively in the outer membrane of mitochondria and chloroplasts. Their presence in these organelles supports the endosymbiotic hypothesis, according to which mitochondria and chloroplasts evolved from prokaryotic ancestors. Indeed, the biogeneses of these proteins in the various systems bear significant similarities (Dolezal et al., 2006; Walther et al., 2009b).
Bacterial β-barrel proteins are synthesized in the cytoplasm with an N-terminal signal sequence for transport across the inner membrane into the periplasm via the SEC system (Bos et al., 2007a). Their later integration into the outer membrane is facilitated by the β-barrel assembly machinery (BAM), the central component of which is the essential protein BamA (Omp85/YaeT) (Voulhoux et al., 2003; Wu et al., 2005). In mitochondria, precursors of β-barrel proteins are synthesized in the cytosol without a cleavable signal sequence. It is currently not clear why bacterial-like cleavable signal sequences were lost during the evolution of bacterial β-barrel proteins to their mitochondrial counterparts. Upon their synthesis, mitochondrial β-barrel precursors are translocated from the cytosol into the intermembrane space (IMS) via the translocase of the outer membrane (TOM) complex (Pfanner et al., 2004; Paschen et al., 2005). Their subsequent assembly into the outer membrane depends on a dedicated translocase, the topogenesis of mitochondrial outer membrane β-barrel proteins (TOB) (also known as sorting and assembly machinery [SAM]) complex. The central member of the latter complex is the essential protein Tob55/Sam50 that bears sequence and functional homology to BamA (Kozjak et al., 2003; Paschen et al., 2003; Gentle et al., 2004). The other two subunits of the TOB complex, Mas37/Sam37 and Tob38/Sam35/Tom38, are peripheral membrane proteins exposed to the cytosol that share no obvious sequence similarity with the accessory lipoproteins of the bacterial Bam complex (Wiedemann et al., 2003; Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004). Thus the biogenesis machineries in bacteria and mitochondria are similar in their central protein component and in an insertion into the outer membrane from the internal side of the membrane. In contrast, they vary with respect to the requirement of a signal sequence, the character of the accessory proteins, and the fact that precursors of mitochondrial β-barrel proteins initially have to cross the outer membrane.
To better understand the assembly process of β-barrel proteins in both bacteria and mitochondria, we expressed bacterial β-barrel proteins in the yeast Saccharomyces cerevisiae and demonstrated that they were imported into mitochondria and formed native-like oligomers. A detailed investigation of the import pathway revealed that it is shared with mitochondrial β-barrel proteins (Walther et al., 2009a). The reciprocal expression approach was also successful, and we observed that expression of mitochondrial porin in Escherichia coli resulted in assembly of the protein into the bacterial outer membrane, where it formed conducting pores (Walther et al., 2010). Taken together, it appears that despite the above-mentioned differences, the basic mechanism of β-barrel assembly in the outer membranes of bacteria and mitochondria is evolutionary conserved and that β-barrel proteins from one system can be dealt with and assembled by the other one.
Although some progress in our understanding of the biogenesis of β-barrel proteins has been made recently, the mitochondrial targeting signal in such proteins is still ill defined. A conserved linear sequence could not be identified, hence it was proposed that the signal is composed by β-barrel–specific structural elements (Walther et al., 2009b). However, neither the character nor the size of such putative structural signal has been identified so far. A crucial question is whether precursor of a full β-barrel structure is required. To shed new light on this issue, we investigated whether mitochondria can recognize and assemble fragments of a β-barrel protein. Such fragments are found in nature in the single subunits of trimeric autotransporter β-barrel proteins. These proteins form a special subfamily of bacterial β-barrel proteins. They have a characteristic arrangement of functional domains, including an N-terminal signal peptide, an internal passenger domain (also called the effector domain), and a relatively short C-terminal β-domain. The passenger moiety mediates the various functions of the autotransporter, and the β-domain forms a β-barrel that anchors the protein to the outer membrane. This anchor is made by a single 12-stranded β-barrel structure to which each monomer contributes four β-strands (Hoiczyk et al., 2000; Linke et al., 2006).
We took advantage of this model system and expressed the β-barrel domain of the bacterial trimeric autotransporter Yersinia adhesin A (YadA) in yeast and analyzed its cellular localization and topology. Our findings demonstrate that four β-strands contain sufficient structural information to be recognized and processed by the mitochondrial assembly machinery. Surprisingly, the bacterial evolutionary origin of mitochondria enables them to assemble even proteins that are absent in modern eukaryotic organisms.
RESULTS
Bacterial signal sequence interferes with the assembly of PhoE in mitochondria
Bacterial β-barrel proteins are synthesized in the cytoplasm with a signal sequence that targets them to the SEC machinery (Bos et al., 2007a). It is commonly believed that mitochondrial β-barrel proteins evolved from their bacterial counterparts. Nevertheless, in the process of developing from endosymbiont to modern time organelle, mitochondrial β-barrel proteins lost such an N-terminal extension. Hence, as part of our efforts to understand the sorting of these proteins in the eukaryotic cell we wanted to understand why during evolution mitochondrial β-barrels lost their bacterial signal sequence. To address this question, we used the capacity of the bacterial β-barrel protein PhoE to be assembled into the mitochondrial outer membrane of yeast cells (Walther et al., 2009a). First, we compared the mitochondrial levels of mature PhoE that lacks the signal sequence to that of a protein containing the N-terminal signal sequence (Sig-PhoE). We observed that Sig-PhoE is present in mitochondria at significantly reduced levels as compared to the form without the signal sequence (Figure 1A). One potential explanation for this decline could be reduced mRNA levels encoding the Sig-PhoE protein. To address this point, we isolated mRNAs from both cell types and performed RT-PCR. The results suggested that the amounts of mRNAs encoding both PhoE forms are comparable (Figure 1B). The bacterial signal sequence has similar characteristics to the eukaryotic signal sequence that directs protein to the secretory pathway. Hence another theoretical possibility that could explain the reduced levels of Sig-PhoE in the mitochondria would be a secretion of a portion of the protein molecules from the yeast cells. However, when we analyzed the medium of the growing culture, we could not detect any PhoE signal (unpublished data).
