The mechanisms of inner nuclear membrane protein trafficking remain mostly unknown. We identified UNC-84 mutants that suggest that moving from the endoplasmic reticulum to the nuclear envelope does not occur by diffusion alone. Three signals need to be disrupted to block localization, suggesting that multiple mechanisms facilitate trafficking to the inner nuclear membrane.
Abstract
Approximately 100 proteins are targeted to the inner nuclear membrane (INM), where they regulate chromatin and nuclear dynamics. The mechanisms underlying trafficking to the INM are poorly understood. The Caenorhabditis elegans SUN protein UNC-84 is an excellent model to investigate such mechanisms. UNC-84 recruits KASH proteins to the outer nuclear membrane to bridge the nuclear envelope (NE), mediating nuclear positioning. UNC-84 has four targeting sequences: two classical nuclear localization signals, an INM sorting motif, and a signal conserved in mammalian Sun1, the SUN—nuclear envelope localization signal. Mutations in some signals disrupt the timing of UNC-84 nuclear envelope localization, showing that diffusion is not sufficient to move all UNC-84 to the NE. Thus targeting UNC-84 requires an initial step that actively transports UNC-84 from the peripheral endoplasmic reticulum to the NE. Only when all four signals are simultaneously disrupted does UNC-84 completely fail to localize and to function in nuclear migration, meaning that at least three signals function, in part, redundantly to ensure proper targeting of UNC-84. Multiple mechanisms might also be used to target other proteins to the INM, thereby ensuring their proper and timely localization for essential cellular and developmental functions.
INTRODUCTION
SUN (Sad1–UNC-84) proteins are a class of integral-membrane proteins that are targeted specifically to the inner nuclear membrane (INM). SUN proteins recruit KASH (Klarsicht, ANC-1 and Syne homology) proteins to the outer nuclear membrane (ONM) to form a bridge across the nuclear envelope (NE) to transfer forces generated in the cytoplasm to the nucleoskeleton. KASH–SUN bridges function in many cellular and developmental events, including nuclear migration and anchorage, centrosome attachment to the NE, telomere movements in meiosis, chromatin regulation, and overall organization of the cytoskeleton (reviewed in Starr, 2009; Starr and Fridolfsson, 2010). Besides SUN proteins, ∼100 integral-membrane proteins are predicted to be enriched at the NE (Schirmer et al., 2003; Wilkie et al., 2011). Together, with the intermediate filament lamin, INM proteins form the nuclear lamina, which provides structural support to the nucleus and is involved in the regulation of chromatin and other cellular processes (Gruenbaum et al., 2005; Akhtar and Gasser, 2007; Stewart et al., 2007). Mutations in lamin or some INM proteins lead to a broad spectrum of >20 human diseases termed laminopathies, which include muscular dystrophies, neuropathies, lipodystrophies, and premature aging disorders (reviewed in Worman and Bonne, 2007). Recently, it was demonstrated that a subset of lamin-A mutations that result in Hutchinson–Gilford progeria syndrome and Emery–Dreifuss muscular dystrophy disrupt the interaction between lamin-A and mammalian Sun1, suggesting that SUN proteins are also involved in the formation or development of laminopathies (Haque et al., 2010). Despite the importance of SUN proteins, the mechanisms of how they are targeted to the INM, the first step in the formation of NE bridges, are unknown. In fact, the mechanisms of how membrane proteins in general are targeted to the INM remain poorly understood.
Proteins destined for the INM must be transported from their sites of synthesis at the peripheral endoplasmic reticulum (ER) to the NE. In some cells, the surface area of the ER is more than 100 times as large as that of the NE (Weibel et al., 1969; Bolender, 1974), making transport from the ER to the ONM unlikely to occur by diffusion alone. Furthermore, INM proteins need to be trafficked across four continuous membrane domains: the ER, the ONM, the highly curved membrane of the nuclear pore, and the INM. Finally, the nuclear pore complex presents a physical barrier to INM protein trafficking. Three nonexclusive mechanisms have been proposed to contribute to trafficking proteins to the INM—diffusion-retention, via classical nuclear localization signals (cNLSs), and using inner nuclear membrane–sorting motifs (INM-SM).
The longest-standing model to explain trafficking of membrane proteins to the INM has been the diffusion-retention model (Powell and Burke, 1990; Holmer and Worman, 2001; Ostlund et al., 2006). In this model, integral membrane proteins destined for the INM are synthesized on the ER membrane and diffuse within the continuous lipid bilayer to the ONM and across the nuclear pore complex to the INM. Once in the INM, the protein binds to lamin, chromatin, or associated proteins and is retained. Three problems with this model have been recently uncovered. First, the size of the nucleoplasmic or lumenal domains of some INM proteins limits diffusion across the nuclear pore (Ohba et al., 2004). Second, even small INM proteins have been shown to require energy for trafficking to the INM (Ohba et al., 2004). Third, a lamin-binding domain in a membrane-bound protein is not sufficient for localization to the INM (Haque et al., 2010).
More recently it has been proposed that INM proteins are actively targeted to the nucleus using the same machinery as soluble proteins destined for the nucleoplasm (King et al., 2006; Lusk et al., 2007). In this model, INM proteins have at least one cNLS in their nucleoplasmic domain that is recognized by the soluble import machinery, which includes importins or karyopherins. The INM protein–importin complex is then transported into the nucleus, using the Ran-guanosine 5’-triphosphate (Ran-GTP) gradient, while remaining membrane bound. This mechanism is used for targeting the Saccharomyces cerevisiae Heh1 and Heh2 to the INM (King et al., 2006). It also plays a role in targeting mammalian Sun2 to the INM (Turgay et al., 2010). Not all INM proteins, however, contain a putative cNLS, and even among those that do, the cNLSs are not always necessary for NE localization. For example, the mammalian INM protein Sun1 lacks a putative cNLS (Lusk et al., 2007), and mutating the putative cNLS in mammalian Sun2 had no effect on its ability to target to the NE (Hodzic et al., 2004; Turgay et al., 2010). An additional possibility is that members of the importin-β family could function independently of importin-α by binding directly to INM proteins (Lu et al., 2010; Xu et al., 2010). Importin-β binding signals, however, are poorly defined, making it difficult to determine their roles in targeting INM proteins. In summary, the widespread use of importins to target INM proteins has yet to be demonstrated in higher eukaryotes.
