Pex34p is a novel peroxisomal protein involved in controlling peroxisome abundance in Saccharomyces cerevisiae. Pex34p acts to control peroxisome numbers both alone and in cooperation with the Pex11 protein family of peroxisome divisional proteins.
Abstract
Peroxisomes are ubiquitous organelles involved in diverse metabolic processes, most notably the metabolism of lipids and the detoxification of reactive oxygen species. Peroxisomes are highly dynamic and change in size and number in response to both intra- and extracellular cues. In the yeast Saccharomyces cerevisiae, peroxisome growth and division are controlled by both the differential import of soluble matrix proteins and a specialized divisional machinery that includes peroxisome-specific factors, such as members of the Pex11 protein family, and general organelle divisional factors, such as the dynamin-related protein Vps1p. Global yeast two-hybrid analyses have demonstrated interactions between the product of the S. cerevisiae gene of unknown function, YCL056c, and Pex proteins involved in peroxisome biogenesis. Here we show that the protein encoded by YCL056c, renamed Pex34p, is a peroxisomal integral membrane protein that acts independently and also in concert with the Pex11 protein family members Pex11p, Pex25p, and Pex27p to control the peroxisome populations of cells under conditions of both peroxisome proliferation and constitutive peroxisome division. Yeast two-hybrid analysis showed that Pex34p interacts physically with itself and with Pex11p, Pex25p, and Pex27p but not with Vps1p. Pex34p can act as a positive effector of peroxisome division as its overexpression leads to increased numbers of peroxisomes in wild type and pex34Δ cells. Pex34p requires the Pex11 family proteins to promote peroxisome division. Our discovery of Pex34p as a protein involved in the already complex control of peroxisome populations emphasizes the necessity of cells to strictly regulate their peroxisome populations to be able to respond appropriately to changing environmental conditions.
INTRODUCTION
Eukaryotic cells have an advantage over prokaryotic cells by having membrane-bound organelles that provide optimized microenvironments for specific metabolic functions. To maintain these advantages, eukaryotes have developed complex mechanisms to regulate the abundance of organelles in response to changing environmental and metabolic stimuli and to partition organelles equitably between mother and daughter cells at cell division.
Peroxisomes are specialized for a variety of metabolic functions, including the oxidation of fatty acids, the elimination of reactive oxygen species, and the synthesis of bile acids and plasmalogens in higher eukaryotes (Wanders and Waterham, 2006; Schrader and Fahimi, 2008). Peroxisomes are essential for normal human development and physiology, as evidenced by the lethality of a spectrum of human diseases collectively known as the peroxisome biogenesis disorders (PBDs) (Steinberg et al., 2006; Schrader and Fahimi, 2008). These inherited disorders arise from an inability to assemble or maintain functional peroxisomes. A better understanding of the causes of the PBDs has been a driving force behind the identification and characterization of the PEX genes involved in peroxisome biogenesis. To date, 33 PEX genes in a number of different organisms have been identified that are involved in the targeting and import of peroxisomal proteins, the formation of the peroxisome membrane, and the control of peroxisome size and abundance (Schrader and Fahimi, 2008; Managadze et al., 2010; Wolfe et al., 2010).
Peroxisome size and abundance are controlled by multiple pathways (for reviews, see Yan et al., 2005; Fagarasanu et al., 2007; Tabak et al., 2008; Hettema and Motley, 2009; Mast et al., 2010; Saraya et al., 2010). One pathway involves the response of cells to specific environmental or metabolic cues, such as growth of yeast on a nonfermentable fatty acid carbon source, which leads to “induction” or up-regulation of the expression of genes encoding peroxisomal proteins and rapid expansion of the peroxisomal compartment through increases in both the number of peroxisomes (i.e., peroxisome proliferation) and their sizes. A second pathway termed peroxisome “constitutive division” functions to maintain the peroxisome population in both the mother cell and the newly forming bud during cell division. The peroxisome population doubles during the cell cycle independently of peroxisome-proliferating stimuli so that essentially equal numbers of peroxisomes can be maintained in the mother cell and apportioned to the daughter cell. In the yeast Saccharomyces cerevisiae, peroxisomes that have doubled in number before cell division are equally partitioned between mother cell and bud through the interplay of Inp2p, the peroxisome-specific receptor for the myosin mediating bud-directed peroxisome transport (Fagarasanu et al., 2006), and Inp1p, which acts in anchoring peroxisomes in both mother cell and bud (Fagarasanu et al., 2005). The third pathway involves the de novo formation of peroxisomes from the endoplasmic reticulum (ER). Cells lacking peroxisomes or their remnants have the ability to reform functional peroxisomes from the ER (Hoepfner et al., 2005; Tam et al., 2005). This process in S. cerevisiae has been shown to be relatively inefficient compared with the process of peroxisome growth and division (Motley and Hettema, 2007). Barring a catastrophic loss of all peroxisomes in a cell, the ER's principal role in peroxisome biogenesis has been proposed to be the contribution of both membrane proteins and lipids to existing peroxisomes for use in their growth and division (Motley and Hettema, 2007).
In S. cerevisiae, the regulation of peroxisome abundance has traditionally been investigated using cells grown in fatty acid–containing medium to permit peroxisome proliferation. Under these conditions, peroxisomes become enlarged and the activity of the peroxisome fission machinery is increased. The Pex11 protein family, consisting of Pex11p, Pex25p, and Pex27p, has been shown to have a major role in peroxisome proliferation (Erdmann and Blobel, 1995; Smith et al., 2002; Rottensteiner et al., 2003; Tam et al., 2003). Cells lacking any of these proteins display fewer and enlarged peroxisomes, whereas their overproduction results in the presence of smaller and more numerous peroxisomes. One major caveat in using yeast grown in fatty acid–containing medium to investigate the regulation of the peroxisome population of a cell is that cells exhibit a drastic reduction in their rate of cellular division, and peroxisome division is uncoupled from cell division. Because of this, relatively little is known about the mechanism of constitutive peroxisome division, which functions in actively growing cells with a normal cell cycle.
Here we report the characterization of a newly recognized peroxisomal integral membrane protein, renamed Pex34p, encoded by the open reading frame (ORF) YCL056c of S. cerevisiae and conserved in several members of the Saccharomycetaceae. We show that Pex34p physically interacts with the Pex11 family of proteins to regulate the peroxisome complement of cells under conditions of both peroxisome proliferation and constitutive peroxisome division.
RESULTS
Pex34p is a peroxisomal integral membrane protein
Large-scale protein interaction studies have provided evidence of interaction of the protein of unknown function encoded by the S. cerevisiae ORF YCL056c and a number of Pex proteins involved in different aspects of peroxisome biogenesis (Yu et al., 2008; Yeast Resource Center [http://www.yeastrc.org./]). In addition, a global analysis of protein localization in S. cerevisiae by fluorescence microscopy showed that a green fluorescent protein (GFP)-tagged version of the Ycl056c protein gave a punctate pattern of fluorescence similar to that exhibited by fluorescent peroxisomes (Huh et al., 2003). These findings prompted us to determine whether the Ycl056c protein is indeed peroxisomal and whether it has a role in peroxisome biogenesis. Data presented herein demonstrate that the Ycl056c protein is localized to peroxisomes and has a role in peroxisome biogenesis. Accordingly, we have designated it a peroxin, Pex34p, and its encoding gene PEX34. Homologues of Pex34p appear to be restricted to members of the Saccharomycetaceae, including Kluveromyces, Zygosaccharomyces, and other species of Saccharomyces (Byrne and Wolfe, 2005).