FIGURE 1:
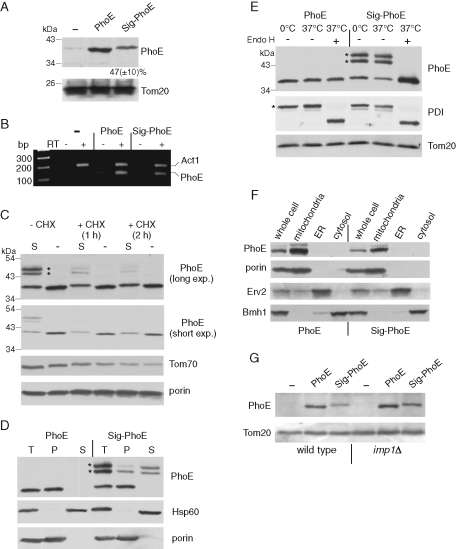
Bacterial signal sequence interferes with assembly of PhoE into mitochondria. (A) Mitochondria isolated from yeast cells transformed with an empty plasmid (–) or with a plasmid encoding either mature PhoE (PhoE) or PhoE with its signal sequence (Sig-PhoE) were analyzed by SDS–PAGE and immunodecoration with antibodies against PhoE and Tom20 as a loading control. The intensity of the PhoE and Sig-PhoE bands in three independent experiments was quantified, and the amount of Sig-PhoE is expressed as mean (±SD)% of the level of PhoE. (B) mRNAs were isolated from the cells just described, and reverse transcriptase (RT) was added to the indicated samples. Then, PCR using primers complementary to PhoE or actin (as a control) was performed using the obtained DNA as template. The PCR-amplified DNA fragments were analyzed on a 2% agarose gel by staining with ethidium bromide. (C) Cells expressing either PhoE (indicated as “–”) or Sig-PhoE (indicated as “S“) were grown in the presence or absence of cycloheximide (CHX). Membrane fraction isolated from these cells was analyzed by SDS–PAGE and immunodecorated with antibodies against PhoE, Tom70, and porin. To better view the different intensities of the various PhoE forms, short and long exposures are presented. Modified forms of Sig-PhoE are indicated with an asterisk. (D) Membrane fractions isolated from cells expressing either PhoE or Sig-PhoE (total, T) were subjected to alkaline extraction and then centrifuged to discriminate between membrane proteins in the pellet (P) and soluble proteins in the supernatant (S). Proteins were analyzed by SDS–PAGE and immunodecorated with antibodies against PhoE, Hsp60 (soluble matrix protein), and porin (embedded in the outer membrane). Modified forms of Sig-PhoE are indicated with an asterisk. (E) Membrane fractions isolated from cells expressing either PhoE or Sig-PhoE were solubilized with a buffer containing 0.5% SDS and 40 mM dithiothreitol in the presence of protease inhibitor cocktail. The samples were then incubated for 1 h at 0°C (as control) or at 37°C in the presence or absence of Endoglycosidase Hf (Endo H). Proteins were analyzed by SDS–PAGE and immunodecorated with antibodies against PhoE, protein disulphide isomerase (PDI, a glycosylated ER protein), and Tom20 (a nonglycosylated mitochondrial protein). Glycosylated forms of Sig-PhoE and PDI are indicated with an asterisk. (F) Lysate of cells expressing either PhoE or Sig-PhoE and fractions corresponding to mitochondria, light microsomal fraction (ER), and cytosol were analyzed by SDS–PAGE and immunodecoration with antibodies against PhoE, the mitochondrial protein porin, the ER protein Erv2, and a marker protein for the cytosol (Bmh1). (G) An empty vector (–) or vector encoding for either PhoE or Sig-PhoE was transformed into either wild type or cells lacking a functional inner membrane peptidase (imp1Δ). Mitochondria isolated from these cells were analyzed by SDS–PAGE and immunodecoration with antibodies against PhoE and Tom20 as a loading control.
We next explored whether an impaired biogenesis or an enhanced degradation of the signal sequence–containing protein can explain the observed reduction. To test the latter option, we added cycloheximide, which blocks protein synthesis, to the yeast culture and compared the levels of PhoE and Sig-PhoE in crude membrane preparations. Surprisingly, we detected in these samples a modified version of Sig-PhoE that had a relatively short half-life, and most of it was degraded after 2 h (Figure 1C). In contrast, the nonmodified form of Sig-PhoE and PhoE, as well as the control β-barrel protein porin, remained stable. Importantly, although nonmodified Sig-PhoE was present in lower levels than PhoE, the turnover rates of both proteins were indistinguishable. Tom70, which exposes a large domain to the cytosol, exhibited an enhanced turnover as compared to the membrane-embedded proteins (Figure 1C). Of note, this membrane fraction contains crude mitochondria and contaminations from other cellular compartments and proteins that are only loosely associated with the organelle. Thus we wondered whether the modified form of Sig-PhoE is membrane embedded. To answer that question, we performed alkaline extraction, after which membrane proteins remain in the pellet and soluble and membrane-peripheral proteins are found in the supernatant. Remarkably, the modified versions of Sig-PhoE were largely extracted under these conditions, whereas the nonmodified species behaved as membrane-embedded proteins (Figure 1D). Thus the modified forms are not integrated into cellular membranes.
We aimed to identify the nature of this modification. Our initial suspicion that the modified forms represent ubiquitination of Sig-PhoE was not confirmed, as an antibody against ubiquitin failed to recognize the modified species (unpublished data). Next we treated the membrane fraction with recombinant endoglycosidase H, which can remove oligosaccharides from N-linked glycoproteins. Surprisingly, this treatment resulted in disappearance of the modified forms concomitantly with an enhancement of the signal of the unmodified Sig-PhoE (Figure 1E). Indeed, analysis of PhoE sequence with the glycosylation sites prediction program, NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) revealed several asparagine residues as candidates for N-glycosylation. As this modification occurs in the lumen of the endoplasmic reticulum (ER), it appears that the similarity between the signal sequence of PhoE to the eukaryotic signal sequence causes a subpopulation of Sig-PhoE to be mistargeted to this latter compartment. Collectively, these results demonstrate that both forms of PhoE are expressed to the same extent. Apparently, a large portion of Sig-PhoE molecules is guided by the signal sequence to the ER, gets glycosylated and then degraded.
Because a portion of Sig-PhoE molecules is getting glycosylated in the ER, we asked whether the membrane-embedded form of Sig-PhoE can also be found in the ER in addition to mitochondria. Upon performing subcellular fractionation, we could not detect Sig-PhoE in the light microsomal fraction (ER), and both PhoE forms were located exclusively in the mitochondria (Figure 1F). Of note, Sig-PhoE is migrating at an apparently higher molecular mass than PhoE is, suggesting that the signal sequence is not processed (Figure 1, A and C–F). Mitochondria contain in their IMS a peptidase (named Imp) belonging to type I signal peptidase family. This peptidase was suggested to share several key features with the bacterial leader peptidase that cleaves the bacterial signal sequence upon its translocation across the inner membrane (Schneider et al., 1991). Thus we wanted to confirm by an additional approach that Imp does not cleave Sig-PhoE upon its import into mitochondria. To that end we used a strain deleted for one of the subunits of the Imp peptidase (imp1) and observed that the migration behavior of Sig-PhoE was not altered (Figure 1G). Collectively, it appears that mitochondrial β-barrels lost their N-terminal extension due to the potential of this signal sequence to wrongly direct them to the ER and, by that, to reduce their assembly into the mitochondrial outer membrane.
The β-barrel domain of YadA is targeted to mitochondria
Next we wanted to narrow down the structural features required for specific targeting to mitochondria and thus asked whether the mitochondrial import machinery can deal with a fragment of a β-barrel structure. To address this point, we used the membrane anchor (MA) domain (amino acids 335 to 422) of YadA, which is located in the C-terminal region of the protein (Figure 2A). Based on our previous and current results, YadA-MA was constructed without the bacterial signal sequence. To allow detection, an HA-tag was introduced at the N terminus of YadA-MA (Figure 2A). The expression of this construct in yeast cells was under the control of the GAL1 promoter. Subcellular fractionation of the transformed cells revealed that YadA-MA was located exclusively in the mitochondrial fraction (Figure 2B). As a control for the specificity of the antibody against the HA-tag, we confirmed the absence of the signal in mitochondria isolated from a nontransformed strain (Figure 2B, left lane).