A third model proposes that truncated membrane-associated importin-α actively facilitates the transport of INM proteins from the ER to the NE (Braunagel et al., 2004, 2007; Saksena et al., 2006; Liu et al., 2010). The truncated, membrane-associated importin-α-16 recognizes an INM-SM on the cargo protein, while it is cotranslationally inserted into the ER membrane. Importin-α-16 is then thought to remain associated with the INM protein until it enters the nucleus (Saksena et al., 2006). The INM-SM consists of two or more positively charged residues within eight residues of the nucleoplasmic face of the transmembrane span (Braunagel et al., 2004). Spodoptera frugiperda importin-α-16 interacts with the INM-SM of viral proteins destined for the INM as well as mammalian INM proteins, lamin-B receptor and nurim (Saksena et al., 2006; Braunagel et al., 2007). The yeast protein Heh2 uses a truncated importin-α and an INM-SM, as well as its cNLS, to localize to the NE (Liu et al., 2010). The mammalian importin-α-16 homologue, KPNA-4–16, interacts with a viral INM-SM in vitro (Braunagel et al., 2007). The presence of predicted INM-SMs in many different INM proteins and the discovery of truncated importin-α isoforms in insect, yeast, and mammalian cells suggest that this mechanism may be conserved to target many different INM proteins (Braunagel et al., 2004; Liu et al., 2010). However, the function of an INM-SM has not been tested in an endogenous metazoan protein.
Here, we test the relative contributions of these three models on the trafficking of the Caenorhabditis elegans protein UNC-84 to the INM. UNC-84 is a founding member of the SUN family of proteins that are conserved across all eukaryotes (Malone et al., 1999; Starr, 2009). UNC-84 localizes to the INM in a lamin-dependent manner (Malone et al., 1999; Lee et al., 2002; McGee et al., 2006). UNC-84 recruits the KASH proteins UNC-83 and ANC-1 to the ONM to mediate nuclear migration and anchorage, respectively (Starr et al., 2001; Starr and Han, 2002; McGee et al., 2006).
UNC-84 is an excellent model in which to study INM protein trafficking for multiple reasons. First, UNC-84 has the largest predicted nucleoplasmic (59 kDa) and lumenal (65 kDa) domains of any known endogenous INM protein. Second, based on the observation that large (>75 kDa) INM protein reporters lacking cNLSs are strongly inhibited from gaining access to the INM (Ohba et al., 2004), it is unlikely that an INM protein of 126 kDa relies solely on diffusion/retention for trafficking to the INM. Third, UNC-84 contains a potential INM-SM (Braunagel et al., 2004) and two potential cNLSs that we hypothesized function to target UNC-84 to the INM.
To identify signals in UNC-84 that function to target it to the INM, we examined the ability of various UNC-84 mutant proteins to localize to the NE during different developmental stages. We also tested the function of our mutants during nuclear migration. Nuclear migration in C. elegans hyp7 cells requires the proper targeting of UNC-84 to the INM; failure of UNC-84 localization results in an easily quantified functional assay in the context of a developing organism (Malone et al., 1999). Importantly, we can express transgenic mutant versions of UNC-84 in an otherwise null background and assay both localization and function (McGee et al., 2006).
Here we show that multiple cytoplasmic/nucleoplasmic signals contribute to the localization of UNC-84 to the NE. A novel NE localization signal conserved among SUN proteins coined the SUN-NELS (nuclear envelope localization signal) and an INM-SM function to efficiently target UNC-84 to the NE during early embryogenesis. There were no observable defects in UNC-84 trafficking in transgenic animals overexpressing our UNC-84 genomic rescuing construct or the rescuing construct with both predicted cNLSs mutated. UNC-84 constructs mutant for the SUN-NELS, the INM-SM, or both were slower to accumulate at the NE during early embryogenesis but nonetheless were able to localize to the NE and function in hyp7 nuclear migration. Only when mutations or small deletions were engineered into all three types of targeting signals did UNC-84 fail to localize to the NE and function during nuclear migration. Thus multiple signals contribute to efficient NE and INM targeting. Furthermore, these data support the model that INM-SMs contribute to active ER-to-NE trafficking of INM proteins.
RESULTS
UNC-84 has one functional transmembrane domain
In our NE bridging model, the N-terminal domain of UNC-84 is in the nucleoplasm and the C-terminal SUN domain is in the perinuclear space, where it can interact with the KASH domains of UNC-83 or ANC-1 during nuclear migration and anchorage, respectively (Starr et al., 2001; Starr and Han, 2002; McGee et al., 2006; Starr, 2009). The topology of UNC-84 in our model is supported by the following observations: (i) UNC-84 does not contain a predicted N-terminal signal sequence (Emanuelsson et al., 2007); (ii) the C-terminal membrane–bound SUN domain of UNC-84 interacts with the KASH domain of UNC-83 in the lumenal compartment of the endomembrane system in a membrane-bound yeast-two hybrid (McGee et al., 2006); (iii) similar topologies have been confirmed for mammalian SUN proteins (Hodzic et al., 2004; Crisp et al., 2006; Haque et al., 2006; Liu et al., 2007). Transmembrane prediction programs identify up to five hydrophobic regions (designated here as H1–H5) as potential membrane-spanning regions in UNC-84 (von Heijne, 1992). More conservative algorithms predict a single transmembrane span at residues 512–532 (H2) (Malone et al., 1999).
To determine which predicted transmembrane regions are real membrane-spanning domains, each of the five hydrophobic regions were systematically deleted from the UNC-84 genomic rescuing fragment (McGee et al., 2006). These deletion constructs were used to create transgenic lines in the null unc-84(n369) background (Malone et al., 1999). Three tests were performed to determine functionality of each UNC-84 mutant construct. First, transgenic lines were assayed for mutant UNC-84 localization to the NE (Lee et al., 2002). Second, UNC-83 localization to the NE, which depends on functional UNC-84 (Starr et al., 2001), was examined. Third, rescue of the nuclear migration phenotype of unc-84(n369) was determined by counting hyp7 nuclei in the dorsal cord. In wild-type animals, hyp7 nuclei migrate across the length of the cells (Sulston et al., 1983), whereas in unc-84(n369) null animals, nuclear migration is disrupted, and 14–15 hyp7 nuclei mislocalize to the dorsal cord of L1 larvae (Horvitz and Sulston, 1980; Malone et al., 1999; McGee et al., 2006). This nuclear migration defect can be efficiently rescued by a wild-type UNC-84 transgene in which only an average of 1–2 nuclei are observed in L1 dorsal cords (McGee et al., 2006). We predicted that, if a deleted hydrophobic domain was a real membrane-spanning domain, the topology of the transgenic UNC-84 construct would be disrupted. This may or may not affect the targeting of UNC-84 to the NE, but would likely disrupt the function of UNC-84 in recruiting UNC-83 to the NE and in nuclear migration.
