We show here that zonula occludens (ZO)-3 acts to sequester cyclin D1 at intercellular tight junctions. This study highlights an important new role for ZO family proteins in regulating epithelial cell proliferation through stabilization of cyclin D1 during mitosis.
Abstract
Coordinated regulation of cell proliferation is vital for epithelial tissue homeostasis, and uncontrolled proliferation is a hallmark of carcinogenesis. A growing body of evidence indicates that epithelial tight junctions (TJs) play a role in these processes, although the mechanisms involved are poorly understood. In this study, we identify and characterize a novel plasma membrane pool of cyclin D1 with cell-cycle regulatory functions. We have determined that the zonula occludens (ZO) family of TJ plaque proteins sequesters cyclin D1 at TJs during mitosis, through an evolutionarily conserved class II PSD-95, Dlg, and ZO-1 (PDZ)-binding motif within cyclin D1. Disruption of the cyclin D1/ZO complex through mutagenesis or siRNA-mediated suppression of ZO-3 resulted in increased cyclin D1 proteolysis and G0/G1 cell-cycle retention. This study highlights an important new role for ZO family TJ proteins in regulating epithelial cell proliferation through stabilization of cyclin D1 during mitosis.
INTRODUCTION
Epithelial tight junctions (TJs) protect an organism from its environment by establishing a barrier between epithelial cells. Transmembrane TJ proteins, such as claudins, occludin, and junctional adhesion molecule (JAM), bridge the extracellular space and form contacts with adjacent cells (Tsukita et al., 2001). Transmembrane cell–cell adhesion molecules bind to cytoplasmic scaffolding proteins forming a protein plaque. This protein scaffold then interacts with the actin cytoskeleton and a host of cell signaling molecules (Paris et al., 2008; Balda and Matter, 2009). The TJ scaffold plaque is mainly composed of the zonula occludens (ZO-1, ZO-2, and ZO-3) and membrane-associated guanylate kinase (MAGUK) family proteins, both of which contain one or more protein–protein interaction domains referred to as PDZ (PSD-95, Dlg, and ZO-1) domains (Beatch et al., 1996; Haskins et al., 1998; Gonzalez-Mariscal et al., 2000). PDZ domains facilitate the formation of multimolecule protein complexes through homotypic association, as well as by binding to short C-terminal residues referred to as PDZ-binding motifs (Harris and Lim, 2001). These highly conserved motifs are categorized as class I (consensus motif = S/T-X-V-COOH), class II (X-φ-X-φ-COOH), or class III (D/E/R/K-X-φ-COOH, where X is any aa and φ is any hydrophobic aa), and confer specificity for protein–protein interactions (Giallourakis et al., 2006).
Recent studies show that TJs also function as reservoirs for cell-signaling components that regulate the proliferative potential of epithelial cells (Matter and Balda, 2007; Guillemot et al., 2008; Balda and Matter, 2009). For example, the TJ plaque protein ZO-1 was found to sequester the transcription factor ZO-1–associated nucleic acid binding protein (ZONAB) at TJs during states of high confluence, thereby suppressing cyclin D1 gene transcription in a cell density–dependent manner (Balda et al., 2003). Similarly, ZO-2 has also been found to regulate cyclin D1 protein levels and increase cyclin D1 protein decay. The molecular mechanisms involved in these processes, however, are incompletely understood (Huerta et al., 2007; Tapia et al., 2009). ZO-3 is an epithelium-specific ZO family protein that, in Madin Darby canine kidney (MDCK) cells, has been shown to regulate Rho GTPase–related cell-signaling events (Inoko et al., 2003; Wittchen et al., 2003). The contribution of ZO-3 in the regulation of cyclin D1–mediated proliferative processes has not been investigated.
Cyclin D1 is a major regulator of cell proliferation. In response to mitogenic signals, high cyclin D1 protein levels drive the cell through the G1-to-S phase transition (Fu et al., 2004). Cyclin D1 forms a complex with cyclin-dependent kinases (Cdks), which together translocate to the nucleus and signal to the cell to proceed through the G1-to-S phase transition (Sherr, 1995). High levels of cyclin D1 are inhibitory with respect to DNA synthesis, however, and S-phase progression requires cyclin D1 degradation. Nuclear export and proteolytic degradation of cyclin D1 proceeds through the cell cycle–dependent phosphorylation of cyclin D1 at a C-terminal threonine residue (Thr-286) (Diehl et al., 1998; Guo et al., 2005). Several kinases have been implicated in this process, notably GSK3-β, ATM/ATR, and IκB α kinases (Diehl et al., 1997; Kwak et al., 2005; Hitomi et al., 2008).
To further investigate the role of ZO family TJ proteins in cyclin D1–mediated cell-cycle progression, we first investigated the subcellular localization of cyclin D1 in colonic enterocytes by immunofluorescence staining. Cyclin D1 and Cdk4 were found to colocalize with TJ proteins in the proliferative zone of mouse intestinal crypt epithelial cells. Additionally, analysis of sucrose density gradient fractions revealed that cyclin D1 cosediments with TJ proteins in the plasma membrane-containing fractions. Consistent with these findings, coimmunoprecipitation studies confirmed a cyclin D1/ZO-3 protein complex, and protein binding assays revealed direct, PDZ-motif–mediated binding between cyclin D1 and ZO-3. Green fluorescent protein (GFP)-cyclin D1 fusion proteins lacking this motif failed to localize at TJs. Surprisingly, cyclin D1 was enriched in TJs during mitosis. Further investigation revealed that siRNA-mediated knockdown of ZO-3 disrupted mitotic cyclin D1 TJ localization, decreased cyclin D1 protein stability, and suppressed epithelial cell proliferation. To our knowledge, these data demonstrate for the first time that cyclin D1 is stabilized during mitosis through TJ sequestration by ZO proteins, representing a novel mechanism of cell-cycle regulation.
RESULTS
Cyclin D1 colocalizes with the TJ plaque protein ZO-1
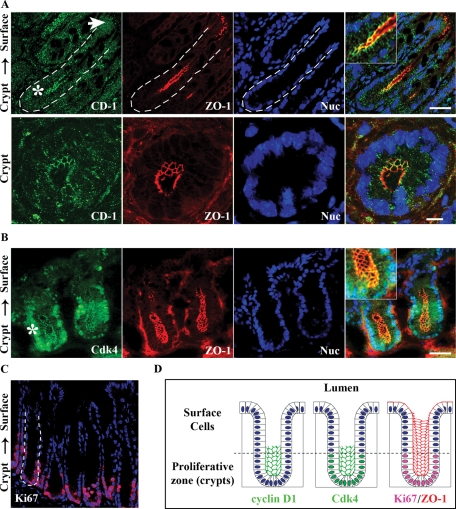
Cyclin D1 is a key promoter of G1-S phase transition and a major regulator of cell proliferation (Malumbres and Barbacid, 2001; Balda et al., 2003; Sherr and Roberts, 2004). Recent reports have linked ZO family TJ proteins to cyclin D1 gene expression and protein stability (Balda et al., 2003; Sourisseau et al., 2006; Tapia et al., 2009). To explore the relationship between TJ scaffolding proteins and cyclin D1 in the intestinal epithelium, we first examined the subcellular distribution of cyclin D1 by immunofluorescence labeling and confocal imaging in native murine colonic epithelial cells. Colocalization of cyclin D1 with the TJ scaffolding protein ZO-1 was observed in the colonic crypt epithelium (Figure 1A). Interestingly, whereas cyclin D1 was localized in TJs of crypt epithelial cells (Figure 1A, bottom panels), epithelial cells at the luminal surface (Figure 1A, white arrow) lacked detectable cyclin D1. Similarly, immunohistochemical staining revealed that cyclin D1 levels decrease along the crypt-to-surface cell axis, indicating that cyclin D1 is differentially expressed within colonic crypts (Supplemental Figure 1). Cdk interactions mediate downstream cyclin D1 function with respect to cell proliferation. Therefore we investigated Cdk4 localization in colonic crypts by immunofluorescence labeling and confocal microscopy. Figure 1B shows Cdk4 localization in TJs in addition to its nuclear localization in crypt epithelial cells. The TJ distribution of Cdk4 further supports localization of its frequent binding partner, cyclin D1, in TJs. Both cyclin D1 and Cdk4 preferentially localize at TJs within the proliferative zone of colonic crypts, as indicated by the proliferation marker, Ki67 (Figure 1C, magenta). Figure 1D summarizes the immunofluorescence data with cyclin D1 and Cdk4 colocalizing with ZO-1 at TJs in colonic crypt epithelial cells. Analogous to cyclin D1, Cdk4 was localized in the Ki67 positive proliferative compartment and was not visualized in epithelial cells at the luminal surface. Together these data show that both cyclin D1 and Cdk4 colocalize with ZO-1 at the cell–cell contacts of proliferative colonic epithelial cells.