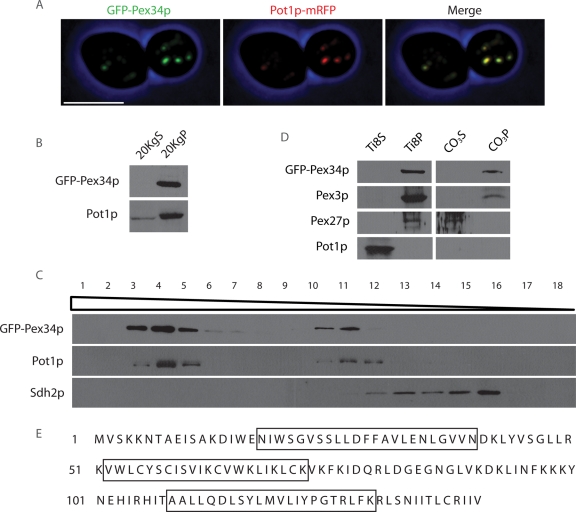
Pex34p tagged at its N terminus with GFP (GFP-Pex34p) colocalized with Pot1p-mRFP, a fluorescent protein fusion between peroxisomal 3-ketoacyl-CoA thiolase (Pot1p) and monomeric red fluorescent protein (mRFP), to punctate structures characteristic of peroxisomes (Figure 1A). Subcellular fractionation was also used to establish that Pex34p is associated with peroxisomes. GFP-Pex34p, like the peroxisomal matrix protein Pot1p, localized preferentially to a 20,000 × g pellet (20KgP) fraction enriched for mature peroxisomes and some forms of immature peroxisomes (Tam et al., 2003; Vizeacoumar et al., 2003, 2004) (Figure 1B). Isopycnic density gradient centrifugation of the 20KgP fraction showed that GFP-Pex34p cofractionated with Pot1p but not with the mitochondrial protein Sdh2p (Figure 1C).
FIGURE 1:
Pex34p is a peroxisomal integral membrane protein. (A) GFP-Pex34p colocalizes with the chimeric peroxisomal marker protein Pot1p-mRFP to punctate structures characteristic of peroxisomes by confocal fluorescence microscopy. Bar, 5 μm. (B) GFP-Pex34p localizes to the 20KgP subcellular fraction enriched for peroxisomes. Immunoblot analysis of equivalent portions of the 20KgS and 20KgP fractions from cells expressing GFP-Pex34p was performed with antibodies to GFP and to the peroxisomal matrix protein, Pot1p. (C) GFP-Pex34p cofractionates with peroxisomes. Organelles in the 20KgP fraction were separated by isopycnic centrifugation on a discontinuous Nycodenz gradient. Fractions were collected from the bottom of the gradient, and equal portions of each fraction were analyzed by immunoblotting. Fractions enriched for peroxisomes and mitochondria were identified by immunodetection of Pot1p and Sdh2p, respectively. (D) The 20KgP fraction from cells expressing GFP-Pex34p was treated with 10 mM Tris-HCl, pH 8.0, to lyse peroxisomes and was then subjected to ultracentrifugation to yield a supernatant (Ti8S) fraction enriched for matrix proteins and a pellet (Ti8P) fraction enriched for membrane proteins. The Ti8P fraction was treated further with 0.1 M Na2CO3, pH 11.3, and separated by ultracentrifugation into a supernatant (CO3S) fraction enriched for peripheral membrane proteins and a pellet (CO3P) fraction enriched for integral membrane proteins. Equal portions of each fraction were analyzed by immunoblotting with antibodies to GFP, the matrix protein Pot1p, the peroxisomal integral membrane protein Pex3p, and the peroxisomal peripheral membrane protein Pex27p. (E) Amino acid sequence of Pex34p. Boxed sequences designate three membrane-spanning regions predicted by SOSUI.
Organelle extraction was used to determine the suborganellar location of Pex34p. Organelles in the 20KgP fraction were subjected to hypotonic lysis in dilute alkali Tris buffer, followed by ultracentrifugation to yield a supernatant (Ti8S) fraction enriched for soluble proteins and a pellet (Ti8P) fraction enriched for membrane proteins (Figure 1D). GFP-Pex34p cofractionated with the peroxisomal integral membrane protein Pex3p and the peroxisomal peripheral membrane protein Pex27p to the Ti8P fraction, whereas the soluble peroxisomal matrix protein Pot1p was found almost exclusively in the Ti8S fraction. The Ti8P fraction was further extracted with alkali Na2CO3 and subjected to ultracentrifugation. This treatment releases proteins associated with, but not integral to, membranes (Fujiki et al., 1982). GFP-Pex34p cofractionated with Pex3p to the pellet (CO3P) fraction enriched for integral membrane proteins but not with Pex27p to the supernatant (CO3S) fraction enriched for peripheral membrane proteins. These data suggest that Pex34p is an integral membrane protein of peroxisomes, consistent with the predictions of three topology prediction programs (SOSUI [http://bp.nuap.nagoya-u.ac.jp/sosui/], HMMTOP [http://www.enzim.hu/hmmtop/], and TMpred [http://www.ch.embnet.org/software/TMPRED_form.html]) that Pex34p contains three transmembrane spanning regions (Figure 1E).
Deletion of the PEX34 gene affects peroxisome abundance under conditions of both peroxisome proliferation and constitutive peroxisome division
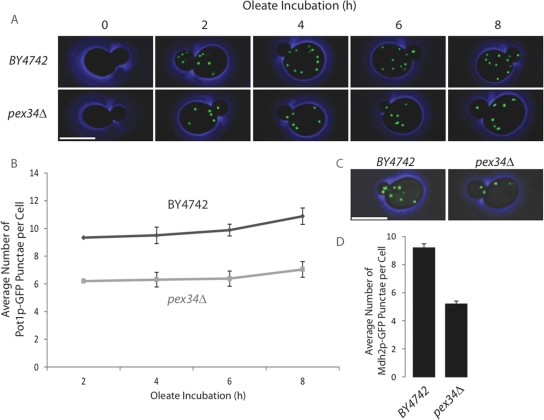
Wild type and pex34Δ cells expressing oleic acid–inducible Pot1p-GFP were grown in glucose-containing medium and then transferred to medium containing oleic acid as the sole carbon source to induce peroxisome proliferation. Cells were imaged by confocal fluorescence microscopy every 2 h (Figure 2A), and the number of Pot1p-GFP–labeled peroxisomes per cell was quantified (Figure 2B). Cells deleted for the PEX34 gene contained fewer peroxisomes than did wild-type cells over the entire time of observation up to 8 h. To determine whether this difference in peroxisome numbers between pex34Δ cells and wild-type cells was dependent on conditions promoting peroxisome proliferation, we analyzed pex34Δ cells and wild-type cells that constitutively express a chimera between GFP and the peroxisomal protein, malate dehydrogenase 2 (Mdh2p-GFP) (Huh et al., 2003; Wolinski et al., 2009), under conditions of constitutive peroxisome division (i.e., growth of cells in glucose-containing medium). pex34Δ cells continued to exhibit reduced numbers of peroxisomes compared with wild-type cells under conditions of constitutive peroxisome division (Figure 2, C and D). Thus, Pex34p plays a role in maintaining the abundance of peroxisomes under conditions of both peroxisome proliferation and constitutive peroxisome division.