FIGURE 2:
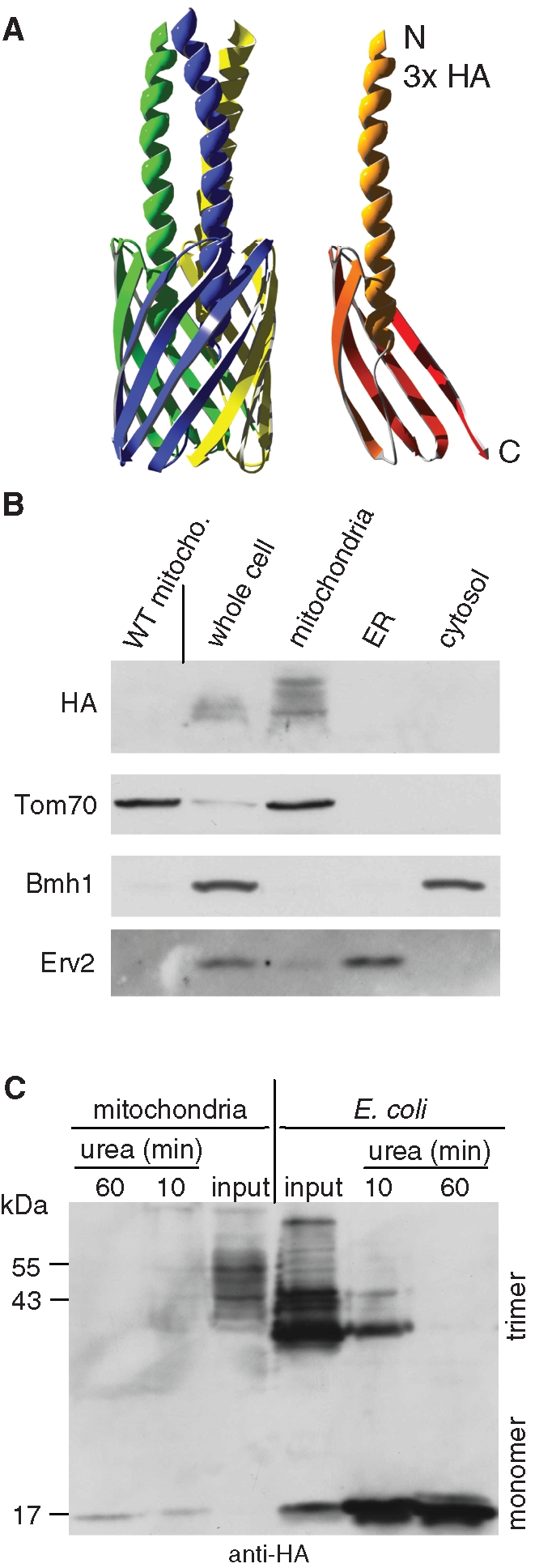
YadA-MA is assembled into mitochondria in a native trimeric conformation. (A) Atomic structure model of YadA-MA monomer with an HA-tag at its N-terminal (right) and trimeric form built from three monomers (left). Each YadA-MA monomer is composed of four β-strands that participate in the β-barrel structure and a linker domain (shown here as a helical structure). (B) YadA-MA is located in mitochondria. Lysate of cells expressing YadA-MA and fractions corresponding to mitochondria, ER, and cytosol were analyzed by SDS–PAGE and immunodecoration with antibodies against HA-tag, the mitochondrial protein Tom70, a marker protein for the cytosol (Bmh1), and the ER protein Erv2. Mitochondria isolated from untransformed wild-type cells were coanalyzed as a control. (C) Monomerization assay of YadA. Mitochondria isolated from yeast cells expressing YadA-MA and envelopes of E. coli–expressing YadA-MA were boiled for 5 min in Laemmli buffer without urea (input) or in Laemmli buffer containing 8 M urea for the indicated time periods. The samples were analyzed by SDS–PAGE and immunodecoration with HA-antibody. Molecular mass markers and the monomeric and trimeric forms of YadA are indicated (left and right, respectively).
YadA-MA migrated in SDS–PAGE as several bands with an apparent molecular weight of 42–50 kDa, a size expected for its trimeric structure (Figure 2C) (Wollmann et al., 2006; Grosskinsky et al., 2007). It is well documented that both full-length YadA and the MA domain build trimeric forms that are stable in SDS–PAGE (Wollmann et al., 2006; Grosskinsky et al., 2007; Ackermann et al., 2008). The 42- to 50-kDa bands can represent various conformations of the native trimeric form. To support this notion, we also expressed HA-tagged YadA-MA in E. coli and heated both E. coli envelopes and mitochondria isolated from transformed yeast cells in a solution containing 1% SDS and 8 M urea. In both expression systems, a shift from the trimeric bands to a single monomeric band was observed (Figure 2C). The detection of a single monomeric band argues against the possibility that the multiple bands behavior reflects a situation in which various trimeric forms harbor different patterns of covalent modifications. Of note, YadA-MA expressed in bacteria also migrates as several bands, suggesting that this phenomenon is not an artifact due to expression in eukaryotic cells. The pattern of the bands differs slightly from bacteria to mitochondria probably due to different membrane composition in these two systems. Collectively, these results confirm the trimeric nature of the 42- to 50-kDa bands observed upon analysis of mitochondria.
We further investigated whether the expression of YadA-MA obstructs the biogenesis of other mitochondrial outer membrane proteins. The levels of outer membrane β-barrel proteins, such as Tob55 and porin, were not affected by the expression of YadA-MA (Figure 3A). Similarly, the growth rate of yeast cells expressing the bacterial protein was similar to that of nontransformed cells under all tested conditions, including growth on a nonfermentable carbon source where yeast cells require fully functional mitochondria (Figure 3B and unpublished data). Next we verified that expressing YadA-MA in yeast cells did not have any effect on the morphology of the organelle (unpublished data). Collectively, it seems that the expression of YadA-MA in yeast cells does not interfere with crucial mitochondrial processes.
FIGURE 3:
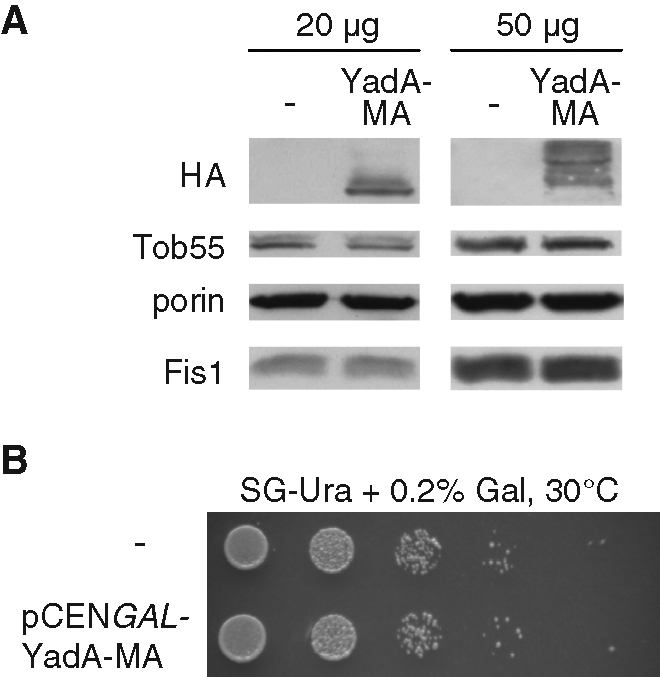
Expression of YadA-MA does not interfere with mitochondrial functions. (A) Mitochondria (20 or 50 μg) isolated from cells transformed with either an empty plasmid (–) or a plasmid encoding YadA-MA were analyzed by SDS–PAGE and immunodecoration with antibodies against HA, mitochondrial β-barrel proteins (Tob55 and porin), and a tail-anchored protein of the outer membrane (Fis1). (B) Expression of YadA-MA does not interfere with growth on a nonfermentable carbon source. Cells harboring either a plasmid encoding YadA-MA under the control of the GAL1 promoter or an empty plasmid (–) as control were tested by drop dilution assay for their ability to grow on synthetic glycerol-containing (SG) medium at 30°C. Small amounts of galactose (0.2%) were added to assure activation of the promoter.