FIGURE 1:
Cyclin D1 and Cdk4 localize at TJs in intestinal epithelial cells in vivo and in vitro. (A) Immunofluorescence imaging of cyclin D1 (green) colocalization with ZO-1 (red) in crypt epithelia of murine distal colon tissue. Nuclei are stained with Topro-3 (blue). White arrow designates epithelial surface cells facing the lumen; the asterisk designates crypt regions. Scale bar = 40 μm. Merged inset at 2× magnification. Bottom panels: Magnified view of murine colonic crypts. Scale bar = 10 μm. (B) Cdk4 (green) colocalizes with ZO-1 (red) in the base of colonic crypts. (C) Mouse distal colon crypts stained for the proliferation marker Ki67 (red) and nuclei (blue). (D) Schematic summarizing the immunofluorescence data with cyclin D1 and Cdk4 colocalizing with ZO-1 in colonic crypt epithelial cells. Cyclin D1 and Cdk4 are not present in lumen-facing surface cell populations, but appear restricted to the proliferative zone as marked by Ki67.
Cyclin D1 cofractionates with TJ proteins in plasma membranes
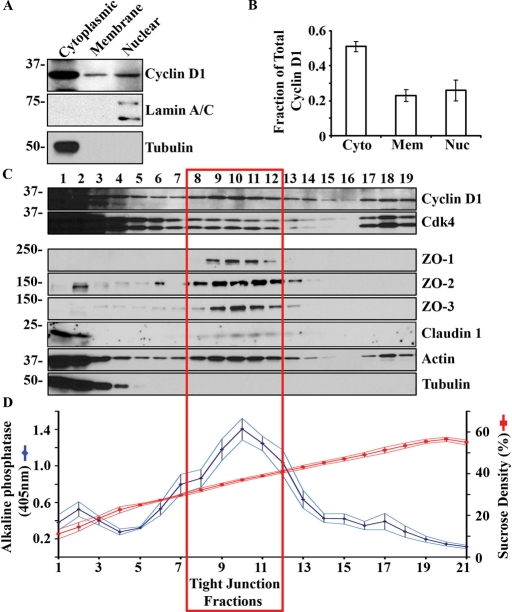
To confirm cyclin D1 membrane association, we performed subcellular fractionation of model human colonic epithelial cells (SKCO-15 cells). Cytoplasmic, membrane, and nuclear fractions were isolated by detergent-free nitrogen cavitation and ultracentrifugation (described in Materials and Methods). The cytosolic and nuclear fractions were identified by immunoblotting for tubulin and lamin A/C, respectively (Figure 2A). Densitometric quantitation revealed that a sizable proportion of cyclin D1 is found in the cell membrane fractions (23% ± 0.2%, Figure 2B) relative to total protein levels, as compared with cytosolic (50% ± 3.0%) and nuclear fractions (26% ± 3.0%).
FIGURE 2:
Cyclin D1 cosediments with TJs in polarized epithelial cells. (A) Representative Western blot of cyclin D1 in isolated SKCO-15 cytosolic, cell membrane, and nuclear fractions. Tubulin is shown as a marker of cytosolic protein and lamin A/C as a nuclear marker. (B) Densitometry analysis of Western blot; n = 3. (C) Sucrose density gradient fractionation of SKCO-15 cell lysate showing cosedimentation of cyclin D1 with tight junction proteins. (D) Sucrose density of indicated fractions (15–60%; red) and ALP activity in the corresponding fractions (blue; n = 8).
The cell membrane preparation described earlier in the text contains a complex mixture of cellular membranes. Therefore, to determine if cyclin D1 is associated with the TJ containing plasma membranes, we performed additional subcellular fractionation using sucrose density gradients. SKCO-15 cell lysates were cleared of nuclear material and applied to a continuous 15–60% sucrose gradient. Alkaline phosphatase (ALP) activity was measured to identify plasma membrane containing fractions (Kaoutzani et al., 1993). Fractions with high ALP activity (Figure 2C, fractions 9–12) contained TJ proteins claudin 1 and ZO-1/2/3. Western blot analysis of cyclin D1 and Cdk4 demonstrated cosedimentation of these proteins with TJ proteins (red box). Additionally, cyclin D1 and Cdk4 cosedimented with high-density, actin-rich fractions (Figure 2C, fractions 17–19). These findings are consistent with immunolocalization studies showing cyclin D1 and Cdk4 in TJs (Figure 1, A and B).
Cyclin D1 binds to ZO-3 through direct PDZ-mediated interactions
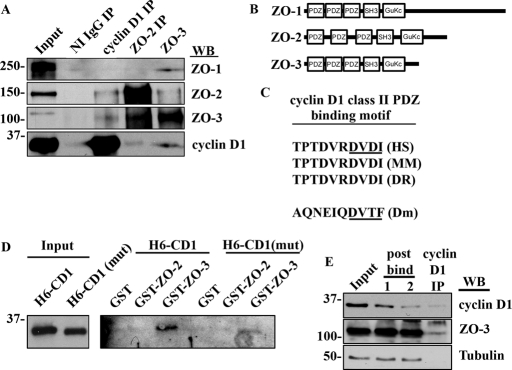
The biochemical data just shown reveal that ZO-2 and ZO-3 cosediment with cyclin D1 in plasma membrane containing fractions suggesting that ZO proteins associate with cyclin D1. Therefore we assessed this possibility by immunoprecipitation (IP) and Western blot analysis of SKCO-15 cell lysates for ZO/cyclin D1 protein complexes. As shown in Figure 3A, both ZO-2 and ZO-3 coimmunoprecipitated with cyclin D1. Interestingly, ZO-3 complexes coimmunoprecipitated with ZO-1, ZO-2, and cyclin D1 (Figure 3A). In addition, ZO-2 immunoprecipitates contained ZO-3 and low levels of cyclin D1. These data reveal that cyclin D1 exists in a protein complex with ZO proteins.