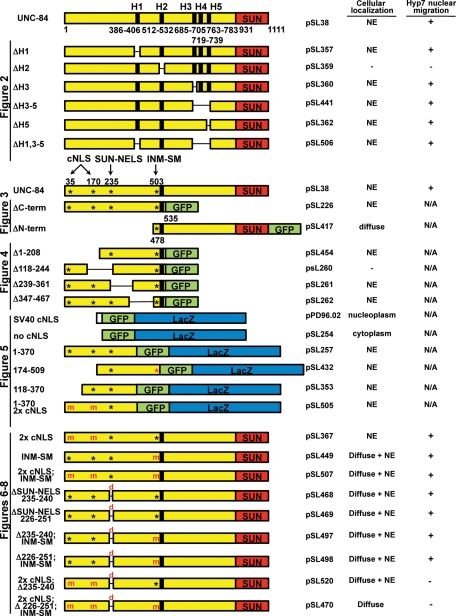
Deleting single, 20-residue hydrophobic regions H1, H3, or H5 or a 97-amino-acid deletion of H3–5 had no effect on the rescuing ability of the UNC-84 transgene (Figures 1 and 2, B and F–L; Table 1). In all four of these cases, both UNC-84 and UNC-83 localized properly to the NE, and the hyp7 nuclear migration defect was rescued (Figure 2A). Deleting residues 512–532, corresponding to H2, however, disrupted the function of the UNC-84 transgene (Figure 2A). In four independent transgenic lines, UNC-84 and UNC-83 failed to localize to the NE, and most hyp7 nuclei were observed in the dorsal cord of transgenic animals (Figure 2, D and E, and Table 1). No UNC-84 staining was observed in the ΔH2 transgenic lines using an antibody that recognizes the extralumenal domain (Figure 2D). To confirm that ΔH2 was expressed, a 6-myc tag was added to the C-terminal, lumenal domain of UNC-84. In two independent transgenic lines, UNC-84 ΔH2::6-myc was found to localize throughout the cytoplasm in embryos but still failed to rescue the unc-84 nuclear migration defect (unpublished data). No UNC-84 ΔH2::6-myc was observed in larvae, suggesting that the mistargeted protein may be unstable. To further confirm the importance of H2, we created an UNC-84 transgene in which all four other potential membrane-spanning regions were deleted (ΔH1,3–5). UNC-84ΔH1,3–5 localized to the NE and rescued unc-84(n369) (Figure 2, A, L, and M; Table 1). We therefore conclude that H2, the hydrophobic span from 512 to 532, is the only membrane-spanning domain and is essential for the function of UNC-84.
FIGURE 1:
UNC-84 constructs used for in vivo localization studies and functional analyses. Each plasmid's common name used in the text is on the left of the schematic. To the right is its official plasmid name, its cellular localization, and its ability to rescue the unc-84(n369) hyp7 nuclear migration defect. Full-length UNC-84 (yellow and red) containing its predicted transmembrane domains (black bars), the location of its potential NLSs (*), and its SUN domain (red) are shown. The figure where the constructs are used is indicated on the left. All membrane-bound fusions were driven by the unc-84 promoter. The soluble fusions for Figure 5 were driven by the unc-54 promoter. Figure 2: UNC-84 mutant constructs with small deletions in hydrophobic regions H1–H5. Figure 3: Two constructs, ΔC-term and ΔN-term, containing membrane-bound halves of UNC-84 were fused to a C-terminal GFP (green). Figure 4: Indicated residues were deleted from the parent ΔC-term UNC-84 construct. Figure 5: Soluble regions of UNC-84's nucleoplasmic domain were fused to GFP::LacZ (green and blue) to test active import in C. elegans muscle cells. Predicted functional NLSs are indicated (black *). In one construct the INM-SM is present (red *) but likely not functional because it is not in the context of a transmembrane span. Control constructs GFP::LacZ and SV40 cNLS::GFP::LacZ are also shown. Figures 6–8: UNC-84 mutant constructs contain mutations (m) in NLSs, where basic residues are converted to alanine or small deletions, 6 or 26 residues (d), which disrupt the SUN-NELS. Analogous constructs were made with C-terminal 6x myc tags for Figure 9.
FIGURE 2:
UNC-84 has one functional transmembrane domain located at residues 512–532. (A) Diagram of UNC-84, including the location of each of its hydrophobic domains (H1–H5) in relation to the SUN domain. The graph shows the number of nuclei in the dorsal cord of an average L1 animal. Error bars show the SD. Only one transgenic line for each construct is shown; full data are in Table 1. All transgenic lines were made in an unc-84(n369) background. (B, D, F, H, J, and L) UNC-84 and (C, E, G, I, K, and M) UNC-83 immunostaining of transgenic lines expressing the following UNC-84 deletions: (B and C) ΔH1; (D–E) ΔH2; (F and G) ΔH3; (H and I) ΔH3–5; (J and K) ΔH5; (L and M) ΔH1, 3–5. Scale bar = 10 μm.
TABLE 1:
Ability of UNC-84 domain deletion transgenic constructs to rescue nuclear migration.
| UNC-84 constructa | Line | Number of nuclei in dorsal cordb |
|---|---|---|
| None (wild-type background) | N2 | 0 ± 0 (11) |
| None (unc-84(n369) background) | MT369 | 14.3 ± 1.5 (25) |
| UNC-84 (+) rescuing construct | UD87 | 0.2 ± 0.5 (25) |
| UNC-84ΔSUN | UD61 | 14.8 ± 1.1 (≥15)c |
| Transmembrane domain analysis (Figure 2) | ||
| ΔH1 (386–406) | 1 | 0.2 ± 0.5 (20) |
| 2 | 0.8 ± 1.9 (10) | |
| ΔH2 (512–532) | 1 | 14.5 ± 1.6 (26) |
| 2 | 14.1 ± 1.5 (17) | |
| 3 | 13.6 ± 1.2 (13) | |
| 4 | 14.5 ± 1.4 (12) | |
| ΔH3 (685–705) | 1 | 1.4 ± 3.4 (25) |
| 2 | 2.6 ± 4.8 (16) | |
| ΔH5 (719–738) | 1 | 0.8 ± 2.2 (44) |
| 2 | 1.5 ± 4.1 (10) | |
| ΔH3–5 (685–782) | 1 | 0.7 ± 2.1 (19) |
| 2 | 1.5 ± 2.3 (11) | |
| 3 | 2.4 ± 2.3 (10) | |
| ΔH1,3–5 (386–406;685–782) | 1 | 2.2 ± 4.3 (6) |
| NLS analysis (Figure 6) | ||
| 2xcNLS (KK 38–39 AA; HRRR 170–173 AAAA) | 1 | 4.8 ± 3.6 (13) |
| 2 | 1.3 ± 1.9 (10) | |
| 3 | 2.9 ± 2.5 (8) | |
| INM-SM (KKSSK 503 AASSA) | 1 | 0.5 ± 1.9 (33) |
| 2 | 0.6 ± 1.8 (8) | |
| 2xcNLS; INM-SM | 1 | 0.7 ± 1.7 (13) |
| ΔSUN-NELS (235–240) | 1 | 0.7 ± 1.7 (9) |
| 2 | 0.1 ± 0.3 (9) | |
| 3 | 0.9 ± 1.9 (7) | |
| ΔSUN-NELS (226–251) | 1 | 1.1 ± 1.8 (14) |
| ΔSUN-NELS (235–240); INM-SM | 1 | 0.9 ± 2.9 (15) |
| 2 | 0.5 ± 1.8 (11) | |
| ΔSUN-NELS (226–251); INM-SM | 1 | 2.9 ± 3.2 (23) |
| 2 | 5.2 ± 3.3 (16) | |
| 3 | 1.8 ± 3.4 (12) | |
| 2xcNLS; ΔSUN-NELS (235–240) | 1 | 13.0 ± 2.0 (66) |
| 2 | 13.7 ± 1.9 (31) | |
| 3 | 13.6 ± 1.4 (17) | |
| 2xcNLS; ΔSUN-NELS (226–251); INM-SM | 1 | 14.0 ± 1.2 (23) |
| 2 | 12.3 ± 3.1 (10) | |
| 3 | 14.4 ± 0.9 (8) | |
| 4 | 13.4 ± 2.3 (7) | |
| 5 | 12.8 ± 1.5 (6) | |
| 6 | 14.5 ± 0.6 (4) |
aAll mutant constructs were constructed in the UNC-84 rescuing construct pSL38 (McGee et al., 2006). The exact residues deleted in the mutant constructs are indicated in parentheses.