FIGURE 2:
Cells deleted for the PEX34 gene have reduced numbers of peroxisomes. (A and B) The wild-type strain BY4742 and the deletion strain pex34Δ expressing Pot1p-GFP were grown in glucose-containing medium and then transferred to medium containing oleic acid as the sole carbon source to promote peroxisome proliferation. Fluorescent images of cells were captured by confocal microscopy every 2 h during oleic acid incubation (A) and scored for the number of Pot1p-GFP–labeled punctae per cell (B). Graphic results present the average number of punctae ± SEM of three independent experiments and 20 budded cells per experiment. (C and D) The wild-type strain BY4742 and the deletion strain pex34Δ expressing Mdh2p-GFP were sampled during exponential growth in glucose-containing medium, imaged by confocal fluorescence microscopy (C), and scored for the number of Mdh2p-GFP–labeled punctae per cell (D). Graphic results present the average number of punctae ± SEM of three independent experiments and 20 cells per experiment. Bar represents 5 μm in panels (A) and (C).
Pex34p interacts with proteins of the Pex11p family to control peroxisome morphology and abundance under conditions of peroxisome proliferation
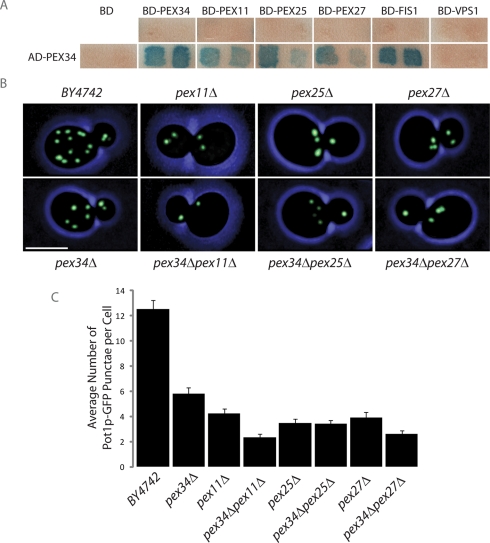
A limited yeast two-hybrid screen was done between Pex34p and other proteins previously implicated in peroxisome division to determine potential physical interactions between them. Chimeric genes were constructed by fusing the ORFs of genes of interest in-frame and downstream of sequences encoding one of two functional domains (transcription-activating domain [AD] or DNA-binding domain [BD]) of the Gal4p transcriptional activator. Pairwise combinations were transformed into S. cerevisiae strain SFY526 and analyzed using a β-galactosidase filter detection assay (Figure 3A). As previously reported, Pex34p was found to interact with Fis1p (Yu et al., 2008), a protein involved in both mitochondrial fission (Mozdy et al., 2000) and peroxisome division (Koch et al., 2005; Motley et al., 2008), and also with Pex11p, Pex25p, and Pex27p, which together make up the Pex11 family of proteins controlling peroxisome proliferation (Erdmann and Blobel, 1995; Smith et al., 2002; Rottensteiner et al., 2003; Tam et al., 2003). These interactions were specific, as the AD and BD fusions showed no self-activation. No interaction between Pex34p and the dynamin-like protein Vps1p, known to play a role in peroxisome fission (Hoepfner et al., 2001), was detected.
FIGURE 3:
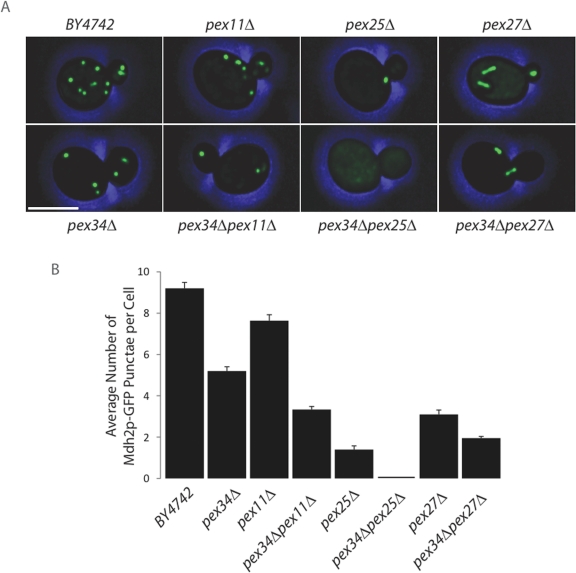
Pex34p acts by itself and together with the proteins of the Pex11 family to control peroxisome size and number under conditions of peroxisome proliferation. (A) β-galactosidase filter assay to test for interaction between Pex34p and Pex34p, Pex11p, Pex25p, Pex27p, Fis1p, and Vps1p by yeast two-hybrid analysis. Two independent transformants for each strain are shown. (B) Wild-type BY4742 cells and cells of the pex34Δ, pex11Δ, pex25Δ, pex27Δ, pex34Δpex11Δ, pex34Δpex25Δ, and pex34Δpex27Δ deletion strains expressing Pot1p-GFP were grown for 16 h in oleic acid–containing SCIM and imaged by confocal fluorescence microscopy. Bar, 5 μm. (C) Cells were scored for the number of Pot1p-GFP–labeled punctae per cell. Graphic results represent the average number of punctae ± SEM of three independent experiments and 20 cells per experiment.
Confocal fluorescence microscopy of pex11Δ, pex25Δ, and pex27Δ cells expressing Pot1p-GFP and grown in oleic acid–containing medium showed reduced numbers of peroxisomes compared with wild-type cells, as has been previously reported (Erdmann and Blobel, 1995; Smith et al., 2002; Rottensteiner et al., 2003; Tam et al., 2003), and confirmed the reduction in peroxisome number in pex34Δ cells (Figure 3, B and C). A greater reduction in the number of peroxisomes was observed in the double deletion strains pex34Δpex11Δ and pex34Δpex27Δ, but not in the pex34Δpex25Δ strain, as compared with the individual pex11Δ, pex27Δ, and pex25Δ strains (Figure 3, B and C).
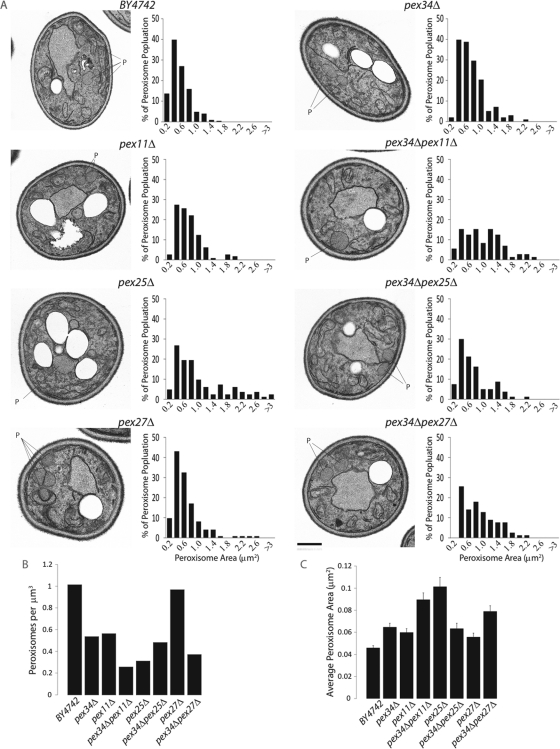
Single and double deletion strains grown in oleic acid–containing medium were further analyzed by electron microscopy for both peroxisome abundance and size (Figure 4). Morphometric analysis (Figure 4, B and C) of electron micrographs (Figure 4A) showed a reduced number of enlarged peroxisomes in pex34Δ cells as compared with wild-type cells, whereas pex34Δpex11Δ and pex34Δpex27Δ cells showed fewer and larger peroxisomes than did cells deleted for the individual genes. Interestingly, deletion of the PEX34 gene in the pex25Δ background often resulted in the single, greatly enlarged peroxisomes frequently observed in pex25Δ cells being replaced with clustered, smaller peroxisomes (Figure 4A).