FIGURE 3:
Cyclin D1 forms a PDZ-dependent complex with ZO-3. (A) Western blot of IP experiments with nonimmune (NI) immunoglobulin (Ig)G, cyclin D1, ZO-2, and ZO-3. (B) Immunodepletion assay showing that only a fraction of ZO-3 is bound to cyclin D1 (C) Schematic diagram of ZO-1, ZO-2, and ZO-3 showing multiple PDZ domains within ZO family proteins. (D) Amino acid sequence alignment of human (HS), murine (MM), zebrafish (DR), and Drosophila (Dm) cyclin D1 C-terminal tails showing a conserved class II PDZ binding motif (underlined). (E) GST–ZO-3 fusion proteins interact preferentially with His6–cyclin D1 (H6 CD-1) containing an intact PDZ binding motif and show attenuated binding to His6–cyclin D1 mutant lacking the putative C-terminal PDZ binding motif [H6-CD1(mut)]. (E) Immunodepletion assay showing retention of the majority of cellular ZO-3 in the postbind fractions after iterative cyclin D1 IP.
ZO-1, ZO-2, and ZO-3 each contain three protein–protein interaction domains, called PDZ binding domains, which provide a scaffolding function in TJs (Figure 3B). To investigate whether ZO proteins interact with cyclin D1 through these domains, we performed an amino acid sequence analysis of the cyclin D1 C-terminal tail. We found that cyclin D1 has an evolutionarily conserved class II PDZ binding motif (consensus site: DVDI-COOH, Figure 3C). Interestingly, the Drosophila cyclin homologue, CycD1, lacks amino acid identity with mammalian cyclin D1, yet contains a C-terminal class II PDZ motif (Figure 3C). These findings suggest that cyclin D1 and ZO family proteins may directly associate through PDZ domain-mediated interactions. To test this hypothesis, cyclin D1 and ZO-2/3 binding assays were performed using bacterially expressed, full-length recombinant proteins. Glutathione S-transferase (GST), GST–ZO-2, or GST–ZO-3 was incubated with His6-labeled cyclin D1 to assay for direct binding. As shown in Figure 3D, His6-cyclin D1 (H6-CD1) binds directly to GST–ZO-3, but not to GST–ZO-2, showing the specificity of ZO-3–cyclin D1 interactions. A His6-cyclin D1 truncation mutant was constructed (H6-CD1(mut)) which lacked the last four amino acids (DVDI) of the PDZ binding motif. A significant decrease in H6-CD1(mut)/ZO-3 association was observed when compared with full-length protein interactions, indicating that PDZ domain interactions are involved in ZO-cyclin D1 complex formation (Figure 3D). We next assessed the proportion of cyclin D1 bound to ZO-3 through immunodepletion assays. SKCO-15 cell lysates were subjected to iterative IPs using cyclin D1 antibodies. The postIP supernatants (post bind) were monitored for both cyclin D1 and ZO-3 protein levels. As shown in Figure 3E, cyclin D1 protein removal from protein lysates results in little change in ZO-3 protein levels. Therefore a significant fraction of ZO-3 in the cell is not bound to cyclin D1.
PDZ-mediated interactions regulate cyclin D1 subcellular localization
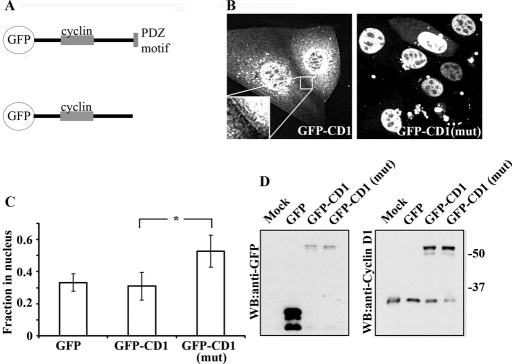
To determine whether ZO protein/cyclin D1 interactions regulate the distribution of cyclin D1 in epithelial cells, we expressed GFP-cyclin D1 fusion proteins either as full-length proteins or constructs lacking the last four amino acids that constitute the PDZ binding motif (Figure 4A). When expressed in SKCO-15 epithelial cells, full-length GFP-cyclin D1 was localized primarily in the cytoplasm and nucleus, with a subpool targeted to the plasma membrane (Figure 4B, inset). In mutant cyclin D1 (which lacks the C-terminal PDZ binding motif), however, we did not observe plasma membrane localization, but rather a striking relocalization of cyclin D1 into the nucleus (GFP-CD1(mut); Figure 4B, right panel). Transient overexpression of GFP, GFP-cyclin D1, or GFP cyclin D1(mut) in SKCO-15 cells revealed altered subcellular localization of GFP-cyclin D1(mut) when compared with GFP-cyclin D1 full-length protein. In contrast, GFP-cyclin D1(mut) protein was found predominantly in the nucleus, with only high-expressing cells exhibiting cytoplasmic GFP-cyclin D1(mut) (Figure 4, B and C). GFP-cyclin D1(mut) protein exhibited significantly higher nuclear localization (50% ± 10% of total fluorescence, p < 0.001) when compared with wild-type controls (30% ± 8%) or GFP alone (33% ± 5%). To ensure that full-length proteins were expressed in transfected cells, GFP-cyclin D1 protein levels were analyzed by Western blotting with both GFP and cyclin D1 antibodies (Figure 4D). Protein expression levels were similar for both full length and GFP-cyclin D1(mut), and the GFP fusion protein levels were comparable to endogenous protein levels. Importantly, GFP–cyclin D1 fusion proteins were expressed as full-length protein, and no GFP cleavage was observed. These results demonstrate that cyclin D1 TJ localization is regulated by the cyclin D1 C-terminal PDZ-binding motif.
FIGURE 4:
The cyclin D1 PDZ motif is required for TJ localization. (A) Schematic representing GFP–cyclin D1 [GFP-CD-1 and GFP-CD-1(mut)] fusion constructs. (B) Live-cell imaging of GFP–cyclin D1, but not GFP–cyclin D1(mut), located to sights of cell–cell contact (see inset). GFP–cyclin D1(mut) is predominantly localized within the nucleus (right panel). (C) Subcellular localization analysis of overexpressed GFP, GFP–cyclin D1, or GFP–cyclin D1(mut) in paraformaldehyde-fixed SKCO-15 cells. Ablation of cyclin D1 C-terminal PDZ binding motif results in redistribution of cyclin D1 to the nucleus (*p < 0.001, n = 40 cells per condition per experiment; n = 3). (D) Western blot of mock transfected, GFP-, GFP–cyclin D1–, or GFP–cyclin D1(mut)–expressing SKCO-15 cells with anti-GFP (left panel) and anti-cyclin D1 (right panel).
Cyclin D1 localizes at TJ predominantly in mitotic cells.