Membrane topology of YadA-MA
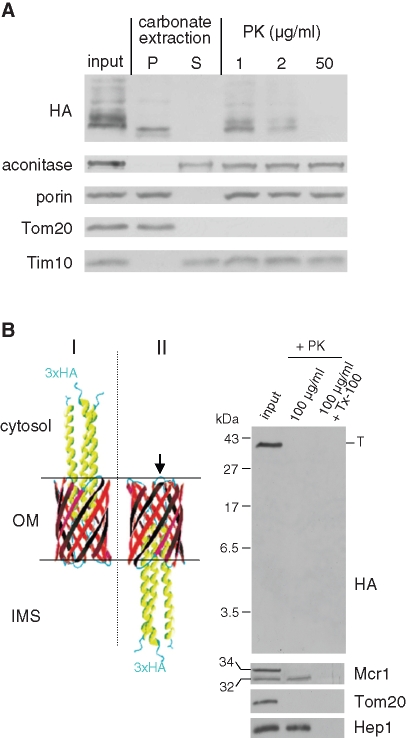
To verify that YadA-MA was embedded within the membrane rather than associated on the surface of the organelle, mitochondria were subjected to alkaline extraction. The YadA-MA protein was found in the pellet fraction together with other membrane-embedded mitochondrial proteins like Tom20 or porin (Figure 4A). In contrast, the soluble proteins aconitase and Tim10 were detected in the supernatant after this treatment (Figure 4A). Moreover, as further support for localization to the outer membrane, treatment of mitochondria isolated from YadA-MA–expressing cells with externally added proteinase K (PK) resulted in disappearance of the HA signal. Of note, the outer membrane was intact under these conditions as verified by the protease resistance of the small Tim10 chaperone residing in the IMS (Figure 4A). The results of the proteolytic assay can be explained by two alternative conformations of the protein. The first one is the native-like conformation with the HA-tags facing the cytosol, thereby being digested by PK (Figure 4B, left panel, I). The second conformation would be an upside-down conformation with the HA-tags facing the IMS (Figure 4B, left panel, II). In the latter case, the HA-tags themselves would be protected, but loops connecting the β-strands of YadA with the linker and the HA-tag might be accessible to the protease. According to an atomic model of the protein (Ackermann et al., 2008) and the crystal structure of the homologous membrane-anchor domain of the autotransporter Hia (Meng et al., 2006), cleavage at this loop would result in a HA-containing fragment with a size of approximately 6 kDa. To completely exclude the upside-down conformation, mitochondria harboring YadA-MA were treated with PK and the samples were analyzed on a urea-containing SDS–PAGE system optimized for detection of small polypeptides. Although a marker protein as small as 3.5 kDa could be detected with this gel system, a band at 6 kDa was not observed after treatment with PK (Figure 4B, right panel). Of note, the IMS isoform of Mcr1 was resistant under these conditions, confirming that the outer membrane was intact (Figure 4B). Taken together, our results demonstrate that, similarly to the topology in bacteria, YadA-MA is integrated into the outer membrane of mitochondria in a conformation in which the N terminus of each monomer is facing the external surface.
FIGURE 4:
YadA-MA is integrated into the mitochondrial outer membrane in the correct topology. (A) Mitochondria isolated from cells expressing YadA-MA were loaded directly on SDS–PAGE gel (input), or were first subjected to carbonate extraction and then centrifuged to discriminate between membrane proteins in the pellet (P) and soluble proteins in the supernatant (S). Additional aliquots of mitochondria were treated with the indicated amounts of PK. Proteins were analyzed by SDS–PAGE and immunodecorated with antibodies against the indicated proteins: aconitase, a mitochondrial matrix protein; porin, a protein embedded in the outer membrane; Tom20, an outer membrane protein exposed to the cytosol; Tim10, a soluble IMS protein. (B) Left, models of two putative conformations of HA-tagged YadA-MA in the mitochondrial outer membrane. The native (I) and the upside-down conformation (II) are displayed with an arrow pointing putative PK-sensitive loops for the upside-down conformation. Right, PK protection assay of mitochondria isolated from yeast cells expressing HA-tagged YadA-MA. Mitochondria were left untreated (input) or were treated with PK in the absence or presence of Triton X-100 (Tx-100). The samples were analyzed by SDS–PAGE on a gel optimized for detection of small polypeptides followed by immunodecoration with antibodies against HA, Tom20 (outer membrane), Hep1 (matrix), and Mcr1 (outer membrane and IMS). The latter protein has two isoforms: a 34 kDa form exposed at the outer membrane and a 32 kDa form in the IMS. Molecular mass markers are indicated on the left. The trimeric form of YadA-MA is indicated to the right with T.
YadA-MA assembly into mitochondria requires the small Tim chaperones and the TOB complex
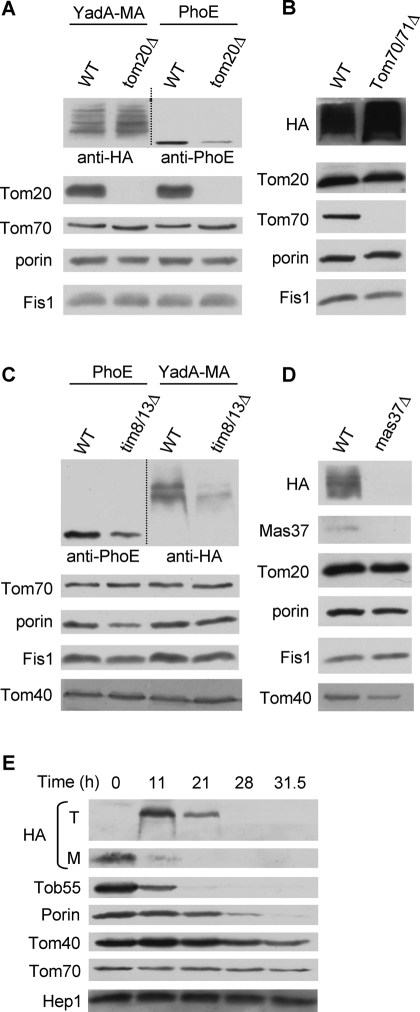
We previously observed that bacterial β-barrel proteins expressed in yeast cells require the import receptor Tom20 for their initial recognition at the organelle (Walther et al., 2009a). This requirement is shared with mitochondrial β-barrel proteins like Tom40, porin, and Tob55 (Rapaport and Neupert, 1999; Krimmer et al., 2001; Model et al., 2001; Habib et al., 2005). Thus we asked whether the import receptors of the TOM complex play a role in the import of YadA-MA. To address this point, we expressed YadA-MA in cells deleted for either Tom20 or Tom70/Tom71 and monitored its level in these cells. Surprisingly, highly pure mitochondria from strains lacking either import receptor had similar (tom20Δ) or even slightly higher (tom70Δ) amounts of YadA-MA as compared to those in wild-type organelles (Figure 5, A and B). As previously reported, the level of bacterial PhoE was reduced in mitochondria lacking Tom20 (Figure 5A). Taken together, it appears that, in contrast to their contribution to the import of precursors of mitochondrial and other bacterial β-barrel proteins, the import receptors are not involved in the membrane integration of YadA-MA.