bMean number of nuclei and the SD are shown. Sample size is in parentheses. Nuclei in dorsal cord were counted blindly, before determining whether the animal was transgenic. podr-1RFP or sur-5::GFP were used as transgenic markers. Nontransgenic animals in each blind count had an average of 13.3–15.3 nuclei (n is between 5 and 44). All lines are in an unc-84(n369) mutant background.
cFrom McGee et al., 2006.
The membrane-bound N terminus of UNC-84 is sufficient for hyp7 NE targeting
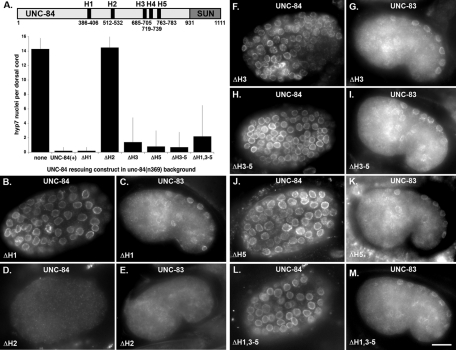
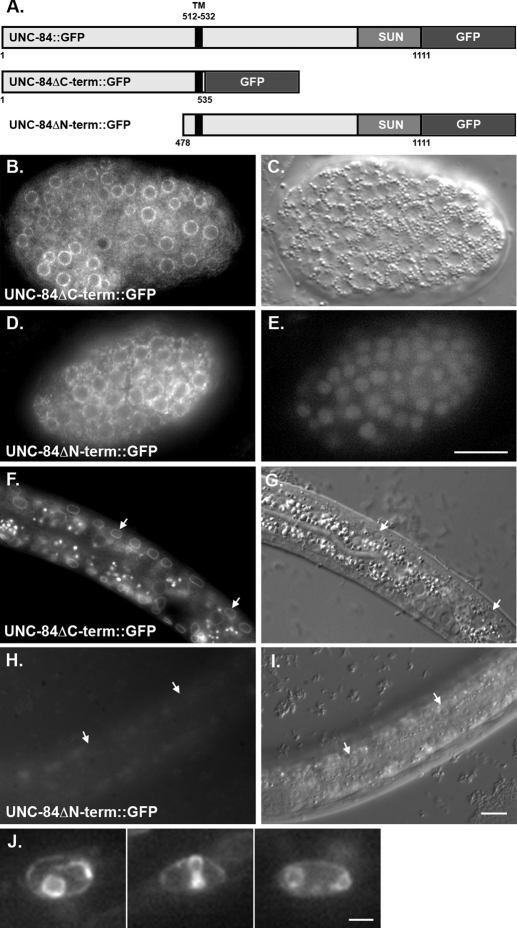
On the basis of the likely topology of UNC-84, we hypothesized that the determinants required for targeting UNC-84 to the INM should reside in its N-terminal, extralumenal domain. To test this hypothesis, two constructs containing either UNC-84's nucleoplasmic domain or lumenal domain, fused to the single membrane-spanning region and a C-terminal GFP, were expressed in transgenic animals under the control of the unc-84 promoter (Figure 3). The N-terminal domain of UNC-84 (UNC-84ΔC::GFP; consisting of residues 1–535) was expressed in an otherwise null unc-84(n369) background to prevent potential nuclear targeting of the transgenic protein through dimerization with endogenous UNC-84 (Malone et al., 1999). It was previously shown that the C-terminal SUN domain of UNC-84 is required for function but not for localization (McGee et al., 2006). Thus UNC-84ΔC::GFP was not functional for nuclear migration. UNC-84ΔC::GFP efficiently trafficked to the NE. Strong GFP was observed at the nuclear periphery in live embryonic and L1 cells (Figure 3, B, C, F, and G). Occasionally, in rare nuclei from animals with the brightest GFP signal (6 of 32 transgenic animals from one transgenic line), UNC-84ΔC::GFP was seen in finger-like and circular projections that extended into the nucleus from the periphery (Figure 3J). The C-terminal domain (UNC-84ΔN::GFP; residues 478–1111) was not detected in live embryos, larva, or adult transgenic animals in a wild-type background (Figure 3, H and I). UNC-84ΔN::GFP was observed in a pattern consistent with the ER in fixed embryonic cells stained with anti-GFP antibodies (Figure 3, D and E). Together, these results demonstrate that the N-terminal, nucleoplasmic half of UNC-84 (residues 1–535) contains all the determinants required for efficient localization of UNC-84 to the NE.
FIGURE 3:
The N-terminal domain of UNC-84 is sufficient for targeting to the NE. (A) Diagram of the constructs. The parent construct, full-length UNC-84 with its endogenous promoter and GFP at its C terminus, rescues unc-84 and localizes to the NE (Malone et al., 1999). UNC-84ΔC-term::GFP (residues 1–535) and UNC-84ΔN-term::GFP (residues 478–1111) are shown. (B, C, F, and G) GFP fluorescence and DIC images of a live embryo (B and C) or an L1 larva (F and G) expressing UNC-84ΔC-term::GFP at the NE. (D and E) Anti-GFP immunofluorescence and DAPI-stained nuclei in a fixed embryo expressing UNC-84ΔN-term::GFP. The GFP was not observed in live embryos. (H and I) GFP fluorescence and DIC images of a live L1 larva expressing UNC-84ΔN-term::GFP. Arrows point out representative hyp7 cell nuclei. (J) Individual nuclei from L1 larvae expressing high levels of UNC-84ΔC-term::GFP. Scale bars for B–E and F–I = 10 μm. Scale bar for J = 1 μm.