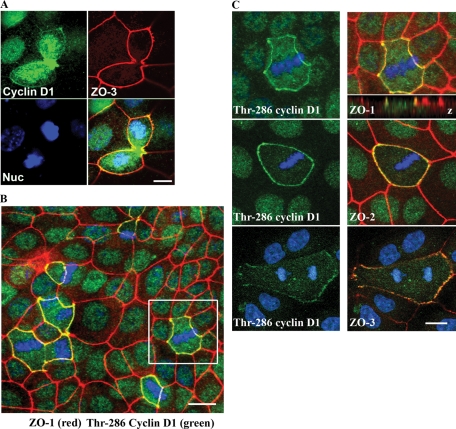
GFP-cyclin D1 was observed at cell contacts in a minority of transfected cells. Furthermore, as shown in Figure 3D, the majority of ZO-3 in the cell does not appear to be bound to cyclin D1. We therefore hypothesized that the cyclin D1/ZO-3 interaction was a transient, cell cycle–dependent interaction. Therefore we screened subconfluent (actively dividing) epithelial cells for cyclin D1 at cell contacts. Surprisingly, we observed that cyclin D1 localized robustly at cell contacts within actively dividing cells, where cyclin D1 cell membrane staining during mitosis colocalized with ZO-3 (Figure 5A). Robust cyclin D1 staining was also seen at the cleavage furrows of late-stage dividing cells, presenting the possibility of additional cyclin D1 interactions with actin structures or adherens junctions. We did not see an increased membrane localization of Cdk4 in mitotic cells, however (Supplemental Figure 8).
FIGURE 5:
Cyclin D1 accumulates at cell junctions during mitosis. (A) Cyclin D1 (green) accumulates at cell–cell contacts (ZO-3; red) during mitosis. Scale bar = 20 μm. (B) Immunofluorescence colocalization of Thr-286 cyclin D1 (green) with ZO-1 (red) at cell contacts during mitosis. Scale bar = 40 μm. (C) Immunofluorescence costaining of ZO family proteins (red) and Thr-286 cyclin (green) D1 at cell contacts during cell division. Z-plane colocalization of cyclin D1 and ZO-1 confirms TJ association (top panel inset). Scale bar = 10 μm.
Phosphorylation of cyclin D1 at Thr-286 has been shown to promote cyclin D1 proteolysis and, therefore, S-phase cell-cycle entry (Diehl and Sherr, 1997; Diehl et al., 1997, 1998; Guo et al., 2005). We therefore asked whether membrane-sequestered cyclin D1 contained Thr-286–phosphorylated cyclin D1. Thr-286 cyclin D1 shows a striking accumulation at cell contacts in all cells undergoing mitosis, and can be seen at all stages of mitosis (Figure 5B). Furthermore, z-stack reconstruction of confocal images confirmed Thr-286 cyclin D1 localization to apical TJ complexes (Figure 5C). Thr-286–phosphorylated cyclin D1 at cell junctions during mitosis also colocalized with ZO family proteins ZO-2 and ZO-3 (Figure 5C). Similar results were found in MDCK cells (Supplemental Figure 3), suggesting a shared mechanism among epithelial cells. Furthermore, we have determined that Thr-286–phosphorylated cyclin D1 is a small fraction of the total cyclin D1 in cell-cycle heterogeneous cultures (Supplemental Figure 3).
Cyclin D1/ZO-3 interactions are required for TJ localization and cyclin D1 protein stability.
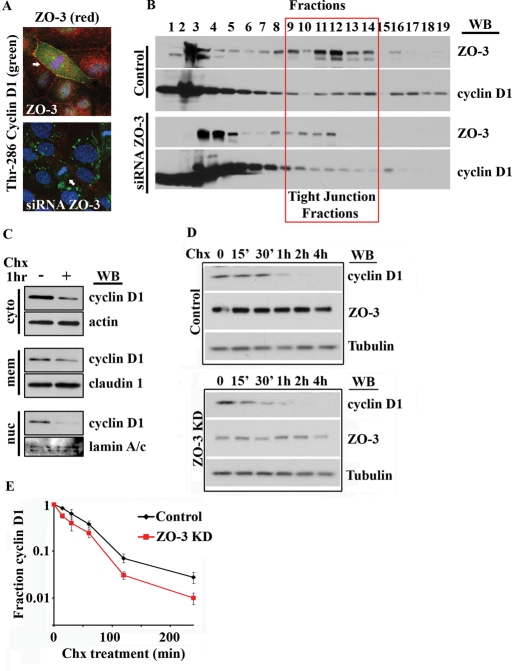
We next asked whether ZO-3 was required for cyclin D1 localization in mitotic cells. We therefore examined cyclin D1 localization in epithelial cells depleted of ZO-3 by immunofluorescence labeling. Given our finding that PDZ-mediated interactions regulate cyclin D1 nuclear localization (Figure 4B), we first investigated whether ZO-3 suppression altered the nuclear distribution of cyclin D1. ZO-3 knockdown cells were identified by immunostaining and scored for the presence of nuclear cyclin D1. No significant difference in the fraction of cells exhibiting nuclear cyclin D1 was detectable between control and siRNA ZO-3–treated cells (Supplemental Figure 6). We next assessed the role of ZO-3 in the localization of cyclin D1 at TJs. As shown in Figure 6A, protein suppression of ZO-3 (red) prevented TJ accumulation of Thr-286 cyclin D1 (green) in mitotic cells. Furthermore, sucrose density cosedimentation of cyclin D1 with TJ proteins was diminished after siRNA-mediated suppression of ZO-3 (Figure 6B). These observations further support a role for ZO-3 in cyclin D1 TJ sequestration.
FIGURE 6:
ZO-3 is required for cyclin D1 TJ localization and cyclin D1 protein stability. (A) SKCO-15 cells treated with siRNA directed against ZO-2 (top panel) or ZO-3 (bottom panel) and immunostained for Thr-286 cyclin D1 (green) and ZO proteins (red). Arrows highlight mitotic cells. (B) Sucrose density gradient of colonic epithelial cells after ZO-3 depletion. Cyclin D1–ZO-3 cosedimentation in TJ fractions is diminished after ZO-3 depletion. (C) Subcellular fractionation reveals differential cyclin D1 protein stability after Chx treatment (50 μM, 1 h). (D) Cyclin D1 shows lower protein stability in ZO-3 siRNA-treated cultures. SKCO-15 cell lysates were harvested at the indicated times up to 4 h posttreatment with Chx (50 μM). (B) Densitometric analysis of Western blot data shows cyclin D1 protein levels in SKCO-15 cells after Chx treatment (n = 3).
Cyclin D1 phosphorylation at Thr-286 has been shown to induce rapid cyclin D1 proteolysis (Diehl et al., 1998). Therefore the accumulation of Thr-286 cyclin D1 at TJs may play a role in regulating cyclin D1 protein stability. To test this hypothesis, we treated SKCO-15 cells with cycloheximide (Chx) to inhibit protein synthesis and then analyzed cyclin D1 protein levels in cytoplasmic, membrane, and nuclear fractions (Figure 6C). We observed that the differential cyclin D1 protein stability is dependent on subcellular localization, with a low-stability nuclear subpool and highly stable, membrane-bound cyclin D1 fraction (Figure 6C). Given the role of ZO-3 in cyclin D1 membrane localization, we next asked whether ZO-3 regulated cyclin D1 protein stability. Cyclin D1 protein levels were monitored in Chx-treated control siRNA cells as well as ZO-3 siRNA–treated cells. Western blot analysis revealed that the protein half-life of cyclin D1 under control conditions is 48 min (±8 min), whereas in the ZO-3 knockdown cells, it is reduced to 32.6 min (±9.4 min) (Figure 6, B and C). These findings demonstrate that, in cells with decreased ZO-3, cyclin D1 is degraded ∼30% faster compared with control epithelial cells.