FIGURE 4:
Deletion of the PEX34 gene results in fewer and larger peroxisomes in oleic acid–grown cells. (A) Ultrastructure and morphometric analysis of cells of the wild-type strain BY4742 and of different deletion strains. Cells were grown for 16 h in oleic acid–containing SCIM, fixed in 3% KMnO4, and processed for electron microscopy. Bar, 1 μm. For morphometric analysis, the cell areas and areas of individual peroxisomes of 300 randomly selected cells from three independent analyses of each strain were determined using Olympus iTEM software. Peroxisomes were then separated into size categories, and a histogram depicting the percentage of total peroxisomes of each size category was generated for each strain. The numbers along the x-axis represent the maximum areas of peroxisomes (in square micrometers) for each category, with the exception of the last number, which represents the minimum area of peroxisomes (in square micrometers) in the last category. (B) Number of peroxisomes per cubic micrometer and (C) average peroxisome area (in square micrometers) for cells of the different strains. Error bars represent the SEM.
Taken together, genetic and microscopic analyses show that Pex34p individually and together with Pex11p, Pex25p, and Pex27p acts to regulate peroxisome number and size under conditions that promote peroxisome proliferation.
Pex34p interacts with proteins of the Pex11 family to control constitutive peroxisome division
Deletion strains expressing Mdh2p-GFP were grown in glucose-containing medium and were analyzed by confocal fluorescence microscopy to determine whether Pex34p acts together with members of the Pex11 protein family to also regulate peroxisome numbers under conditions of constitutive peroxisome division (Figure 5A). Like cells grown under conditions promoting peroxisome proliferation, deletion of the PEX34 gene led to reduced numbers of peroxisomes as compared with wild-type cells (Figure 5B). Deletion of the PEX25 and PEX27 genes led to even greater reductions in the numbers of peroxisomes as compared with wild-type levels than did deletion of PEX34 (Figure 5B), demonstrating a role for Pex25p and Pex27p in controlling peroxisome numbers also under conditions of constitutive peroxisome division. Combining the deletion of PEX34 with deletion of PEX25 led to dramatic reductions in the numbers of peroxisomes per cell, whereas cells of the pex34Δpex27Δ double deletion strain showed more modest, yet still significant, reductions in the number of peroxisomes per cell when compared with cells deleted for PEX27 alone (Figure 5B). Interestingly, both pex27Δ and pex34Δpex27Δ cells often exhibited elongated vermiform peroxisomes (Figure 5A), similar to those previously described for cells lacking the dynamin-related protein, Vps1p (Hoepfner et al., 2001; Kuravi et al., 2006). This peroxisome phenotype was observed only in cells without Pex27p and only under conditions of constitutive peroxisome division.
FIGURE 5:
Pex34p functions with the proteins of the Pex11 family to control peroxisome numbers under conditions of constitutive peroxisome division. (A) Wild-type BY4742 cells and cells of different deletion strains expressing Mdh2p-GFP were harvested during exponential growth in glucose-containing YEPD medium and imaged by confocal fluorescence microscopy. Bar, 5 μm. (B) Cells were scored for the number of Mdh2p-GFP–labeled punctae per cell. Graphic results represent the average number of punctae ± SEM of three independent experiments and 20 cells per experiment.
Deletion of the PEX11 gene did not lead to a dramatic reduction in the number of peroxisomes compared with peroxisome numbers in wild-type cells under conditions of constitutive peroxisome division (Figure 5B), possibly because the expression of the PEX11 gene is extremely low under conditions of cell growth in glucose but is greatly induced when cells are incubated in medium containing a carbon source like oleic acid that promotes peroxisome proliferation (Karpichev and Small, 1998; Smith et al., 2002; Knoblach and Rachubinski, 2010). Deletion of PEX34 in combination with PEX11, however, led to dramatically reduced numbers of peroxisomes, suggesting a role for Pex34p and the limited amounts of Pex11p present during glucose growth of cells in controlling peroxisome numbers during constitutive peroxisome division.
Together, our data demonstrate a role for Pex34p, alone and in conjunction with the Pex11p family of peroxisome divisional proteins, in controlling peroxisome numbers during both peroxisome proliferation and constitutive peroxisome division.
PEX34 acts with PEX25 to maintain mature peroxisomes in actively dividing cells
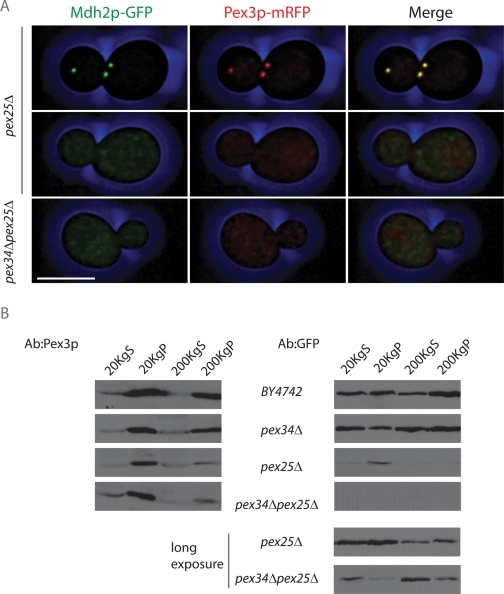
A significant proportion of pex25Δ cells (unpublished data) and all pex34Δpex25Δ cells (Figure 5A) were observed to be devoid of Mdh2p-GFP–labeled punctae. Cells deleted for PEX25 have been reported to be impaired in the import of PTS1-containing matrix proteins (Smith et al., 2002). Although Mdh2p does not contain a readily identifiable peroxisome-targeting signal type 1 (PTS1), its import into peroxisomes is dependent on the PTS1 receptor, Pex5p (unpublished data). We asked whether pex25Δ cells and pex34Δpex25Δ cells are truly devoid of peroxisomes or whether they are simply impaired in matrix protein import. pex25Δ cells and pex34Δpex25Δ cells expressing Mdh2p-GFP were additionally labeled by a genomically expressed chimera of the peroxisomal membrane protein Pex3p and mRFP (Pex3p-mRFP) and grown in glucose-containing medium to permit robust cell growth. Cells were imaged by confocal fluorescence microscopy (Figure 6A). When present, Mdh2p-GFP–labeled punctae in pex25Δ cells colabeled with Pex3p-mRFP, whereas pex34Δpex25Δ cells and pex25Δ cells lacking definitive Mdh2p-GFP-labeled punctae also lacked any definitive Pex3p-mRFP punctae, suggesting that a lack of Mdh2p-GFP punctae in cells with these genetic backgrounds is not simply the result of impaired matrix protein import but is due, at least in part, to compromised assembly of the peroxisomal membrane. Both Mdh2p-GFP and Pex3p-mRFP showed a generalized pattern of fluorescence in pex34Δpex25Δ cells and in those pex25Δ cells lacking definitive punctae; neither chimeric protein exhibited preferential localization in the perinuclear region or at the cell periphery characteristic of an ER-localized protein.