Identifying regions necessary for targeting UNC-84 to the NE
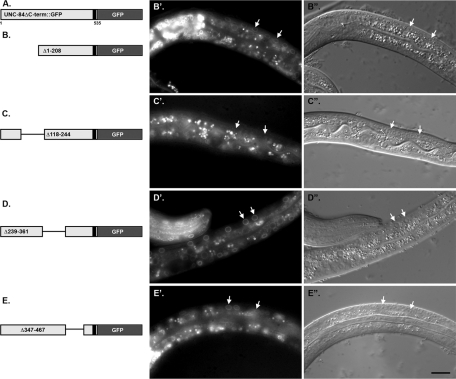
To further identify regions that are required for targeting UNC-84 to the NE, a deletion analysis was performed. Transgenic lines were made expressing various deletions of the membrane-bound UNC-84ΔC::GFP construct (consisting of residues 1–535 driven by the unc-84 promoter) in an unc-84(n369) background (Figure 4). We examined the localization of the GFP constructs in postmitotic larval hyp7 nuclei. When residues 1–208, 239–361, or 367–467 were deleted, UNC-84 localized to the NE of hyp7 nuclei (Figure 4B′, D′, and E′). Deleting residues 118–244, however, severely disrupted UNC-84 localization to the NE in hyp7 cells (Figure 4C′). It remains unclear whether mistargeting of the 118–244 deletion is a result of protein misfolding or represents an important region in UNC-84 that allows it to interact with the nuclear lamina and/or that acts as a nuclear import signal. We therefore hypothesize that residues 118–244 are important for the localization of UNC-84 to the NE.
FIGURE 4:
Residues 118–244 of UNC-84 are necessary for targeting the membrane-bound N-terminal domain of UNC-84 to the NE. (A) Schematic of the parent construct. (B–E) Individual deletions in UNC-84ΔC-term::GFP were expressed in transgenic unc-84(n369) animals. The deleted residues are indicated. (B′–E′) GFP fluorescence (B′′–E′′) and corresponding DIC images of live larvae. Arrows mark representative hyp7 nuclei. Scale bar = 10 μm.
UNC-84 is actively transported into the nucleus as a soluble fusion protein
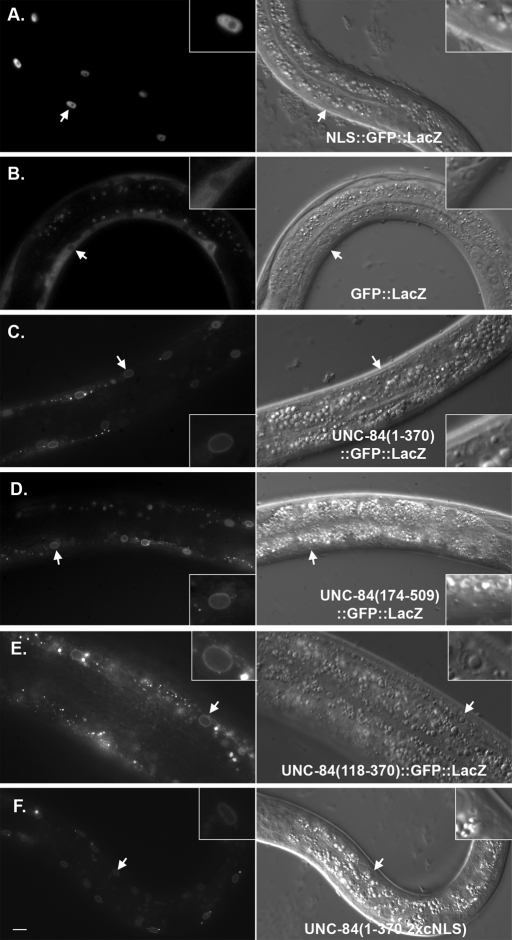
The experiments just mentioned tested the necessity of UNC-84 domains for NE targeting. They failed, however, to discriminate between simple diffusion and active import across the nuclear pore. Soluble proteins greater than ∼40–60 kDa are not able to diffuse through the nuclear pore and must be actively imported through nuclear pores and into the nucleus using importins and the Ran-GTP gradient (Nigg, 1997). For example, in C. elegans, a 70-kDa dextran does not diffuse through the nuclear pore (Galy et al., 2003). A GFP::LacZ fusion protein (∼145 kDa) remained in the cytoplasm (Figure 5B). The addition of a classical simian virus 40 (SV40) cNLS, however, was sufficient to drive the large fusion protein into the nucleoplasm (Figure 5A). To test whether regions in the cytoplasmic domain of UNC-84 are capable of soluble active transport across the nuclear pore, pieces of the extralumenal domain of UNC-84, without the membrane-spanning domain, were fused to GFP::LacZ and expressed in transgenic muscles under control of the unc-54 promoter. The unc-54 promoter turns on postmitotically in 81 of the 95 body wall muscle cells born in the early embryo (Epstein et al., 1993). Regions 1–370 or 174–509 of UNC-84 were sufficient to target GFP::LacZ to the NE (Figure 5, C and D). Our imaging was not able to formally distinguish between outer and inner NE localization. Given that UNC-84 is normally localized to the INM (McGee et al., 2006), however, our data are consistent with the hypothesis that regions 1–370 and 174–509 of UNC-84 contain signals for active import into the nucleus. Therefore these regions of UNC-84 are likely sufficient for both active targeting of the large fusion protein into the nucleus and for retention of UNC-84 at the NE, presumably through an interaction with a component of the nuclear lamina. To further map nuclear localization signals (NLSs) and lamina-interacting domains of UNC-84, a smaller portion of the soluble N-terminal domain of UNC-84 was expressed as a GFP::LacZ fusion protein. Residues 118–370 targeted the fusion protein to the NE (Figure 5E). Thus residues 118–370 of UNC-84 contain both nuclear import signals and lamina-interacting domains.
FIGURE 5:
UNC-84 is actively imported into the nucleus. Live larvae were imaged for both GFP fluorescence (left) and DIC (right). Transgenes are under the control of a muscle-specific promoter. Arrows point to muscle nuclei that are enlarged in the insets. (A) The SV40-NLS::GFP::LacZ control is targeted to the nucleoplasm. (B) The GFP::LacZ control is cytoplasmic and excluded from the nucleus. (C–E) Soluble fragments of UNC-84 fused to GFP::LacZ were examined for nuclear localization. Residues 1–370 (C), 174–509 (D), and 118–370 (E) of UNC-84 were sufficient for NE targeting. UNC-84(1–370; 2xcNLS)::GFP::LacZ, with mutations in both predicted cNLSs, also localizes to the NE. Scale bar = 10 μm.