ZO-3/cyclin D1 interactions regulate G1-to-S-phase cell-cycle transition
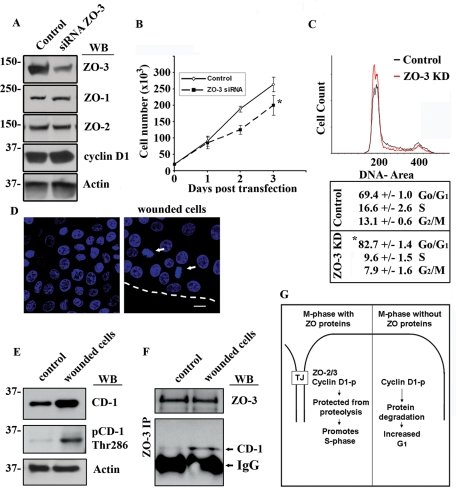
We next hypothesized that the shorter cyclin D1 half-life observed in ZO-3 knockdown cells would induce a decrease in the proportion of replicating cells. Figure 7A highlights siRNA-induced ZO-3 down-regulation as determined by Western blotting. The suppression of ZO-3 had no detectable effects on the protein expression of ZO-1 or ZO-2 (Figure 7A). To determine whether the disruption of cyclin D1–ZO-3 protein interactions altered epithelial cell proliferation, subconfluent SKCO-15 monolayers transiently transfected with ZO-3 siRNA were monitored for cell growth for up to 3 d after transfection (Figure 7B). We observed a significant decrease in cell number in epithelial cells that had depleted ZO-3 protein levels when compared with control siRNA transfected cells (day 3, control = 2.6 × 105 ± 0.2 vs. siRNA ZO-3 = 2.0 × 105 ± 0.3 cells, p < 0.001, n = 4). This response was not due to increased apoptosis, as ZO-3 suppression did not increase terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining (Supplemental Figure 4). Cyclin D1 acts to promote G1-to-S-phase transition and cell proliferation. To determine the effect of ZO-3 depletion on cell-cycle status, cells were transiently transfected with control siRNA (Control) or ZO-3 siRNA (ZO-3) and analyzed by flow cytometry. First, we identified cell-cycle status of SKCO-15 cells using propidium iodide staining (Figure 7B), and siRNA knockdown of ZO-3 was confirmed by flow cytometry (unpublished data). Indeed, ZO-3–depleted cells contain a significantly larger pool of cells in G1 (82.7 ± 1.0%) as compared with control cells (69.4 ± 1.0%) (Figure 6B; p < 0.01). The increase in G1 cells was accompanied by a commensurate decrease in S and G2/M phase cell populations (Figure 6B).
FIGURE 7:
ZO-3–cyclin D1 interaction regulates cell proliferation in epithelial cells by promoting S-phase transition. (A) Western blot analysis of SKCO-15 cell lysates treated with control siRNA (Control) or ZO-3 directed siRNA. (B) Growth curve showing cell number over 3 d posttransfection with control siRNA (Control, ◊) or ZO-3 directed siRNA (▪) (*p < 0.001, n = 4). (C) Propidium iodide–stained SKCO-15 cells, treated with either scrambled siRNA (Control, black) or ZO-3–directed siRNA (red) were assayed for cell-cycle status by flow cytometry (n = 2). (D) Confluent SKCO-15 cell monolayers (control) were scratch wounded to stimulate cell proliferation (dotted line marks wound edge; mitotic cells are marked with white arrows). Scale bar = 20 μm. (E) Scratch-wounded monolayers show increased cyclin D1 as well as increased phospho-Thr-286–cyclin D1 when compared with confluent cultures (control). (F) Western blot showing enhanced ZO-3 coimmunoprecipitation with cyclin D1 from lysates of scratch-wounded cells. (G) Hypothetical model whereby ZO-3 acts to sequester cyclin D1 at TJs during mitosis. This prevents cyclin D1 degradation during M-phase and promotes S-phase transition. Without ZO-3, cyclin D1 is degraded during M-phase, delaying cells in G1.
In vivo, the intestinal epithelium is continuously regenerating. Indeed, in response to mucosal damage or inflammation, reparative wound-healing processes are associated with increased epithelial cell replication (Nava et al., 2010). To determine whether increased proliferation enhanced ZO-3–cyclin D1 interactions, we performed an in vitro scratch wound assay (see Materials and Methods). Mechanical wounding of epithelial monolayers was found to increase the number of mitotic cells, particularly in the second or third row of cells behind the wound edge (Figure 6C, arrows). Immunoblot analysis of whole-cell lysates from confluent or wounded SKCO-15 monolayers showed an increase in cyclin D1 as well as Thr-286 phospho-cyclin D1 in wounded monolayers (Figure 7D). IP of ZO-3, from lysates derived from resealing wounds, revealed an increase in the amount of cyclin D1 that coprecipitated with ZO-3 (Figure 7E). These data show that the ZO-3–cyclin D1 complex is enhanced in actively proliferating cells (Figure 7F). Taken together, these data show that cyclin D1–ZO-3 interactions are important for appropriate cyclin D1 subcellular localization, protein stability, and, ultimately, cell proliferation.
DISCUSSION
Correct spatial and temporal regulation of cyclin D1 is crucial for its function as a mediator of cell-cycle advancement. Indeed, aberrant cyclin D1 protein levels and localization have been reported to contribute to carcinogenesis (Gladden and Diehl, 2005). As a case in point, a spontaneously occurring mutation in the cyclin D1 gene generates an alternate splice variant (cyclin D1b) that has enhanced transformative properties due to its constitutive nuclear localization (Solomon et al., 2003; Fu et al., 2004). Importantly, the cyclin D1b splice variant results in a truncated protein that lacks the conserved C-terminal PDZ motif characterized in this report. We showed earlier in the text that deletion of the C-terminal PDZ-binding motif is sufficient to displace cyclin D1 from TJs and promote nuclear translocation. Furthermore, our demonstration of a direct, PDZ-mediated interaction between cyclin D1 and ZO-3 provides strong evidence for ZO-3 regulation of cyclin D1 localization. siRNA-mediated suppression of ZO-3, however, did not detectably affect the nuclear localization of endogenous cyclin D1 (Supplemental Figure 4). These studies suggest that ZO-3 regulates cyclin D1 TJ localization, whereas other cyclin D1 binding partners regulate nuclear localization.
Our data indicate that cyclin D1 is sequestered at TJs to prevent its degradation during mitosis. Recent studies in NIH3T3 cells reveal that iterative cell proliferation requires that cyclin D1, as well as Thr-286–phosphorylated cyclin D1, accumulate throughout G2, M-phase, and G1 (Guo et al., 2005). DNA synthesis then requires rapid S-phase degradation of cyclin D1 (Pagano et al., 1994; Fukami-Kobayashi and Mitsui, 1999). These findings would indicate that cyclin D1 levels are maintained throughout mitosis. Phosphorylation at Thr-286, however, is considered a requirement for cyclin D1 protein degradation. Therefore we propose that formation of the ZO-3/cyclin D1 complex has a protective effect against protein degradation during mitosis (Figure 7G). Consistent with this hypothesis, siRNA-mediated suppression of ZO-3 leads to lower cyclin D1 stability, loss of Thr-286 cyclin D1 at TJs, and decreased overall cell proliferation. These findings correlate with an accumulation of Go/G1 cells in ZO-3–depleted epithelial cells and indicate that ZO-3 promotes cell-cycle progression.