FIGURE 6:
PEX34 and PEX25 function in maintaining mature peroxisomes in actively dividing cells. (A) pex25Δ and pex34Δpex25Δ cells expressing the fluorescent peroxisomal matrix protein chimera Mdh2p-GFP and the fluorescent peroxisomal membrane protein chimera Pex3p-mRFP were grown in glucose-containing medium to promote active cell division. Exponentially growing cells were imaged by confocal fluorescence microscopy. Bar, 5 μm. (B) Cells of the wild-type BY4742 strain and of the pex34Δ, pex25Δ, and pex34Δpex25Δ deletion strains expressing Mdh2p-GFP and Pex3p-mRFP were grown in glucose-containing medium, harvested during exponential growth, and subjected to subcellular fractionation to yield 20KgS and 20KgP fractions. The 20KgS fraction was subjected to ultracentrifugation at 200,000 × g to yield a cytosolic 200KgS fraction and a 200KgP fraction containing small vesicles. Equivalent portions of each fraction were analyzed by immunoblotting with antibodies to Pex3p and GFP. The bottom two panels at right are a longer exposure of the corresponding pex25Δ and pex34Δpex25Δ panels (top).
To determine whether any of the Mdh2p-GFP pool in pex34Δpex25Δ cells is present in membrane-bound compartments not visible by fluorescence microscopy, glucose-grown cells of the wild-type strain BY4742 and of the deletion strains pex34Δ, pex25Δ, and pex34Δpex25Δ were fractionated to yield 20KgS and 20KgP fractions. In addition, the 20KgS fraction was subjected to ultracentrifugation at 200,000 × g to yield a pellet (200KgP) fraction enriched for small vesicles and a cytosolic supernatant (200KgS) fraction. Equivalent portions of the 20KgS and 20KgP and of the 200KgS and 200KgP were analyzed by immunoblotting with anti-Pex3p antibodies to detect Pex3p-mRFP and anti-GFP antibodies to detect Mdh2p-GFP (Figure 6B). In agreement with the results of fluorescence microscopy (Figure 6A), immunoblotting confirmed that Mdh2p-GFP was present in reduced amounts in pex25Δ and pex34Δpex25Δ cells in comparison to wild-type BY4742 or pex34Δ cells and required longer exposure for its ready detection (Figure 6B). In all strains, Pex3p-mRFP could be found in the 20KgP fraction, which contains both mature and some forms of immature peroxisomes (Tam et al., 2003; Vizeacoumar et al., 2003, 2004), and in the 200KgP fraction containing vesicular structures. Importantly, only small amounts of Mdh2p-GFP were present in the 20KgP fraction of pex34Δpex25Δ cells as compared with cells of the wild type and pex34Δ and pex25Δ strains, although some Mdh2p-GFP from pex34Δpex25Δ cells could be found in the 200KgP fraction containing small vesicles. Therefore pex34Δpex25Δ cannot readily assemble mature peroxisomes but can assemble vesicular structures containing the peroxisomal membrane marker protein chimera, Pex3p-mRFP. Our findings suggest that Pex34p acts in conjunction with Pex25p to maintain the population of mature peroxisomes in actively dividing cells.
Epistatic analysis of Pex34p and Pex11 protein family members in peroxisome division
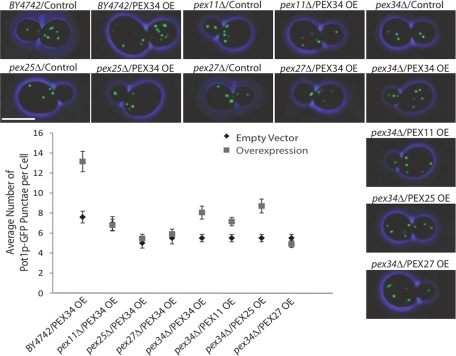
An epistatic analysis was done to investigate the interplay of PEX34 and the genes of the PEX11 family in peroxisome division. Plasmids expressing PEX11, PEX25, PEX27, or PEX34 under the regulation of the galactose-inducible GAL1 promoter were introduced into cells deleted for PEX34 or for a gene of the PEX11 family and containing Pot1p-GFP to fluorescently label peroxisomes. Cells carrying the empty pBY011 parental vector served as controls. Cells were first grown in oleic acid–containing medium, and galactose was then added to induce gene overexpression. Cells were imaged by confocal fluorescence microscopy, and GFP punctae were quantified (Figure 7). Wild-type cells or pex34Δ cells, but not pex11Δ, pex25Δ, or pex27Δ cells, overexpressing PEX34 showed greater numbers of Pot1p-GFP punctae compared with their corresponding empty vector controls. These data suggest that Pex34p acts as a positive factor of peroxisome division and requires members of the Pex11 protein family to promote peroxisome division. Moreover, overproduction of Pex11p or Pex25p could rescue the abnormal peroxisome phenotype of pex34Δ cells, reestablishing essentially wild-type levels of peroxisomes in these cells. In contrast, overproduction of Pex27p could not substitute for a lack of Pex34p in cells, as peroxisome numbers remained unchanged from what was observed in pex34Δ cells containing the empty pBY011 vector.
FIGURE 7:
Pex34p acts as a positive effector of peroxisome division. PEX34, PEX11, PEX25, and PEX27 were overexpressed from a galactose-inducible promoter in wild-type BY4742 and mutant pex34Δ, pex11Δ, pex25Δ, and pex27Δ cells grown in oleic acid–containing medium and expressing the peroxisomal marker Pot1p-GFP. The number of GFP punctae were scored and plotted against cells carrying the empty expression vector pBY011 as a control. Values are the average number of punctae ± SEM of three independent experiments and 20 cells per experiment. Bar, 5 μm.
DISCUSSION
Global studies of protein–protein interactions have become key resources for predicting the possible functions of uncharacterized proteins in S. cerevisiae through their interactions with proteins of known biological function. These studies have demonstrated interactions between the uncharacterized protein, which we have designated as Pex34p, encoded by the ORF YCL056c and peroxins required for peroxisome biogenesis. Pex34p is a peroxisomal integral membrane protein that functions in controlling peroxisome abundance. Pex34p works in concert with the three members of the Pex11 protein family of peroxisome divisional factors, Pex11p, Pex25p, and Pex27p, to control peroxisome abundance. Pex34p interacts with itself and with Pex11p, Pex25p, and Pex27p, implicating Pex34p homo-oligomerization and Pex34p hetero-oligomerization with Pex11 protein family members in regulating peroxisome division. The PEX34 gene is expressed under conditions of yeast growth in glucose-containing medium and at slightly reduced levels in oleic acid–containing medium (Smith et al., 2002).