The role of predicted nuclear import signals in targeting UNC-84 to the INM
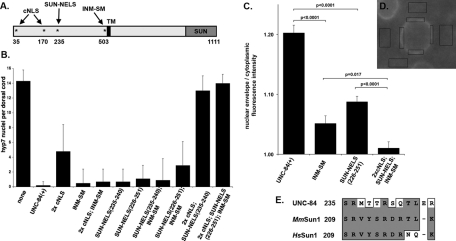
Having established that the extralumenal domain of UNC-84 contains regions that are necessary for NE localization and sufficient to target soluble pieces of UNC-84 to the NE in postmitotic body-wall muscle cells, we asked whether signals that have previously been shown to play a role in the active targeting of other INM proteins (the INM-SM and cNLSs) function in trafficking UNC-84. To test whether these potential signals are required for UNC-84 localization, mutations were created in our unc-84 rescuing construct (Figures 1 and 6A). The resulting constructs were expressed in unc-84(n369) null animals. Transgenic animals were assayed as mentioned earlier in the text for UNC-84 function by quantifying hyp7 nuclear migration and by immunofluorescence to examine UNC-84 and UNC-83 localization at the NE. As a control, the wild-type unc-84 rescuing construct was shown to restore both nuclear migration in hyp7 cells and UNC-84 and UNC-83 localization to the NE (Table 1 and Figures 6B and 7C).
FIGURE 6:
Signals in the nucleoplasmic domain of UNC-84 that function for localization and migration. (A) Schematic of full-length UNC-84 showing the location of the cNLSs, SUN-NELS, and INM-SM (*). Transgenic lines were made in an unc-84(n369) background. (B) The number of nuclei in the dorsal cord of an average L1 animal. Standard deviation bars are shown. Only one transgenic line for each construct is shown; complete data are in Table 1. (C and D) Quantification of transgenic UNC-84 protein localization. (C) The ratio of the average pixel intensity of anti-UNC-84 antibody localization to the NE over the cytoplasm is shown for nuclei expressing various unc-84 transgenes. Standard error bars are shown. Sample sizes include at least a total of 25 nuclei from at least four different embryos. Significant p values from statistical Student's t tests are indicated. (D) To determine the ratio, immunofluorescence images as in Figure 7 were analyzed. Four regions of the NE (as determined by DAPI staining; unpublished data) and four neighboring regions of the cytoplasm were measured for each nucleus. (E) Alignment of the SUN-NELS conserved between UNC-84 and its mammalian homologues MmSun1(GenBank: EDL19166.1) and HsSun1 (GenBank: EAW87177.1). Residue positions are from the largest isoforms. Similar residues are boxed and identical residues are shaded. The dash represents a gap in the alignment.
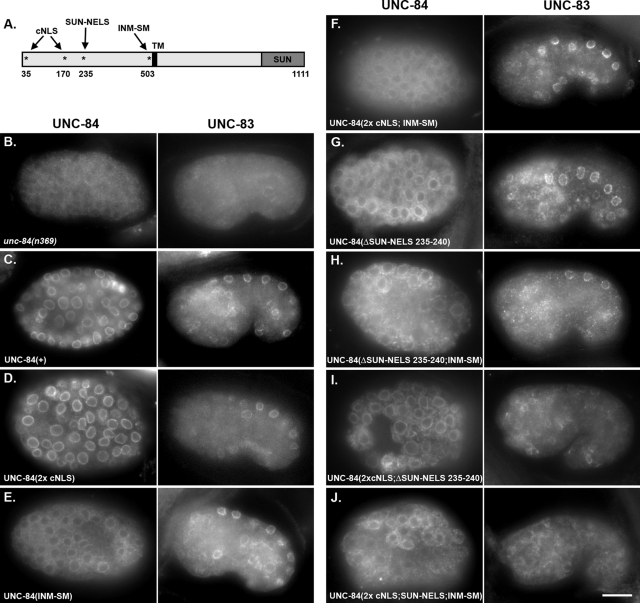
FIGURE 7:
Multiple signals in the nucleoplasmic domain of UNC-84 are required for INM targeting. (A) Schematic of full-length UNC-84 as in Figure 6A. (B–J) Immunofluorescence showing localization of UNC-84 (left) and UNC-83 (right). The right panels are from comma-stage embryos after hyp7 nuclear migration, and the left panels are from embryos approximately 1 h earlier when the cells were larger. Anterior is left and dorsal is up. All transgenic lines were made in an unc-84(n369) background. The following forms of UNC-84 were expressed from transgenes: (B) no transgene in the unc-84(n369) background; (C) rescuing UNC-84(+); (D) UNC-84(2x cNLS); (E) UNC-84(INM-SM); (F) UNC-84(2x cNLS; INM-SM); (G) UNC-84(ΔSUN-NELS 235–240); (H) UNC-84(ΔSUN-NELS 235–240; INM-SM); (I) UNC-84(2xcNLS;ΔSUN-NELS 235–240); (J) UNC-84(2xcNLS;ΔSUN-NELS 226–251; INM-SM). Scale bar = 10 μm.
cNLSs, such as those originally identified in the SV40 T antigen (Kalderon et al., 1984), bind the soluble import machinery, importins/karyopherins, for transport across nuclear pores and into the nucleus. cNLSs consist of short stretches of basic residues, but are often difficult to predict based on primary sequence alone (Cook et al., 2007; Lange et al., 2007; Stewart, 2007). Two short stretches of basic residues (35KVRRK39 and 170HRRR173) that could potentially act as cNLSs were identified in the N-terminal domain of UNC-84 by manual scanning or PSORT II, respectively (Horton and Nakai, 1997). Mutating the potential cNLS 170HRRR173 to 170AAAA173 or mutating both potential cNLSs (35KVRAA39 and 170AAAA173) had no effect on NE localization of UNC-84ΔC::GFP in L1 larvae (unpublished data). In addition, mutating both potential cNLSs in the UNC-84(1–370)::GFP::LacZ soluble fusion protein had no effect on localization of the fusion protein to the NE (Figure 5F). These data suggest that an independent, unidentified NE localization signal resides within residues 1–370. To further test the role of predicted cNLSs, both potential cNLSs were mutated in full-length UNC-84 and expressed in transgenic animals. In such animals, UNC-84 and UNC-83 localized normally to the NE and most hyp7 nuclei migrated properly (Table 1, Figures 6B and 7D). We therefore concluded that the putative cNLSs play little or no role of their own in the targeting of UNC-84 to the INM but they may still function redundantly to other signals.