Although we were able only to confirm direct binding between ZO-3 and cyclin D1, both endogenous ZO-2 and ZO-3 were found to coimmunoprecipitate with cyclin D1 in a protein complex (Figure 3A). Therefore we cannot, as yet, rule out the possibility of in vivo interactions between ZO-2 and cyclin D1. ZO-1, ZO-2, and ZO-3 are closely related proteins, and several recent reports have found that ZO proteins are negative regulators of cyclin D1 gene expression and protein stability (Huerta et al., 2007; Balda and Matter, 2009; Tapia et al., 2009). ZO-1 is known to suppress cell growth through the sequestration of ZONAB, thereby suppressing cyclin D1 transcription (Balda and Matter, 2009). ZO-1 may also play a role in the regulation of cyclin D1 membrane localization, although we believe this function is secondary to ZO-1's ability to act as a ZO-3 scaffold (Supplemental Figure 7). ZO-2 nuclear localization has been previously reported to reduce cyclin D1 gene expression and protein stability (Diehl et al., 1998; Islas et al., 2002; Betanzos et al., 2004; Huerta et al., 2007; Traweger et al., 2008). Indeed, overexpression of ZO-2 in MDCK cells inhibits cell proliferation due to Go/G1 cell-cycle block (Tapia et al., 2009). Conflicting studies have reported, however, that ZO-2 plays a proproliferative role (Traweger et al., 2008). What could account for the apparently contradictory findings? First, in MDCK cells, ZO protein nuclear localization preferentially occurs in low-density cultures and is diminished in confluent monolayers (Gottardi et al., 1996; Islas et al., 2002). These observations may point to a role for nuclear ZO proteins during conditions of relatively increased cell proliferation, before monolayer confluence. Second, ZO proteins may perform a dual role in regulating epithelial proliferation, with proliferative effects during sparse cell states and inhibitory effects in highly differentiated monolayers. Last, the subcellular localization and protein stability of cyclin D1 fluctuates during the cell cycle to promote cell division (Gladden and Diehl, 2005; Guo et al., 2005). Indeed, enhanced protein stability of cyclin D1 due to ZO protein binding would promote G1-to-S phase progression, as high cyclin D1 protein levels initiate S-phase (Sherr, 1995). Conversely, low cyclin D1 levels promote DNA synthesis during S-phase. Therefore cell-cycle phase is highly relevant in assessing the role of cyclin D1 inhibitors with respect to cell proliferation. Further studies will be required to determine the mechanism through which ZO family proteins regulate cell growth. Elucidating the role of cyclin D1/ZO protein interactions in cell-cycle progression will require careful attention to both subcellular localization and cell-cycle status. Based on our data, however, we conclude that ZO-3/cyclin D1 interactions promote cell-cycle progression through inhibition of cyclin D1 proteolysis during mitosis.
This study highlights a novel role for ZO-3 in the regulation of cell proliferation. Recent studies have shown that ZO-3 function is not required for effective TJ function, as RNAi-mediated suppression of ZO-3 had no discernable consequences on TJ formation or maintenance (Adachi et al., 2006). Similarly, ZO-3 knockout mice are viable and show no apparent phenotype (Adachi et al., 2006; Xu et al., 2008). Suppression of ZO-3 in developing zebrafish embryos, however, results in epithelial barrier defects, decreased body length, and reduced tailfin size (Kiener et al., 2008). While it is tempting to speculate that the reduction in body length and tailfin size is due to ZO-3 regulation of cell proliferation, Kiener et al. contend that this phenotype is due in large part to osmotic imbalance during embryogenesis. ZO-3 expression is restricted to epithelial tissues, and elucidating its biological function may require investigation of ZO-3 function during periods of iterative cell proliferation, such as epithelial wound restitution.
Overall, our findings show that ZO-3 promotes cell-cycle progression through the sequestration of cyclin D1 at TJs during mitosis. In conclusion, plasma membrane localization of cyclin D1 and its interaction with the TJ protein ZO-3 represent a novel pathway that mediates cross-talk between epithelial TJs and the nucleus.
MATERIALS AND METHODS
Cell culture, transfection conditions, and mouse tissue
SKCO-15 human colonic epithelial cell lines were cultured as previously described (Mandell et al., 2005). For transfection studies, DharmaFECT (Thermo Fisher Scientific, Waltham, MA) was used for siRNA transfection, and Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used for expression of GFP and GFP–cyclin D1 fusion proteins. To visualize cyclin D1 in vivo, mouse distal colon tissue was excised from 8- to 10-wk-old C57BL/6 mice and processed for cryosectioning. All procedures using animals were reviewed and approved by the Emory University Institutional Animal Care and Use Committee and were performed according to National Institutes of Health criteria.
Western blot and IP
Cells were scraped into RIPA lysis buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris, pH 8.0) containing protease and phosphatase inhibitors (Sigma, St. Louis, MO) and 1 mM phenylmethylsulfonyl fluoride (PMSF), sonicated, and cleared by centrifugation. Protein concentration was determined using a bicinchoninic acid protein assay, and samples were boiled in SDS sample buffer with 50 mM dithiothreitol. Equal amounts of protein were separated by SDS–PAGE and transferred onto polyvinylidene fluoride membranes. Membranes were blocked for 1 h with 5% wt/vol dry milk or bovine serum albumin in Tris-buffered saline containing 0.1% Tween-20, and incubated with primary antibodies in blocking buffer overnight at 4°C. Antibodies were detected using horseradish peroxidase (HRP)-linked secondary antibodies (Jackson ImmunoResearch, West Grove, PA) and chemiluminescent substrate (Denville Scientific, Metuchen, NJ). Image quantitation was performed by densitometry using National Institutes of Health (NIH) ImageJ. To determine the protein half-life of cyclin D1, relative intensity was determined as a proportion of signal at time 0. Multiple exposures were examined in order to exclude overexposed bands. All bands were normalized to actin loading control. IPs were performed using DSP (dithiobis[succinimidyl propionate]) (#22586; Thermo Fisher Scientific, Waltham, MA), a thiol-cleavable, cell-permeable cross-linking agent with a 12.0 A spacer arm at 1 mM for 30 min at room temperature. Cell were then scraped into RIPA lysis buffer containing protease and phosphatase inhibitors (Sigma) and 1 mM PMSF. Antibody was added to the lysate at 1 μg, and IPs were incubated overnight at 4°C. The following day, IPs were incubated with G-coupled Sepharose (GE Healthcare, Piscataway, NJ) for 30 min at 4°C and then washed three times in cold phosphate-buffered saline (PBS). Protein complexes were assessed by Western blot.
Antibodies and reagents
Antibodies used were as follows: rabbit polyclonal anti-ZO-1/2/3 (Zymed, South San Francisco, CA), rabbit polyclonal cyclin D1 and Cdk4 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal phospho-Thr-286 cyclin D1 (3300S; Cell Signaling Technology, Danvers, MA), mouse monoclonal anti-ZO-1 (Zymed), cyclin D1 (DCS-6; Sigma), and lamin A/C (Santa Cruz). Secondary antibodies used were as follows: Alexa-conjugated secondary antibodies were obtained from Molecular Probes (Alexa dye series; Eugene, OR). HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Nuclei were stained with ToPro-3 (Molecular Probes). Chx (Sigma) was used at 50 μM for the times indicated before cell lysis. siRNA reagents include ZO-3 (sc-43538), ZO-2 (sc-29833; Santa Cruz Biotechnology, Santa Cruz, CA), and control nontargeting siRNA (pool #1; Thermo Fisher Scientific, Waltham, MA).