FIGURE 5:
The assembly of YadA-MA depends on Tim8/Tim13 and Mas37. (A and C) Mitochondria isolated from either tom20Δ (A) or tim8Δ/tim13Δ (C) and their corresponding wild-type strains transformed with either YadA-MA or PhoE were analyzed by SDS–PAGE and immunodecoration with antibodies against either HA-tag or PhoE, respectively. In addition, immunodecoration with antibodies against the indicated mitochondrial proteins was performed. (B and D) Mitochondria isolated from either tom70Δ/tom71Δ (B) or mas37Δ (D) and their corresponding wild-type cells transformed with YadA-MA were analyzed as in (A). (E) YadA-MA was transformed into cells expressing Tob55 under the control of the GAL10 promoter. Cells were harvested at the indicated time points after a shift from galactose- to glucose-containing medium. Crude mitochondria were isolated, and proteins were analyzed by SDS–PAGE and immunodecoration with antibodies against HA-tag and the indicated mitochondrial proteins. The monomeric and trimeric forms of YadA-MA are indicated with M and T, respectively. Tob55, Tom40, and porin are β-barrel proteins.
Next we investigated whether YadA-MA requires the small chaperones in the IMS for its assembly in mitochondria. To that end, both PhoE and YadA-MA were transformed into a strain lacking both Tim8 and Tim13. Crude mitochondria were isolated from these cells and subjected to SDS–PAGE and immunodecoration. It can be observed that the steady-state levels of both PhoE and YadA-MA are reduced in cells lacking the Tim8/Tim13 complex (Figure 5C). Hence it seems that these small chaperones are playing an important role in the assembly of YadA-MA in mitochondria.
Does the TOB complex facilitate the membrane insertion of YadA-MA? Cells lacking the peripheral subunit of the TOB complex, Mas37, were transformed with a plasmid encoding YadA-MA. Mitochondria were isolated from these cells and subjected to SDS–PAGE and immunoblotting. Whereas the wild-type control shows significant expression of YadA-MA, the protein is hardly detectable in mas37Δ cells (Figure 5D). Interestingly, the effect of the absence of Mas37 on endogenous β-barrel proteins like porin or Tom40 is less severe (Figure 5D). Nevertheless, as the steady-state levels of Tom40 are also reduced in mas37Δ mitochondria, we wanted to exclude the possibility that the compromised insertion of YadA-MA is solely due to reduced levels of this central Tom component. To that goal we transformed a plasmid for the expression of YadA-MA under the control of the TPI promoter into cells in which the essential component Tob55 is under the control of the inducible GAL promoter (Paschen et al., 2003). When these cells are grown on glucose, the level of Tob55 is gradually reduced and, as a result, the cells' growth is slowed down (Paschen et al., 2003; Walther et al., 2009a). Mitochondria were isolated at various time points from these Tob55-depleted cells, and the levels of various mitochondrial proteins were analyzed. Noticeably, Tob55 was gradually depleted upon growth on glucose-containing medium. The compromised amounts of Tob55 caused a clear reduction in the levels of additional mitochondrial β-barrel proteins like porin and Tom40 as well as in those of YadA-MA (Figure 5E). Importantly, the decline in the amounts of YadA-MA preceded that of Tom40, suggesting that the depletion of YadA-MA is not initiated by the reduction in the levels of Tom40.
Of note, cells grown in galactose-containing medium contain excess amounts of Tob55 molecules (Figure 5E, time 0). These unassembled surplus molecules interfere with the assembly of YadA-MA; therefore we initially observed only YadA-MA monomers. Similarly, we previously observed that overexpression of Tob55 resulted in severely compromised assembly of newly synthesized Tom40 and porin molecules into isolated organelles (unpublished data). Upon shifting the cells to glucose-containing medium, Tob55 is gradually depleted and returns to its normal levels. This initial reduction resulted in assembly of YadA-MA, whereas further depletion of Tob55 caused diminished assembly of the former protein. Collectively, these results demonstrate the involvement of the TOB complex in the membrane integration of YadA-MA.
A eukaryotic-like β-signal improves the stability but not the overall assembled levels of YadA-MA
Recently a signature motif termed β-signal, located at the C-terminal β-strand of mitochondrial β-barrel precursors, was identified. This signal, which contains a highly conserved glycine residue, was suggested to be important for the interaction of β-barrel substrates with the TOB complex (Kutik et al., 2008; Figure 6A). To resemble this eukaryotic β-signal and to test the importance of such a signal for the assembly of YadA-MA in mitochondria, a serine residue in position 417, which resides within the last of the four encoded β-strands, was replaced by a glycine residue resulting in the S417G variant (Figure 6A). We verified by subcellular fractionation that, similarly to the native protein, the YadA-MA-S417G variant is localized to mitochondria (Figure 6B). Furthermore, proteolytic assay and alkaline extraction revealed that the mutated protein is embedded in the correct topology in the mitochondrial outer membrane (Figure 6C). Next we compared the stability of both forms. The wild-type trimeric form was partially converted after 5 min of boiling in the presence of 8 M urea to the monomeric one. In contrast, the trimeric form of the variant was turned into the smaller form much more slowly, and, even after 60 min, the monomeric band could hardly be observed (Figure 6D). These experiments show that the point mutation S417G leads to the formation of a trimer that is even more stable than the native protein. When we compared the steady-state levels of both proteins in yeast cells, however, we observed lower amounts of the mutant protein (Figure 6E). Hence it seems that increased stability of a β-barrel structure does not necessarily lead to overall improved biogenesis of such a protein.
FIGURE 6:
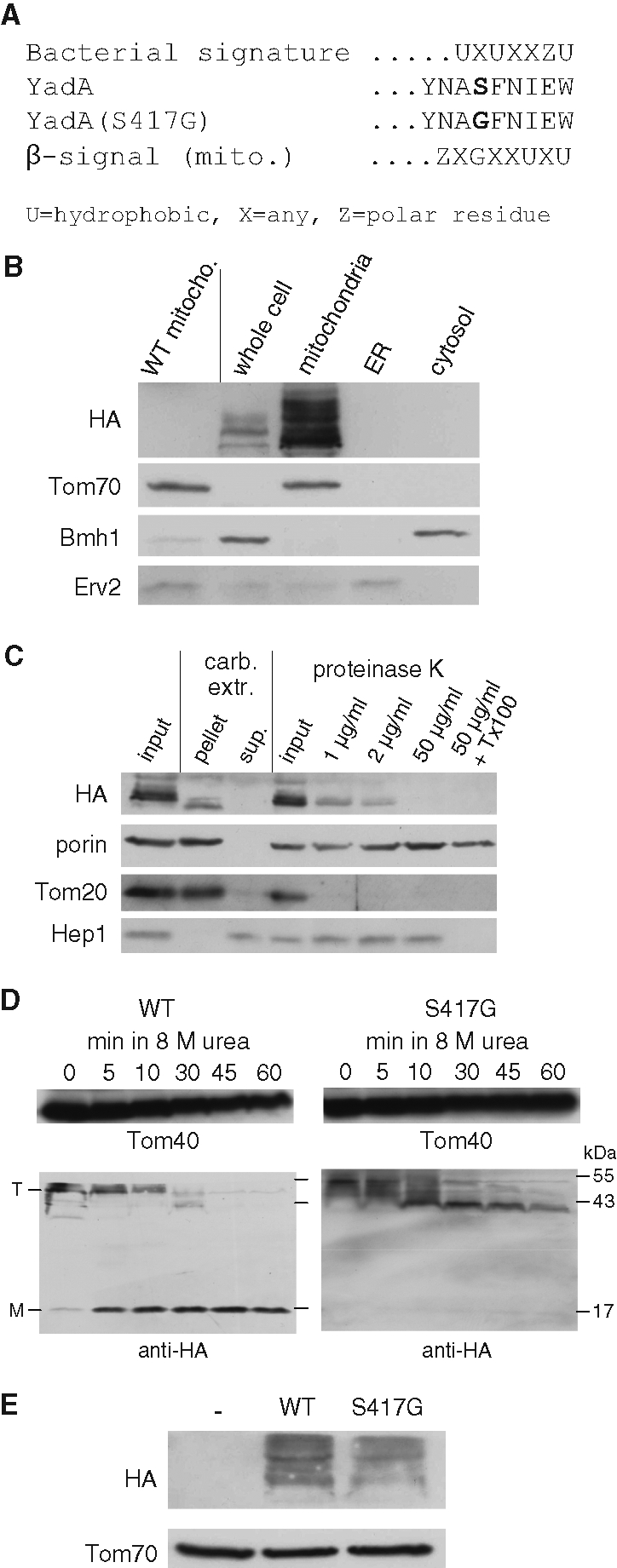