The consensus sequence of an INM-SM is loosely defined to consist of at least two positively charged residues within five to eight residues of the nucleoplasmic face of the transmembrane domain (Braunagel et al., 2004). UNC-84 has a predicted INM-SM consisting of amino acids KKSSK five to nine residues from the N-terminal end of the transmembrane span (Braunagel et al., 2004). To test the requirement of this predicted INM-SM for targeting UNC-84, the three codons for the positively charged residues were mutated to encode alanines in the unc-84 rescuing construct. The INM-SM mutation (503KKSSK507 to 503AASSA507) did not disrupt the localization of UNC-84 to the NE in comma-stage embryos shortly after nuclear migration and was indistinguishable from wild type (Figure 8B, compare to 8A). The INM-SM mutation, however, disrupted efficient transport of UNC-84 to the NE about 1–2 h earlier in embryogenesis. Although there appeared to be some UNC-84 at the NE of earlier embryos, most of the anti-UNC-84 immunofluorescence was instead distributed throughout the cytoplasm (Figure 7E). We quantified this localization defect by measuring the ratio of the UNC-84 staining intensities at the NE over the cytoplasm. The NE/cytoplasmic ratio of UNC-84 fluorescence intensity was significantly higher in UNC-84(+) transgenics than in the UNC-84(INM-SM) (Figure 6, C–D), showing a significant decrease in localization at the NE. Surprisingly, given this trafficking defect, UNC-84 localized normally to comma-stage nuclei (Figure 8B), and the INM-SM mutant construct fully rescued the unc-84(n369) nuclear migration defect in hyp7 cells (Table 1, Figures 6B and 7E). We conclude that the INM-SM is required for efficient trafficking of UNC-84 but is not essential for a minimally required amount of UNC-84 to traffic to the INM and function to rescue the hyp7 nuclear migration defect.
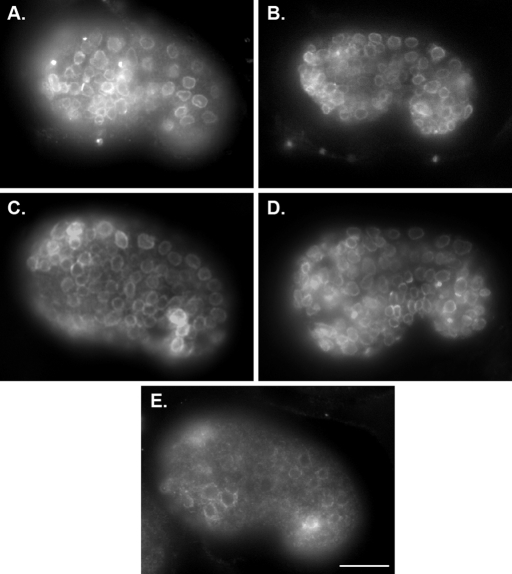
FIGURE 8:
Localization of UNC-84 with NLS mutations in older, comma-stage embryos. Transgenic animals from lines shown in Figure 7 showing mutant UNC-84 localization at the NE of older, comma-stage embryos. Anterior is left and dorsal is up. All transgenic animals were in an unc-84(n369) null background. The following wild-type or mutant forms of UNC-84 were expressed from transgenes: (A) wild-type rescuing UNC-84(+); (B) UNC-84(INM-SM); (C) UNC-84(ΔSUN-NELS 235–240); (D) UNC-84(2xcNLS; ΔSUN-NELS 235–240); (E) UNC-84(2xcNLS;ΔSUN-NELS 226–251; INM-SM). Scale bar = 10 μm.
To test whether the cNLSs and INM-SM of UNC-84 function, in part, redundantly, we created an UNC-84(2× cNLS; INM-SM) triple mutant. This mutant looked similar to the UNC-84 INM-SM mutant; it was defective in localizing UNC-84 to the NE during early embryogenesis but was able to rescue the nuclear migration defect (Table 1, Figures 6B and 7F). One interpretation of these data is that a third unidentified signal can function in the absence of the putative cNLSs and the INM-SM to mediate UNC-84's targeting to the INM.
A novel SUN-NELS contributes to the targeting of UNC-84 to the NE
Because the UNC-84(2× cNLS; INM-SM) triple mutant eventually localized and rescued nuclear migration, we hypothesized that there is at least one additional NE localization signal in UNC-84. Moreover, we hypothesized that it should reside between residues 174 and 370 of UNC-84, which represent the overlap of the soluble portions of UNC-84 that were actively imported and retained at the NE (Figure 5). Analysis of the UNC-84 mammalian homologue, Sun1, revealed that it does not contain a cNLS. Residues 200–917 of human Sun1 are sufficient for NE targeting in transfected HeLa cells, but residues 300–917 are not (Hasan et al., 2006). We therefore searched for conserved motifs between residues 200 and 300 of mammalian Sun1 and between residues 174 and 370 of UNC-84 by manual scanning. A short motif, coined SUN-NELS, corresponding to residues 235–244 in UNC-84, was found to be conserved between UNC-84 and Sun1 (Figure 6E). No regions of similarity were found outside of this short region. To test the role of the SUN-NELS on UNC-84 trafficking and function, two UNC-84 mutants were created, UNC-84(Δ235–240) and UNC-84(Δ226–251), and analyzed as mentioned earlier in the text. Similar to the INM-SM mutant, deletion of the SUN-NELS quantitatively disrupted UNC-84 trafficking during early embryogenesis (Figures 6C and 7G), but was able to later localize normally (Figure 8C). Moreover, the INM-SM mutant had no effect on its ability to rescue hyp7 nuclear migration (Table 1 and Figure 6B). These results suggest that the SUN-NELS plays a role in efficient UNC-84 targeting, but is not the sole determinant for INM protein localization.
Testing for functional redundancy among UNC-84 NE-targeting signals
The data just presented show that neither the cNLSs, the INM-SM, nor the SUN-NELS alone are necessary for the localization and function of UNC-84. We hypothesized, on the basis of single mutant localization data, that only the SUN-NELS and INM-SM play a role in UNC-84 INM localization. To test this hypothesis, we created an UNC-84 construct mutant for both the SUN-NELS and the INM-SM and expressed it in unc-84(n369). The UNC-84(SUN-NELS; INM-SM) double mutant was inefficient in NE targeting in early embryogenesis (Figure 7H) but nonetheless functioned normally in hyp7 nuclear migration (Table 1, Figure 6B).
To test the extent to which the putative cNLSs and the SUN-NELS function redundantly, a construct containing only a wild-type INM-SM, the UNC-84(2× cNLS; SUN-NELS) mutant, was expressed in an unc-84(n369) background. This construct inefficiently trafficked to the NE in the early embryo (Figure 7I); there was some NE localization, but more remained in the cytoplasm (compare to Figure 7C). Later in embryogenesis at the time of nuclear migration, this triple mutant protein efficiently localized to the NE (Figure 8D), suggesting the INM-SM was sufficient for at least partial localization of UNC-84 to the NE. The 2× cNLS; SUN-NELS triple mutant also recruited UNC-83 to the NE, but at significantly lower levels than wild type (Figure 7I). In wild-type embryos, the mean fluorescence intensity of UNC-83 staining at the NE of hyp7 nuclei was 865 ± 178 arbitrary units (SD shown; n = 230 nuclei from 45 embryos) as compared with 485 ± 156 arbitrary units in the UNC-84(2× cNLS; SUN-NELS) triple mutant embryos (n = 115 nuclei in 28 embryos; p = 2.2 × 10−16). Despite this low amount of UNC-83 that was recruited to the NE, the 2× cNLS; SUN-NELS triple mutant failed to rescue the unc-84 nuclear migration defect (Table 1 and Figure 6B). These data demonstrate that multiple signals mediate efficient targeting of UNC-84 to the NE.