Immunofluorescence labeling
Immunofluorescence detection of proteins of interest was performed as described previously (Severson et al., 2009). Briefly, cells were grown as confluent or subconfluent cultures as indicated and fixed for immunofluorescence as follows: For the visualization of cyclin D1 at TJs (SKCO-15 and mouse colon tissue), cells were fixed in absolute ethanol for 20 min at 20°C. For the visualization of Cdk4 and ZO family proteins at TJ, monolayers were prepermeabilized in 0.1% Triton X-100 for 2 min before fixation. Samples were blocked in 1% BSA for 1 h before incubation with the indicated primary antibody for 1 h at room temperature. Incubation with Alexa-secondary antibodies followed for 45 min at room temperature. To visualize ZO family proteins simultaneously with phospho-Thr-286 cyclin D1, primary antibodies were labeled directly using Zenon Tricolor Rabbit Antibody Labeling Kit (Z25370; Invitrogen, Carlsbad, CA). To visualize both nuclei and apical TJs in columnar epithelial cultures, images were compressed from multiple consecutive z-stack confocal sections (LSM 5 image browser; Zeiss Microimaging, Thornwood, NY). Fluorescence images were acquired using an LSM 510 and META image analysis software (version 4.2; Zeiss). Image quantitation was performed using NIH ImageJ.
TUNEL assay
In vitro cell viability after ZO-3 siRNA treatment was assessed using an In Situ Cell Death Detection Kit, Fluorescein (Roche, Mannheim, Germany), according to the manufacturer's instructions. Briefly, 48 h post-siRNA transfection, cells were air-dried and fixed in 4% paraformaldehyde for 1 h at room temperature. Samples were washed in PBS and permeabilized in 0.1% Triton X-100, 0.1% sodium citrate for 2 min at 4°C. As a positive control, cells were treated with DNase I (Sigma) in 50 mM Tris, pH 7.4, and 1 mg/ml BSA for 10 min. Cells were incubated with labeling reagent for 1 h at 37°C before visualization.
Propidium iodide cell-cycle assessment
Cell-cycle analysis was performed as follows: Adherent SKCO-15 cells were washed in Ca2+/Mg2+–free PBS and detached using Trypsin-EDTA (Sigma) followed by a 15-min incubation in Ca2+/Mg2+–free PBS, 2 mM EDTA at 4°C. The resulting single-cell suspension was pelleted and fixed in absolute ethanol overnight at 4°C. Fixed cells were incubated in 50 μg/ml propidium iodide (Sigma), 0.1 mg/ml RNase A (Sigma), and 0.05% Triton X-100 for 40 min at 37°C. Cell cycle was detected with a 488-nm argon-ion laser line using FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) and analyzed with WinMDI 2.9 software.
Subcellular fractionation and sucrose density gradient analysis
SKCO-15 cell lysis and sucrose density fractionation were performed as previously described (Kaoutzani et al., 1993). Briefly, epithelial cells were grown to confluence and harvested in 10 mM HEPES, pH 7.4, 100 mM KCl, 3 mM NaCl, 1 mM Na2ATP, 3.5 MgCl2, Protease Inhibitor Cocktail (Sigma), Phosphatase Inhibitor Cocktail I and II (Sigma), and 1 mM PMSF. Cell lysis was achieved by nitrogen cavitation, at 200 psi for 15 min, 4°C. Lysates were then cleared of nuclear material by 1000 × g spin for 10 min at 4°C. Supernatants were then applied to a 36 ml, 15–60% continuous sucrose gradient and separated by centrifugation at 180,000 × g for 1 h at 4°C in a VTi50 rotor (Beckman Coulter, Brea, CA). Fractions (2 ml) were collected and analyzed by SDS–PAGE after methanol/chloroform protein extraction. ALP activity was assessed in each fraction as described (Kaoutzani et al., 1993).
Fusion constructs and protein expression
GST–ZO-2/3 constructs were a gift of the Stevenson lab. GST fusion protein expression was induced with 0.2 mM iIsopropyl-β-d-thio-galactoside for 4 h at 37°C, and proteins were purified according to the manufacturer's protocol. His6-cyclin D1 constructs were cloned into pet24a and expressed using a wheat germ extract TNT system (Promega, Madison, WI). Cyclin D1 constructs were cloned into pCDNA3.1 GFP and expressed for 24 h before imaging.
Supplementary Material
Acknowledgments
We thank Caroline Addis and Susan Voss for expert technical assistance. This work was supported by grants from the National Institutes of Health (DK72564, DK61379, and DK79392 to C.P.; DK53202, DK55679, and DK59888 to A.N.; and DK64399 [NIH Digestive Diseases Research Consortium tissue culture and morphology grant]) and the Crohn's and Colitis Foundation of America (fellowship award to S.K., and C.T.C.).
Abbreviations used:
- ALP
alkaline phosphatase
- Cdk
cyclin-dependent kinase
- Chx
cycloheximide
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HRP
horseradish peroxidase
- IP
immunoprecipitation
- JAM
junctional adhesion molecule
- MAGUK
membrane-associated guanylate kinase
- MDCK
Madin Darby canine kidney
- PBS
phosphate-buffered saline
- PDZ
PSD-95, Dlg, and ZO-1
- PMSF
phenylmethylsulfonyl fluoride
- TJ
tight junction
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- ZO
zonula occludens
- ZONAB
ZO-1–associated nucleic acid binding protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0677) on March 16, 2011.