A eukaryotic-like β-signal improves the stability but not the overall assembly of YadA-MA. (A) Comparison of the bacterial and mitochondrial β-barrel assembly signals and the C termini of the YadA-MA variants used in this study. X, any amino acid; U, hydrophobic residue; Z, polar residue. (B) YadA-MA-S417G is located in mitochondria. Whole cell lysate of cells expressing the YadA-MA-S417G variant and fractions corresponding to highly pure mitochondria, ER, and cytosol were analyzed by SDS–PAGE and immunodecoration with antibodies against HA-tag, the mitochondrial protein Tom70, a marker protein for the cytosol (Bmh1), and the ER protein Erv2. To demonstrate the specificity of the HA antibody, crude mitochondria isolated from wild-type, untransformed cells were coanalyzed as a control. (C) Mitochondria isolated from cells expressing YadA-MA-S417G were loaded directly on SDS–PAGE gel (input), or were subjected first to carbonate extraction and then centrifuged to discriminate between membrane proteins in the pellet and soluble proteins in the supernatant (sup). Additional aliquots of mitochondria were left intact (input) or were treated with the indicated amounts of PK in the absence or presence of Triton X-100 (Tx-100). Samples were analyzed by SDS–PAGE and immunodecoration with antibodies against the indicated proteins. Porin, protein embedded in the outer membrane; Tom20, outer membrane protein exposed to the cytosol; Hep1, a mitochondrial matrix protein. (D) Monomerization assay of YadA-MA. Mitochondria were isolated from yeast cells expressing either native YadA-MA (WT) or the S417G variant. Mitochondria were boiled for 5 min in Laemmli buffer without urea (0) or in Laemmli buffer containing 8 M urea for the indicated time periods. The samples were analyzed by SDS–PAGE and immunodecoration with antibodies against the HA-tag and against Tom40 as a loading control. The monomeric and trimeric forms of YadA-MA are indicated with M and T, respectively. (E) The S417G variant is present in lower steady-state levels. Mitochondria isolated from yeast cells transformed with an empty plasmid (–) or with a plasmid encoding either native YadA-MA (WT) or its variant (S417G) were analyzed by SDS–PAGE and immunodecoration with antibodies against HA and Tom70 as a loading control.
DISCUSSION
Mitochondrial β-barrel proteins are synthesized in the cytosol and therefore must bear targeting signals to direct them to the right organelle. Their bacterial counterparts contain an N-terminal signal sequence that mediates their translocation from the bacterial cytoplasm across the inner membrane. This signal shows some similarity to signal sequences that direct eukaryotic proteins to the ER. During evolution mitochondrial β-barrel proteins lost such an extension, and our results show that indeed bacterial PhoE with a signal sequence is assembled in reduced levels into mitochondria as compared to a construct without this extension. The presence of a signal sequence results in a protein with two competing targeting signals, one for the mitochondria (within the β-barrel domain) and one for the ER (signal sequence). Neither of these signals is dominant, resulting in a dual localization of the protein. Those molecules that reach the mitochondria integrate into the outer membrane in a stable manner. In contrast, we propose a scenario in which the signal sequence directs the other population to the SEC system in the ER, where Sig-PhoE is translocated into the lumen because there is no hydrophobic membrane-spanning segment that stops the translocation. This process is similar to the transport of the protein into the periplasm through the bacterial SEC machinery in the inner membrane (Bos et al., 2007a). Because there is no BAM complex (or eukaryotic equivalent) in the ER, these molecules cannot get assembled into the membrane and remain in the ER lumen. Comparable accumulation of β-barrel precursors is observed in the periplasm of BamA-depleted bacterial cells (Bos et al., 2007a). In the ER lumen, PhoE can become glycosylated and eventually destined for degradation because the yeast cell probably recognizes it as an unfolded, nonfunctional protein. Analogously, unassembled β-barrel precursors are degraded in the bacterial periplasm (Bos et al., 2007a). Taken together, as the signal sequence appears to be counterproductive for the assembly into the mitochondrial outer membrane, these observations provide an experimental explanation for the absence of bacterial-like signal sequences in precursors of modern mitochondrial β-barrel proteins.
Rather than the presence of a linear sequence, it was suggested that the ability of a protein to adopt a membrane-embedded β-barrel-like conformation could be sufficient for its specific targeting to mitochondria (Rapaport, 2003). Recent results supported this hypothesis by demonstrating that bacterial β-barrel proteins, like PhoE, expressed in yeast cells are targeted to mitochondria, although these proteins show no significant sequence similarity with mitochondrial β-barrel proteins (Walther et al., 2009a). To better understand this putative structural signal, we tested if specific targeting to mitochondria requires a complete β-barrel precursor structure or whether even a fragment of such a structure would be sufficient. For this purpose, we used YadA, a member of the class of trimeric autotransporters that is found only in bacteria. These proteins are synthesized in the cytoplasm as monomers and form β-barrel-like trimers with their membrane-embedded, C-terminal domain. Recent work demonstrated that BamA, similarly to its function in the biogenesis of other β-barrel proteins, interacts directly with YadA and is essential for its membrane integration (Lehr et al., 2010).
Our data demonstrate that YadA was exclusively targeted to mitochondria where it formed native trimeric structure. Thus it appears that even fragments of a β-barrel structure are sufficient for the recognition of a β-barrel protein and its correct targeting to mitochondria. The usage of the heterologous expression system can also help to address the yet open question: In which step of the protein biogenesis is the trimeric structure formed? To investigate whether YadA monomers can form a trimeric structure already in the eukaryotic cytosol, we performed cell-free translation experiments using rabbit reticulocyte lysate. Our results suggest that a formation of cytosolic trimer is unlikely because only signals corresponding to monomeric YadA-MA were observed under these conditions (unpublished data).
The finding that YadA-MA is specifically targeted to mitochondria raised this question: Which components of the mitochondrial import machinery are used? The initial interaction between endogenous β-barrel proteins like porin or Tom40 and the general entry gate, the TOM-complex is mediated by Tom20 (Rapaport and Neupert, 1999; Krimmer et al., 2001; Yamano et al., 2008). The same appears to be true for β-barrel proteins of bacterial origin (this study and Walther et al., 2009a), but surprisingly we found that this is not the case for YadA-MA. Similarly, Tom70 is also not required for the import of YadA, and even a slight increase in YadA-MA levels was observed in its absence. Tom70 exposes a large domain on the cytosolic surface of the outer membrane. As this receptor is part of the TOM holo complex, this bulky domain can be in the vicinity of the import pore and thus form a steric hindrance for precursor proteins that are translocated via this pore. Thus, for those proteins that are not recognized by Tom70, the absence of this receptor can even result in a slight improvement of their import efficiency. A similar observation was made by Hines et al. regarding the import of CoxIV-dihydrofolate reductase (DHFR) into mitochondria lacking Tom70 (Hines et al., 1990).