Cells deleted for the PEX34 gene exhibit fewer peroxisomes under conditions of both peroxisome proliferation and constitutive peroxisome division. During growth in oleic acid–containing medium, which promotes peroxisome proliferation, pex34Δ cells were observed by electron microscopy to have larger peroxisomes than wild-type cells. No firm conclusion on the sizes of peroxisomes in pex34Δ cells grown in glucose-containing medium, in which peroxisomes divide constitutively, could be made by electron microscopy because peroxisomes under these conditions do not exhibit characteristic “peroxisome morphology.” Cells deleted for PEX34 and for either PEX11 or PEX27 showed fewer peroxisomes than cells deleted individually for the genes under conditions of both peroxisome proliferation and constitutive peroxisome division. Under conditions of peroxisome proliferation, peroxisomes in pex34Δpex11Δ cells and pex34Δpex27Δ cells were larger than the peroxisomes in cells deleted for only one of the genes.
Interestingly, cells deleted for both the PEX34 and PEX25 genes and grown in glucose-containing medium to promote constitutive peroxisome division showed no evidence of characteristic peroxisomes by fluorescence microscopy. Mdh2p-GFP and Pex3p-mRFP did not form discrete, punctate foci but instead produced a generalized fluorescence throughout the cell. Subcellular fractionation confirmed that, although these cells do not have mature peroxisomes, they do have structures that are pelletable at high centrifugal force and contain peroxisomal membrane and matrix proteins. At present, it is unknown whether these structures are some form of immature peroxisome or are bona fide functional peroxisomes that are uncharacteristically small. Our data suggest that Pex34p may have a role in peroxisome biogenesis outside of its role in peroxisome division, as has been postulated previously for the PEX11 family proteins (Rottensteiner et al., 2003). Interestingly, Pex34p has been shown to interact with proteins involved in peroxisomal protein import, including Pex7p, Pex10p, and Pex13p (Yu et al., 2008; Yeast Resource Center [http://www.yeastrc.org/]), suggesting that Pex34p could have a regulatory role in peroxisomal protein import through its interaction with the import machinery.
Electron microscopy showed that, under conditions of peroxisome proliferation, pex34Δpex25Δ cells contain more and smaller peroxisomes than do pex34Δ or pex25Δ cells. The reason for the increased numbers of smaller peroxisomes in pex34Δpex25Δ cells grown in oleic acid is unknown, although the lack of mature peroxisomes in these cells under conditions of constitutive peroxisome division may be a contributing factor. Nevertheless, our results confirm that the PEX34 gene, either alone or together with the members of the PEX11 gene family, functions in controlling peroxisome abundance in cells that are proliferating peroxisomes in response to the presence of a carbon source requiring peroxisomes for its metabolism, or are dividing peroxisomes constitutively to respond to the rapid cell division that occurs in a rich glucose-containing medium.
Pex11p, Pex25p, and Pex27p have been reported to act as positive effectors of peroxisome division, as their overproduction leads to increased numbers of peroxisomes in cells (Rottensteiner et al., 2003; Tam et al., 2003). Similarly, Pex34p acts as a positive effector of peroxisome division as its overproduction leads to increased numbers of peroxisomes in wild-type and pex34Δ cells. Pex34p, however, requires the members of the Pex11 protein family to function as a positive effector of peroxisome division. At this time, it is unknown whether Pex34p must interact physically with the Pex11 protein family members to promote peroxisome division or whether there is some form of intramolecular signaling between these proteins that results in Pex34p's capacity to act as a positive effector of peroxisome division.
Cells deleted for the PEX27 gene showed a high proportion of elongated peroxisomes under conditions of constitutive peroxisome division. This observation suggests that Pex27p may act downstream of peroxisome elongation, whereas Pex34p, Pex11p, and Pex25p act upstream of, or play a role in, this elongation step. Elongated peroxisomes were previously observed in cells lacking the dynamin-related protein, Vps1p, in which the myosin motor-dependent inheritance of the single enlarged peroxisome present in these cells results in the elongation of the peroxisome into a tubular structure that passes through the neck region between mother cell and bud (Hoepfner et al., 2001; Kuravi et al., 2006; Fagarasanu et al., 2009). Whether the elongated peroxisomes observed in pex27Δ cells are also dependent on the peroxisome inheritance machinery or they represent an intermediate in the peroxisome division process remains to be determined.
Unlike PEX34, PEX25, and PEX27, deletion of the PEX11 gene was found to affect peroxisome abundance only under conditions of peroxisome proliferation. The levels of Pex11p have been shown to be extremely low in glucose-grown cells and become elevated only when cells are grown in oleic acid–containing medium promoting peroxisome proliferation (Karpichev and Small, 1998; Smith et al., 2002; Knoblach and Rachubinski, 2010). Therefore cells have adapted to maintaining their peroxisome number during rapid cell division with little requirement for Pex11p. It is interesting to speculate that, under conditions of constitutive peroxisome division, the proposed peroxisome elongation function of Pex11p (Schrader et al., 1998; Thoms and Erdmann, 2005; Koch et al., 2010) may be substituted for, at least in part, by the pulling force applied by the inheritance machinery that has been shown to elongate peroxisomes in cells lacking Vps1p (Hoepfner et al., 2001; Fagarasanu et al., 2009).
In closing, we have shown that Pex34p is a peroxisomal protein involved in controlling peroxisome abundance under conditions of both peroxisome proliferation and constitutive peroxisome division. Pex34p acts in controlling peroxisome numbers both alone and in cooperation with the Pex11 protein family of peroxisome divisional proteins. We also have provided new insight into the roles of these proteins in matrix protein import and in peroxisome stability and elongation. The discovery of Pex34p as a newly recognized peroxisomal protein involved in the already complex control of the peroxisome population of S. cerevisiae emphasizes the importance that cells place on strictly regulating their peroxisome population and ensuring that they have sufficient numbers of peroxisomes to thrive under a variety of conditions.
MATERIALS AND METHODS
Strains and cultures conditions
The S. cerevisiae strains used in this study are listed in Supplemental Table S1. All strains were cultured at 30°C. Strains containing plasmids were cultured in synthetic minimal (SM) medium. Media components were as follows: YPD, 1% yeast extract, 2% peptone, 2% glucose; YPBO, 0.3% yeast extract, 0.5% peptone, 0.5% K2HPO4, 0.5% KH2PO4, 0.2% (wt/vol) Tween 40, 1% (vol/vol) oleic acid; SM, 0.67% yeast nitrogen base without amino acids, 2% glucose, 1× complete supplement mixture (Bio 101, Vista, CA) without uracil or leucine; SCIM, 0.67% yeast nitrogen base without amino acids, 0.5% yeast extract, 0.5% peptone, 0.2% (wt/vol) Tween 40, 0.3% glucose, 0.3% (vol/vol) oleic acid, 1× complete supplement mixture or 1× complete supplement mixture without uracil, as appropriate.
GFP and mRFP tagging of genes
Genes were genomically tagged with sequence encoding an improved version of GFP from Aequoria victoria (Scholz et al., 2000) or mRFP by homologous recombination with a PCR-based integrative transformation of parental BY4742 haploid cells (Dilworth et al., 2001) and selection for the Streptomyces noursei NAT gene conferring resistance to the antibiotic neurseothricin (Krügel et al., 1988).
Gene overexpression
Individual PEX genes were cloned into the vector pBY011 (HIP FLEXGene S. cerevisiae ORF collection; Harvard Proteomics Institute, Cambridge, MA) for overexpression. PEX34 was cloned into the vector pGREG576 using drag-and-drop cloning (Jansen et al., 2005) for overexpression. For gene overexpression, cells were grown in SCIM for 16 h, at which time glucose and galactose were added to 0.2 and 1%, respectively. Images were acquired 1.5 h after addition of galactose.