REFERENCES
- Adachi M, Inoko A, Hata M, Furuse K, Umeda K, Itoh M, Tsukita S. Normal establishment of epithelial tight junctions in mice and cultured cells lacking expression of ZO-3, a tight-junction MAGUK protein. Mol Cell Biol. 2006;26:9003–9015. doi: 10.1128/MCB.01811-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Garrett MD, Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol. 2003;160:423–432. doi: 10.1083/jcb.200210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Matter K. Tight junctions and the regulation of gene expression. Biochim Biophys Acta. 2009;1788:761–767. doi: 10.1016/j.bbamem.2008.11.024. [DOI] [PubMed] [Google Scholar]
- Beatch M, Jesaitis LA, Gallin WJ, Goodenough DA, Stevenson BR. The tight junction protein ZO-2 contains three PDZ (PSD-95/Discs-Large/ZO-1) domains and an alternatively spliced region. J Biol Chem. 1996;271:25723–25726. doi: 10.1074/jbc.271.42.25723. [DOI] [PubMed] [Google Scholar]
- Betanzos A, Huerta M, Lopez-Bayghen E, Azuara E, Amerena J, Gonzalez-Mariscal L. The tight junction protein ZO-2 associates with Jun, Fos and C/EBP transcription factors in epithelial cells. Exp Cell Res. 2004;292:51–66. doi: 10.1016/j.yexcr.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Sherr CJ. A dominant-negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (CDK4) and its phosphorylation by CDK-activating kinase. Mol Cell Biol. 1997;17:7362–7374. doi: 10.1128/mcb.17.12.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi J, Mitsui Y. Cyclin D1 inhibits cell proliferation through binding to PCNA and cdk2. Exp Cell Res. 1999;246:338–347. doi: 10.1006/excr.1998.4306. [DOI] [PubMed] [Google Scholar]
- Giallourakis C, Cao Z, Green T, Wachtel H, Xie X, Lopez-Illasaca M, Daly M, Rioux J, Xavier R. A molecular-properties-based approach to understanding PDZ domain proteins and PDZ ligands. Genome Res. 2006;16:1056–1072. doi: 10.1101/gr.5285206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladden AB, Diehl JA. Location, location, location: the role of cyclin D1 nuclear localization in cancer. J Cell Biochem. 2005;96:906–913. doi: 10.1002/jcb.20613. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L, Betanzos A, Avila-Flores A. MAGUK proteins: structure and role in the tight junction. Semin Cell Dev Biol. 2000;11:315–324. doi: 10.1006/scdb.2000.0178. [DOI] [PubMed] [Google Scholar]
- Gottardi CJ, Arpin M, Fanning AS, Louvard D. The junction-associated protein, zonula occludens-1, localizes to the nucleus before the maturation and during the remodeling of cell-cell contacts. Proc Natl Acad Sci USA. 1996;93:10779–10784. doi: 10.1073/pnas.93.20.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot L, Paschoud S, Pulimeno P, Foglia A, Citi S. The cytoplasmic plaque of tight junctions: a scaffolding and signalling center. Biochim Biophys Acta. 2008;1778:601–613. doi: 10.1016/j.bbamem.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Guo Y, Yang K, Harwalkar J, Nye JM, Mason DR, Garrett MD, Hitomi M, Stacey DW. Phosphorylation of cyclin D1 at Thr 286 during S phase leads to its proteasomal degradation and allows efficient DNA synthesis. Oncogene. 2005;24:2599–2612. doi: 10.1038/sj.onc.1208326. [DOI] [PubMed] [Google Scholar]
- Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- Haskins J, Gu L, Wittchen ES, Hibbard J, Stevenson BR. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi M, Yang K, Stacey AW, Stacey DW. Phosphorylation of cyclin D1 regulated by ATM or ATR controls cell cycle progression. Mol Cell Biol. 2008;28:5478–5493. doi: 10.1128/MCB.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta M, Munoz R, Tapia R, Soto-Reyes E, Ramirez L, Recillas-Targa F, Gonzalez-Mariscal L, Lopez-Bayghen E. Cyclin D1 is transcriptionally down-regulated by ZO-2 via an E box and the transcription factor c-Myc. Mol Biol Cell. 2007;18:4826–4836. doi: 10.1091/mbc.E07-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoko A, Itoh M, Tamura A, Matsuda M, Furuse M, Tsukita S. Expression and distribution of ZO-3, a tight junction MAGUK protein, in mouse tissues. Genes Cells. 2003;8:837–845. doi: 10.1046/j.1365-2443.2003.00681.x. [DOI] [PubMed] [Google Scholar]
- Islas S, Vega J, Ponce L, Gonzalez-Mariscal L. Nuclear localization of the tight junction protein ZO-2 in epithelial cells. Exp Cell Res. 2002;274:138–148. doi: 10.1006/excr.2001.5457. [DOI] [PubMed] [Google Scholar]
- Jaramillo BE, Ponce A, Moreno J, Betanzos A, Huerta M, Lopez-Bayghen E, Gonzalez-Mariscal L. Characterization of the tight junction protein ZO-2 localized at the nucleus of epithelial cells. Exp Cell Res. 2004;297:247–258. doi: 10.1016/j.yexcr.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Kaoutzani P, Parkos CA, Delp-Archer C, Madara JL. Isolation of plasma membrane fractions from the intestinal epithelial model T84. Am J Physiol. 1993;264:C1327–C1335. doi: 10.1152/ajpcell.1993.264.5.C1327. [DOI] [PubMed] [Google Scholar]
- Kiener TK, Selptsova-Friedrich I, Hunziker W. Tjp3/zo-3 is critical for epidermal barrier function in zebrafish embryos. Dev Biol. 2008;316:36–49. doi: 10.1016/j.ydbio.2007.12.047. [DOI] [PubMed] [Google Scholar]
- Kwak YT, Li R, Becerra CR, Tripathy D, Frenkel EP, Verma UN. IkappaB kinase alpha regulates subcellular distribution and turnover of cyclin D1 by phosphorylation. J Biol Chem. 2005;280:33945–33952. doi: 10.1074/jbc.M506206200. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- Mandell KJ, Babbin BA, Nusrat A, Parkos CA. Junctional adhesion molecule 1 regulates epithelial cell morphology through effects on beta1 integrins and Rap1 activity. J Biol Chem. 2005;280:11665–11674. doi: 10.1074/jbc.M412650200. [DOI] [PubMed] [Google Scholar]
- Matter K, Balda MS. Epithelial tight junctions, gene expression and nucleo-junctional interplay. J Cell Sci. 2007;120:1505–1511. doi: 10.1242/jcs.005975. [DOI] [PubMed] [Google Scholar]
- Nava P, et al. Interferon-gamma regulates intestinal epithelial homeostasis through converging beta-catenin signaling pathways. Immunity. 2010;32:392–402. doi: 10.1016/j.immuni.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M, Theodoras AM, Tam SW, Draetta GF. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 1994;8:1627–1639. doi: 10.1101/gad.8.14.1627. [DOI] [PubMed] [Google Scholar]
- Paris L, Tonutti L, Vannini C, Bazzoni G. Structural organization of the tight junctions. Biochim Biophys Acta. 2008;1778:646–659. doi: 10.1016/j.bbamem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Severson EA, Lee WY, Capaldo CT, Nusrat A, Parkos CA. Junctional adhesion molecule A interacts with Afadin and PDZ-GEF2 to activate Rap1A, regulate beta1 integrin levels, and enhance cell migration. Mol Biol Cell. 2009;20:1916–1925. doi: 10.1091/mbc.E08-10-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- Solomon DA, Wang Y, Fox SR, Lambeck TC, Giesting S, Lan Z, Senderowicz AM, Conti CJ, Knudsen ES. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278:30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- Sourisseau T, Georgiadis A, Tsapara A, Ali RR, Pestell R, Matter K, Balda MS. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol Cell Biol. 2006;26:2387–2398. doi: 10.1128/MCB.26.6.2387-2398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia R, Huerta M, Islas S, Avila-Flores A, Lopez-Bayghen E, Weiske J, Huber O, Gonzalez-Mariscal L. Zona occludens-2 inhibits cyclin D1 expression and cell proliferation and exhibits changes in localization along the cell cycle. Mol Biol Cell. 2009;20:1102–1117. doi: 10.1091/mbc.E08-03-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traweger A, Lehner C, Farkas A, Krizbai IA, Tempfer H, Klement E, Guenther B, Bauer HC, Bauer H. Nuclear zonula occludens-2 alters gene expression and junctional stability in epithelial and endothelial cells. Differentiation. 2008;76:99–106. doi: 10.1111/j.1432-0436.2007.00227.x. [DOI] [PubMed] [Google Scholar]
- Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- Wittchen ES, Haskins J, Stevenson BR. NZO-3 expression causes global changes to actin cytoskeleton in Madin-Darby canine kidney cells: linking a tight junction protein to Rho GTPases. Mol Biol Cell. 2003;14:1757–1768. doi: 10.1091/mbc.E02-08-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Kausalya PJ, Phua DC, Ali SM, Hossain Z, Hunziker W. Early embryonic lethality of mice lacking ZO-2, but not ZO-3, reveals critical and nonredundant roles for individual zonula occludens proteins in mammalian development. Mol Cell Biol. 2008;28:1669–1678. doi: 10.1128/MCB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.