Of note, Tom import receptors are not absolutely required for the translocation in vitro of bona fide mitochondrial precursor proteins. Import can still occur, albeit with low efficiency, after destroying protease-sensitive receptors (Pfaller et al., 1989). The import via this so-called “bypass” route occurs most probably by a direct interaction of the precursor proteins with the Tom40 import pore. Alternatively, Tom22 can function as a secondary receptor and thus might be involved in the recognition of the YadA precursor. The receptor domain of Tom22 was shown recently to be required for the in vitro import of porin. Furthermore, Tom22 and Tom20 were suggested to be involved in the same step or sequential steps in similar import pathways (Yamano et al., 2008). Hence we propose that YadA is recognized on the surface of the organelle either by Tom22 or directly by Tom40. Naturally, these two alternatives are not mutually exclusive.
The finding that the import of YadA-MA is independent of the import receptors could have evolutionary reasons. Whereas the TOB complex is most probably derived from a bacterial translocase, the TOM complex has no bacterial ancestor (Dolezal et al., 2006) and only three of the TOM-complex components (Tom40, Tom7, and Tom22) are commonly found in eukaryotes (Macasev et al., 2004). It is thought that the TOM complex developed on the way of converting the endosymbiont into an organelle. Thus, although it is not clear when trimeric autotransporters emerged, it could be hypothesized that the class of these proteins was lost in early eukaryotes before the development of the primary import receptors (Tom20 and Tom70). In such a scenario, there was never a need for the import receptors to recognize such proteins, and thus import of YadA is independent of the two receptors just mentioned. Astonishingly, the evolutionary origin of mitochondria from bacteria allows the organelle to assemble a class of proteins that are not present in modern eukaryotic organisms.
Upon leaving the TOM complex, YadA is probably exposed in its assembly pathway to the IMS as its overall import efficiency is reduced in cells lacking the small chaperones Tim8/Tim13. This reduction, however, is somewhat less significant as compared to that observed for PhoE. One possible explanation of this difference is the smaller size of hydrophobic elements in YadA as compared to those in PhoE. This proposal is supported by a previous report that larger bacterial β-barrel proteins were more dependent on the presence of all five polypeptide-transport-associated (POTRA) domains of Neisseria meningitidis BamA as compared to small β-barrel proteins (Bos et al., 2007b). From the IMS, precursor molecules of YadA-MA are most likely relayed to the TOB complex, and our results clearly show a strong dependence of YadA-MA assembly on the TOB subunits, Tob55 and Mas37. These findings are in accordance with our previous findings for PhoE the import of which into mitochondria is also severely affected by the deletion of Mas37 or the depletion of Tob55 (Walther et al., 2009a). Although both PhoE and YadA-MA can be assembled by the TOB complex, they probably represent suboptimal substrates for this complex. Hence an efficient membrane integration of these proteins necessitates most likely the presence of a fully functional TOB complex. Therefore in the absence of Mas37, the Tob55-Tob38 subcomplex cannot deal efficiently with bacterial precursors, whereas it can still process mitochondrial β-barrel substrates.
Assembly of mitochondrial β-barrel proteins appears to be facilitated by the presence of a eukaryotic-specific β-signal present in the most C-terminal β-strand (Kutik et al., 2008). Interestingly we found that mutation of Ser-417 to glycine, a mutation that allows the last β-strand of YadA-MA to resemble the eukaryotic β-signal, led to a much higher stability of the trimer. This mutation enhances also the stability of the bacterially expressed trimeric form of full-length YadA (Lehr et al., 2010). Nevertheless, wild-type YadA-MA was present in higher steady-state levels than was the mutant construct. Thus it can be speculated that, although β-signal-like sequences improve the final stability of β-barrel proteins, some structural flexibility is actually an advantage in other stages in the assembly pathway of these proteins, most probably in the integration into the lipid core of the membrane.
In conclusion, our findings shed new light on the biogenesis of mitochondrial β-barrel proteins. They demonstrate that rather than a specific linear sequence, the structural information contained in four β-strands is sufficient for it to be recognized and processed by the mitochondrial import machinery.
MATERIALS AND METHODS
Yeast strains and growth methods
Standard genetic techniques were used for growth and manipulation of yeast strains. The wild-type strains YPH499 and W303 were used. For construction of the tom20Δ mutant strain, the TOM20 gene was deleted by replacement with a HIS3 gene cassette. The mas37Δ and tim8Δ/tim13Δ strains were described before (Habib et al., 2005, and Paschen et al., 2000, respectively). The tom70Δ/tom71Δ double deletion strain was a gift from K. Okamoto (Kondo-Okamoto et al., 2008). The imp1Δ strain was purchased from Euroscarf (Frankfurt, Germany). For drop-dilution assays, yeast cells were grown to an OD600 of 1.0 in synthetic medium and diluted in 10-fold increments, and then 5 μl of each dilution was spotted onto solid medium with different carbon sources. In some experiments, cycloheximide (100 μg/ml) was added to the yeast culture.
Recombinant DNA techniques
Sequences encoding E. coli PhoE with or without its signal sequence (first 21 amino-acid residues) were cloned by PCR amplification from a plasmid encoding the full-length protein. An additional N-terminal methionine was added in constructing PhoE without its signal sequence. The PCR products were inserted into the yeast expression vector pYX113, in which the GAL1 promoter was replaced by the S. cerevisiae POR1 promoter.
Sequences encoding YadA-MA or YadA-MA-S417G were obtained by PCR amplification using pASK-IBA2 encoding the Yersinia enterocolitica YadA as a template. The S417G mutation was introduced by using a reverse primer containing the desired mutation. Both sequences were inserted into either pYX113 or pYX242 vectors using EcoRI and SalI restriction sites. For constructing HA-tagged YadA-MA, the 3xHA cassette was PCR amplified from pFA6a-3HA-KanMX4 plasmid and inserted into the target vectors pYX113-GAL1pro-URA or pYX242-TPIpro-LEU using EcoRI and NcoI restriction sites. For expression of HA-tagged YadA-MA in E. coli, we modified our published procedure for expressing Strep-tagged YadA-MA (Wollmann et al., 2006). In short, HA-tagged YadA-MA was PCR-amplified using the yeast expression vector as template. The PCR product was then cloned using the BsaI restriction site into the pASK-IBA2 vector, which already encodes an N-terminal signal sequence derived from the E. coli outer membrane protein OmpA.
RT-PCR
Total RNA from yeast was isolated by phenol/chloroform/isoamylalcohol (ratio 25:24:1) extraction and subsequent ethanol precipitation. Isolated RNA (2 μg) was treated with RQ1-DNase (Promega, Madison, WI). The samples were split in half and used for RT-PCR in the presence or absence of RevertAid Premium Reverse Transcriptase (Fermentas, Glen Burnie, MD) using oligo-dT and random hexamer primers. PCR amplification from the cDNA was performed using Taq-Polymerase (Fermentas) and primers specific for phoE or ACT1 (as a control).
Biochemical procedures and computational biology
Mitochondria were isolated from yeast cells by differential centrifugation as described (Daum et al., 1982). Subcellular fractionation was performed according to published procedures (Walther et al., 2009a). Treatment of samples with Endoglycosidase Hf (New England BioLabs, Ipswich, MA) was for 1 h at 37°C according to the manufacturer's recommendations and in the presence of a cocktail of protease inhibitors (Roche, Basel, Switzerland). Radiolabeled YadA-MA was synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine (Perkin-Elmer, Rodau, Germany) after in vitro transcription by SP6 polymerase from pGEM4 vectors (Promega). A monomerization assay was performed by resuspending 50 μg of isolated mitochondria in sample buffer containing 8 M urea. Samples were then boiled at 95°C for various time periods before their analysis by SDS–PAGE.