Microscopy
Strains expressing GFP and/or mRFP fusion proteins were grown to midlog phase in YPD medium or SM medium, and then for 8 h in YPBO medium or for 16 h in SCIM, if required. Images were captured and analyzed essentially as described (Fagarasanu et al., 2009). Specifically, 2 μl of culture was combined with 8 μl of warmed nonfluorescent medium (in 1 l, 0.90 g KH2PO4, 0.23 g K2HPO4, 0.50 g MgSO4, 3.52 g (NH4)2SO4, 20 g glucose, 1× complete supplement mixture) containing 1.5% low-melting agarose and spread on a slide with two 18-mm square wells (Cel-line Brand, Thermo Scientific, Waltham, MA). Cells were incubated at room temperature for image capture, which was as described (Hammond and Glick, 2000) using a modified LSM 510 META confocal microscope equipped with a 63× 1.4 NA Plan-Apo objective (Carl Zeiss, Thornwood, NY). A piezoelectric actuator was used to drive continuous objective movement, allowing for the rapid collection of z-stacks. Stacks of 37 optical sections spaced 0.16 μm apart were captured.
Acquired images were deconvolved using algorithms provided by Huygens Professional Software (Scientific Volume Imaging, Hilversum, The Netherlands). For this method, three-dimensional (3D) data sets were processed to remove noise and reassign blur through an iterative Classic Maximum Likelihood Estimation algorithm and an experimentally derived point spread function. The transmission image was treated differently. In Huygens, a Gaussian filter was applied to the transmission image, and blue color was applied to the transmission image using Imaris 7.0 software (Bitplane, South Windsor, CT). The level of the transmission image was modified, and the image was processed until only the circumference of the cell was visible. To prevent interference of internal structures captured in the transmission images, the internal structures were removed in Adobe Photoshop. Imaris 7.0 was subsequently used to display the deconvolved 3D data set with the processed transmission image and to prepare the image files before final figure assembly in Adobe Photoshop and Adobe Illustrator. All images shown are representative, maximum intensity projections. Quantification was done using the surface measure function in Imaris 7.0.
Electron microscopy of whole yeast cells (Eitzen et al., 1997) and morphometric analysis of images (Tam et al., 2003) were performed as described.
Yeast two-hybrid analysis
Physical interactions between Pex34p, Pex11p, Pex25p, Pex27p, Fis1p, and Vps1p were determined as described (Tam et al., 2003). Chimeric genes were generated by amplifying the ORFs by PCR and ligating them in-frame and downstream of the DNA encoding the AD and the BD of the GAL4 transcriptional activator in the plasmids pGAD424 and pGBT9, respectively. Cells of the S. cerevisiae strain SFY526 were cotransformed with a pGAD424-derived plasmid and a pGBT9-derived plasmid. Transformants were grown as patches on filter paper overlaying selective medium agar plates overnight at 30°C and tested for activation of the integrated lacZ construct using a colorimetric assay for β-galactosidase activity.
Subcellular fractionation and isolation of peroxisomes
Subcellular fractionation and peroxisome isolation were done essentially as described (Smith et al., 2002; Tam et al., 2003). Cells grown to midlog phase in YPD medium or for 16 h in SCIM were harvested and converted to spheroplasts by digestion with Zymolyase 100T (MP Biomedicals, Solon, OH). Spheroplasts were disrupted by homogenization in buffer H (0.6 M sorbitol, 2.5 mM MES, pH 5.5, 1 mM EDTA) containing 1× complete protease inhibitor cocktail (Roche, Basel Switzerland). The homogenate was subjected to five repeated centrifugations for 7 min each at 1800 × g to yield a postnuclear supernatant (PNS) fraction. The PNS fraction was subjected to further differential centrifugation at 20,000 × g for 35 min to yield a pellet (20KgP) fraction enriched for peroxisomes and mitochondria and a supernatant (20KgS) fraction enriched for cytosol. The 20KgP fraction was resuspended in buffer H containing 11% Nycodenz and 1× complete protease inhibitor cocktail, and a volume containing 5 mg of protein was overlaid onto a 30-ml discontinuous gradient consisting of 17%, 25%, 35%, and 50% (wt/vol) Nycodenz, both in buffer H containing 1× complete protease inhibitor cocktail. Organelles were separated by centrifugation at 100,000 × g for 90 min in a VTi50 rotor (Beckman Coulter, Brea, CA). Fractions of 2 ml were collected from the bottom of the gradient.
In some experiments, the 20KgS fraction was subjected to ultracentrifugation at 200,000 × g for 1 h in a TLA 120.2 rotor (Beckman) to yield a pellet (200KgP) fraction enriched for small vesicles and a cytosolic supernatant (200KgS) fraction.
Extraction of peroxisomes
Peroxisomes were extracted as described previously (Fujiki et al., 1982; Tam et al., 2003). Essentially, organelles in the 20KgP fraction (50 μg of protein) were lysed by incubation in 10 volumes of ice-cold Ti8 buffer (10 mM Tris-HCl, pH 8.0) containing 2× complete protease inhibitor cocktail on ice for 1 h and separated by centrifugation at 200,000 × g for 1 h at 4°C in a TLA 120.2 rotor into pellet (Ti8P) and supernatant (Ti8S) fractions. The Ti8P fraction was resuspended in Ti8 buffer, and a portion was extracted with 0.1 M Na2CO3, pH 11.3, for 45 min on ice and then separated by centrifugation at 200,000 × g for 1 h at 4°C in a TLA 120.2 rotor into pellet (CO3P) and supernatant (CO3S) fractions. Proteins in fractions were precipitated by addition of trichloroacetic acid, and precipitates were washed with acetone. Proteins in equal portions of each fraction were separated by SDS–PAGE and analyzed by immunoblotting.
Antibodies
Antibodies to thiolase (Eitzen et al., 1996) and Sdh2p (Dibrov et al., 1998) have been described. Antibodies to full-length GFP were raised in rabbit and affinity-purified against full-length GFP for use in immunoblot analysis. Horseradish peroxidase–conjugated donkey anti–rabbit immunoglobulin (Ig)G and horseradish peroxidase–conjugated goat anti–guinea pig IgG secondary antibodies (Amersham Biosciences, Pittsburgh, PA) were used to detect primary antibodies in immunoblot analysis.
Acknowledgments
We thank Richard Poirier, Hanna Kroliczak, Elena Savidov, and Dwayne Weber for expert technical assistance. We also thank Fred Mast for help with confocal microscopy and subcellular fractionation and Barbara Knoblach and Jenny Chang for helpful discussion. R.J.T. is the recipient of an Alberta Advanced Education and Technology Advanced Student Scholarship. This work was supported by grant 53326 from the Canadian Institutes of Health Research to R.A.R and grant GM075152 from the U.S. National Institutes of Health to J.D.A. R.A.R. is an International Research Scholar of the Howard Hughes Medical Institute.
Abbreviations used:
- AD
transcription-activating domain
- BD
DNA-binding domain
- 3D
three-dimensional
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- mRFP
monomeric red fluorescent protein
- ORF
open reading frame
- PBD
peroxisome biogenesis disorder
- PEX
gene encoding a peroxin
- PNS
postnuclear supernatant
- PTS1
peroxisome-targeting signal type 1
- SM
synthetic minimal
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-01-0084) on March 25, 2011.
REFERENCES
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrov E, Fu S, Lemire BD. The Saccharomyces cerevisiae TCM62 gene encodes a chaperone necessary for the assembly of the mitochondrial succinate dehydrogenase (complex II) J Biol Chem. 1998;273:32042–32048. doi: 10.1074/jbc.273.48.32042. [DOI] [PubMed] [Google Scholar]
- Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153:1465–1478. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen GA, Szilard RK, Rachubinski RA. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane peroxin. J Cell Biol. 1997;137:1265–1278. doi: 10.1083/jcb.137.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen GA, Titorenko VI, Smith JJ, Veenhuis M, Szilard RK, Rachubinski RA. The Yarrowia lipolytica gene PAY5 encodes a peroxisomal integral membrane protein homologous to the mammalian peroxisome assembly factor PAF-1. J Biol Chem. 1996;271:20300–20306. doi: 10.1074/jbc.271.34.20300. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol. 1995;128:509–523. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, et al. Myosin-driven peroxisome partitioning in S. cerevisiae. J Cell Biol. 2009;186:541–554. doi: 10.1083/jcb.200904050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, Fagarasanu M, Eitzen GA, Aitchison JD, Rachubinski RA. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev Cell. 2006;10:587–600. doi: 10.1016/j.devcel.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Fagarasanu A, Fagarasanu M, Rachubinski RA. Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- Fagarasanu M, Fagarasanu A, Tam YYC, Aitchison JD, Rachubinski RA. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J Cell Biol. 2005;169:765–775. doi: 10.1083/jcb.200503083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond AT, Glick BS. Raising the speed limits for 4D fluorescent microscopy. Traffic. 2000;1:935–940. [PubMed] [Google Scholar]
- Hettema EH, Motley AM. How peroxisomes multiply. J Cell Sci. 2009;122:2331–2336. doi: 10.1242/jcs.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Hoepfner D, Van Den Berg M, Philippsen P, Tabak HF, Hettema EH. A role for Vps1p, actin and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J Cell Biol. 2001;155:979–990. doi: 10.1083/jcb.200107028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:568–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jansen G, Wu C, Schade B, Thomas DY, Whiteway M. Drag and drop cloning in yeast. Gene. 2005;344:43–51. doi: 10.1016/j.gene.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Karpichev IV, Small GM. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6560–6570. doi: 10.1128/mcb.18.11.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B, Rachubinski RA. Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J Biol Chem. 2010;285:6670–6680. doi: 10.1074/jbc.M109.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J, Pranjic K, Huber A, Ellinger A, Hartig A, Kragler F, Brocard C. PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J Cell Sci. 2010;123:3389–3400. doi: 10.1242/jcs.064907. [DOI] [PubMed] [Google Scholar]
- Krügel H, Fiedler G, Haupt I, Sarfert E, Simon H. Analysis of the nourseothricin-resistance gene (nat) of Streptomyces noursei. Gene. 1988;62:209–217. doi: 10.1016/0378-1119(88)90559-8. [DOI] [PubMed] [Google Scholar]
- Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, Van Der Klei IJ. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- Managadze D, Würtz C, Wiese S, Schneider M, Girzalsky W, Meyer HE, Erdmann R, Warscheid B, Rottensteiner H. Identification of PEX33, a novel component of the peroxisomal docking complex in the filamentous fungus Neurospora crassa. Eur Cell Biol. 2010;89:955–964. doi: 10.1016/j.ejcb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Mast FD, Fagarasanu A, Knoblach B, Rachubinski RA. Peroxisome biogenesis: something old, something new, something borrowed. Physiology. 2010;25:347–356. doi: 10.1152/physiol.00025.2010. [DOI] [PubMed] [Google Scholar]
- Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A, Ward G, Hettema E. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci. 2008;121:1633–1640. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–379. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner H, Stein K, Sonnenhol E, Erdmann R. Conserved function of Pex11p and the novel Pex25p and Pex27p in peroxisome biogenesis. Mol Biol Cell. 2003;14:4316–4328. doi: 10.1091/mbc.E03-03-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraya R, Veenhuis M, Van Der Klei IJ. Peroxisomes as dynamic organelles: peroxisome abundance in yeast. FEBS J. 2010;277:3279–3288. doi: 10.1111/j.1742-4658.2010.07740.x. [DOI] [PubMed] [Google Scholar]
- Scholz O, Thiel A, Hillen W, Niederweis M. Quantitative analysis of gene expression with an improved green fluorescent protein. Eur J Biochem. 2000;267:1565–1570. doi: 10.1046/j.1432-1327.2000.01170.x. [DOI] [PubMed] [Google Scholar]
- Schrader M, Fahimi HD. The peroxisome: still a mysterious organelle. Histochem Cell Biol. 2008;129:421–440. doi: 10.1007/s00418-008-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Reuber BE, Morrell JC, Jimenez-Sanchez G, Obie C, Stroh TA, Valle D, Schroer TA, Gould SJ. Expression of PEX11β mediates peroxisome proliferation in the absence of extracellular stimuli. J Biol Chem. 1998;273:29607–29614. doi: 10.1074/jbc.273.45.29607. [DOI] [PubMed] [Google Scholar]
- Smith JJ, et al. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J Cell Biol. 2002;158:259–271. doi: 10.1083/jcb.200204059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biovchim Biophys Acta. 2006;1763:1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Tabak HF, Van Der Zand A, Braakman I. Peroxisomes: minted by the ER. 2008;20:393–400. doi: 10.1016/j.ceb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Tam YYC, Fagarasanu A, Fagarasanu M, Rachubinski RA. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem. 2005;280:34933–34939. doi: 10.1074/jbc.M506208200. [DOI] [PubMed] [Google Scholar]
- Tam YYC, Torres-Guzman JC, Vizeacoumar FJ, Smith JJ, Marelli M, Aitchison JD, Rachubinski RA. Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:4089–4102. doi: 10.1091/mbc.E03-03-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S, Erdmann R. Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 2005;272:5169–5181. doi: 10.1111/j.1742-4658.2005.04939.x. [DOI] [PubMed] [Google Scholar]
- Vizeacoumar FJ, Torres-Guzman JC, Bouard D, Aitchison JD, Rachubinski RA. Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:665–677. doi: 10.1091/mbc.E03-09-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar FJ, Torres-Guzman JC, Tam YYC, Aitchison JD, Rachubinski RA. YHR150w and YDR479c encode peroxisomal integral membrane proteins involved in the regulation of peroxisome number, size, and distribution in Saccharomyces cerevisiae. J Cell Biol. 2003;161:321–332. doi: 10.1083/jcb.200210130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJA, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Bichem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- Wolfe J, Schliebs W, Erdmann R. Peroxisomes as dynamic organelles: peroxisomal matrix protein import. FEBS J. 2010;277:3268–3278. doi: 10.1111/j.1742-4658.2010.07739.x. [DOI] [PubMed] [Google Scholar]
- Wolinski H, Petrovicˇ U, Mattiazi M, Petschnigg J, Heise B, Natter K, Kohlwein SD. Imaging-based live cell yeast screen identifies novel factors involved in peroxisome assembly. J Proteome Res. 2009;8:20–27. doi: 10.1021/pr800782n. [DOI] [PubMed] [Google Scholar]
- Yan M, Naganand R, Subramani S. The control of peroxisome number and size during division and proliferation. Curr Opin Cell Biol. 2005;17:376–383. doi: 10.1016/j.ceb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]