Three-dimensional models of YadA-MA were produced using the Swiss PDB Viewer in combination with Persistence of Vision Raytracer (PovRay) rendering software, based on published model coordinates (Grosskinsky et al., 2007).
Acknowledgments
We thank K. Rehn and E. Kracker for technical support. This work was supported by the Deutsche Forschungsgemeinschaft (RA 1048/4–1 to D.R., SFB766/B1 to I.A., and SFB766/B4 to D.L.), the Evolution and Ecology Forum Tübingen (to J.E.N.M.), a postdoctoral fellowship from the Carl Zeiss Stiftung (to K.S.D.), and the Max Planck Society (to D.L. and I.G.).
Abbreviations used:
- BAM
β-barrel assembly machinery
- DHFR
dihydrofolate reductase
- ER
endoplasmic reticulum
- IMS
intermembrane space
- MA
membrane anchor
- PK
proteinase K
- SAM
sorting and assembly machinery
- TOB
topogenesis of mitochondrial outer membrane β-barrel proteins
- TOM
translocase of the outer membrane
- YadA
Yersinia adhesin A
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-12-0943) on April 1, 2011.
REFERENCES
- Ackermann N, Tiller M, Anding G, Roggenkamp A, Heesemann J. Contribution of trimeric autotransporter C-terminal domains of oligomeric coiled-coil adhesin (Oca) family members YadA, UspA1, EibA, and Hia to translocation of the YadA passenger domain and virulence of Yersinia enterocolitica. J Bacteriol. 2008;190:5031–5043. doi: 10.1128/JB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007a;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007b;8:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria: cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskinsky U, Schütz M, Fritz M, Schmid Y, Lamparter MC, Szczesny P, Lupas AN, Autenrieth IB, Linke D. A conserved glycine residue of trimeric autotransporter domains plays a key role in Yersinia adhesin A autotransport. J Bacteriol. 2007;189:9011–9019. doi: 10.1128/JB.00985-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib SJ, Waizenegger T, Lech M, Neupert W, Rapaport D. Assembly of the TOB complex of mitochondria. J Biol Chem. 2005;280:6434–6440. doi: 10.1074/jbc.M411510200. [DOI] [PubMed] [Google Scholar]
- Hines V, Brandt A, Griffiths G, Horstmann H, Brütsch H, Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiczyk E, Roggenkamp A, Reichenbecher M, Lupas A, Heesemann J. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 2000;19:5989–5999. doi: 10.1093/emboj/19.22.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Shaw JM, Okamoto K. Tetratricopeptide repeat proteins Tom70 and Tom71 mediate yeast mitochondrial morphogenesis. EMBO Rep. 2008;9:63–69. doi: 10.1038/sj.embor.7401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- Krimmer T, et al. Biogenesis of the major mitochondrial outer membrane protein porin involves a complex import pathway via receptors and the general import pore. J Cell Biol. 2001;152:289–300. doi: 10.1083/jcb.152.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Lehr U, Schütz M, Oberhettinger P, Ruiz-Perez F, Donald JW, Palmer T, Linke D, Henderson IR, Autenrieth IB. C-terminal amino acid residues of the trimeric autotransporter adhesin YadA of Yersinia enterocolitica are decisive for its recognition and assembly by BamA. Mol Microbiol. 2010;78:932–946. doi: 10.1111/j.1365-2958.2010.07377.x. [DOI] [PubMed] [Google Scholar]
- Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 2006;14:264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Macasev D, Whelan J, Newbigin E, Silva-Filho MC, Mulhern TD, Lithgow T. Tom22′, an 8-kDa trans-site receptor in plants and protozoans, is a conserved feature of the TOM complex that appeared early in the evolution of eukaryotes. Mol Biol Evol. 2004;21:1557–1564. doi: 10.1093/molbev/msh166. [DOI] [PubMed] [Google Scholar]
- Meng G, Surana NK, St Geme JW 3rd, Waksman G. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 2006;25:2297–2304. doi: 10.1038/sj.emboj.7601132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D, Kozjak V, Wiedemann N, Lohaus C, Meyer HE, Guiard B, Pfanner N, Meisinger C. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J Biol Chem. 2004;279:22781–22785. doi: 10.1074/jbc.C400120200. [DOI] [PubMed] [Google Scholar]
- Model K, Meisinger C, Prinz T, Wiedemann N, Truscott KN, Pfanner N, Ryan MT. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat Struct Biol. 2001;8:361–370. doi: 10.1038/86253. [DOI] [PubMed] [Google Scholar]
- Paschen SA, Neupert W, Rapaport D. Biogenesis of β-barrel membrane proteins of mitochondria. Trends Biochem Sci. 2005;30:575–582. doi: 10.1016/j.tibs.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Paschen SA, Rothbauer U, Kaldi K, Bauer MF, Neupert W, Brunner M. The role of the TIM8–13 complex in the import of Tim23 into mitochondria. EMBO J. 2000;19:6392–6400. doi: 10.1093/emboj/19.23.6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Pfaller R, Pfanner N, Neupert W. Mitochondrial protein import—bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J Biol Chem. 1989;264:34–39. [PubMed] [Google Scholar]
- Pfanner N, Wiedemann N, Meisinger C, Lithgow T. Assembling the mitochondrial outer membrane. Nat Struct Mol Biol. 2004;11:1044–1048. doi: 10.1038/nsmb852. [DOI] [PubMed] [Google Scholar]
- Rapaport D. How to find the right organelle—targeting signals in mitochondrial outer membrane proteins. EMBO Rep. 2003;4:948–952. doi: 10.1038/sj.embor.embor937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Neupert W. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J Cell Biol. 1999;146:321–331. doi: 10.1083/jcb.146.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Behrens M, Scherer P, Pratje E, Michaelis G, Schatz G. Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J. 1991;10:247–254. doi: 10.1002/j.1460-2075.1991.tb07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Waizenegger T, Habib SJ, Lech M, Mokranjac D, Paschen SA, Hell K, Neupert W, Rapaport D. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep. 2004;5:704–709. doi: 10.1038/sj.embor.7400183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Bos MP, Rapaport D, Tommassen J. The mitochondrial porin, VDAC, has retained the ability to be assembled in the bacterial outer membrane. Mol Biol Evol. 2010;27:887–895. doi: 10.1093/molbev/msp294. [DOI] [PubMed] [Google Scholar]
- Walther DM, Papic D, Bos MP, Tommassen J, Rapaport D. Signals in bacterial b-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc Natl Acad Sci USA. 2009a;106:2531–2536. doi: 10.1073/pnas.0807830106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Rapaport D, Tommassen J. Biogenesis of β-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cell Mol Life Sci. 2009b;66:2789–2804. doi: 10.1007/s00018-009-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Kozjak V, Chacinska A, Schönfish B, Rospert S, Ryan MT, Pfanner N, Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- Wimley WC. The versatile β-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- Wollmann P, Zeth K, Lupas AN, Linke D. Purification of the YadA membrane anchor for secondary structure analysis and crystallization. Int J Biol Macromol. 2006;39:3–9. doi: 10.1016/j.ijbiomac.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Yamano K, Yatsukawa Y, Esaki M, Hobbs AE, Jensen RE, Endo T. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J Biol Chem. 2008;283:3799–3807. doi: 10.1074/jbc.M708339200. [DOI] [PubMed] [Google Scholar]