Mutating all four UNC-84 NE targeting signals disrupts UNC-84 localization to the NE
A mutant lacking all four of the signals identified as contributing to the localization of UNC-84 to the INM, UNC-84(2× cNLS; SUN-NELS; INM-SM), was expressed in unc-84(n369) animals. This quadruple mutant failed to localize to the NE (Figures 6C and 7J), failed to recruit UNC-83 (Figure 7J), and failed to rescue the nuclear migration defect (Table 1 and Figure 6B). Mutant protein was localized throughout the cytoplasm and failed to properly target to the NE (Figure 7J). Quantification of the NE/cytoplasmic ratio of UNC-84 fluorescence was significantly reduced when compared with either UNC-84(INM-SM) or UNC-84(SUN-NELS) single mutants (Figure 6C). Moreover, the protein failed to accumulate at the NE later in development (Figure 8E). Together, these data suggest that the predicted cNLSs, the SUN-NELS, and the INM-SM each contribute to the transport of UNC-84 to the INM.
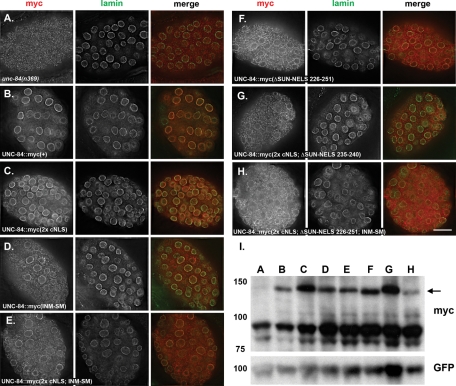
To exclude the possibility that the staining observed in the transgenic animals was only N-terminal fragments instead of full-length protein, the UNC-84 NLS mutants were reengineered with a C-terminal 6x myc tag. Western blotting confirmed that all UNC-84::6x myc NLS mutants, even the quadruple mutant that failed to localize, were stably expressed and the correct size (Figure 9I).
FIGURE 9:
UNC-84::6x myc mutants are stably expressed and phenocopy untagged UNC-84 mutants. All UNC-84::6x myc mutants were expressed in an unc-84(n369) background. (A–H) The left panel shows UNC-84 (anti-myc) immunolocalization, the middle panel shows lamin immunolocalization, and the right panel shows the merged UNC-84 (red) and lamin (green) staining. The mutant form of UNC-84 (as in Figure 7) is indicated. Scale bar = 10 μm. (I) The top anti-myc blot show expression of each of the UNC-84::6x myc mutants. Lanes A–H correspond to each of the mutants shown in (A–H). The arrow points to UNC-84::6x myc mutant proteins. The lower bands are background because they are seen in lane A, which is not expressing a myc-tagged construct. The bottom anti-GFP blot is a loading control for transgenic animals expressing SUR-5::GFP.
The UNC-84::6x myc tag lines were further used to confirm our conclusions that the same targeting defect observed in untagged mutants could be obtained with independent transgenic lines probed with a commercially available antibody. Furthermore, our conclusions that many of the untagged versions localized, at least in part, to the NE were confirmed by colocalization with lamin by deconvolution microscopy (Figure 9, A–H, compared with Figure 7). Together, data from the myc-tagged lines confirm the localization defects of the various UNC-84 mutants.
DISCUSSION
The molecular mechanisms of how SUN and other large, integral-membrane proteins are trafficked from their site of membrane insertion in the ER to the INM are poorly understood. At least three mechanisms have been proposed: the diffusion-retention model, active transport using a cNLS, and active trafficking using an INM-SM (Holmer and Worman, 2001; Braunagel et al., 2007; Lusk et al., 2007).
The C. elegans SUN protein UNC-84 was used as a model to study the mechanisms of trafficking proteins to the INM. By deleting each of the predicted membrane-spanning regions of UNC-84, we concluded that UNC-84 likely has only one membrane-spanning domain that separates its 59 kDa cytoplasmic/nucleoplasmic domain from its 65 kDa lumenal domain (Figure 2). This topology is consistent with that of mammalian Sun1 and Sun2 (Hodzic et al., 2004; Crisp et al., 2006; Haque et al., 2006; Liu et al., 2007) and suggests that the signals to target UNC-84 to the NE and INM reside in the N-terminal, extralumenal domain. UNC-84 is required for nuclear migration in embryonic hypodermal cells (Malone et al., 1999), thereby allowing us to test the localization and function of various UNC-84 mutant proteins in the context of a developing organism. Thus we were able to identify signals in the extralumenal domain of UNC-84 that, when mutated, delayed the targeting of UNC-84 to the NE but had no effect on the function of UNC-84 during nuclear migration. Additionally, we were able to create UNC-84 mutants that failed to target to the NE, or that targeted to the NE but failed to rescue the unc-84 nuclear migration phenotype. Therefore the extralumenal domain of SUN proteins not only play a role in localization to the INM, but may also function to regulate the formation of SUN–KASH bridges.
The extralumenal domain of UNC-84 contains multiple NE localization signals
The cytoplasmic/nucleoplasmic domain of UNC-84 contains at least four possible signals that contribute to trafficking UNC-84 to the INM. We identified two possible SV40-like cNLSs (35KVRRK39 and 170HRRR173), the SUN-NELS (235SRMTTRSQTLER246), and an INM-SM (503KKSSK507). These signals function, in part, redundantly to traffic UNC-84 to the INM.
Examination of both soluble and membrane-bound portions of UNC-84 implicated regions of UNC-84 that contain signals for trafficking to the NE. The membrane-bound extralumenal domain of UNC-84 was necessary and sufficient for trafficking to the NE, suggesting that it contained all the determinants required for proper localization (Figure 3). Moreover, soluble fragments UNC-84(118–370) and UNC-84(174–509) fused to GFP::LacZ localized to the NE of postmitotic muscle cells (Figure 5), suggesting that these regions of UNC-84 contain signals for active transport into the nucleus and domains that associate with the nuclear lamina. In a reciprocal experiment, deleting residues 118–244 (including predicted cNLS 170HRRR173 and part of the SUN-NELS) of membrane-bound UNC-84 disrupted NE localization (Figure 4).
To examine the relative contribution of each potential signal, we mutated or deleted small sequences encoding each signal in our unc-84–rescuing transgene and assayed for UNC-84 localization and function (Figures 6–8; Table 1). Disruption of both predicted cNLSs had no obvious effect. Mutating either the INM-SM or the SUN-NELS led to a delay in targeting UNC-84 efficiently to the NE. Despite this delay, UNC-84 still functioned later in embryogenesis to recruit UNC-83 to the ONM for nuclear migration. Surprisingly, all three types of signals—cNLSs, SUN-NELS, and INM-SM—needed to be mutated before localization or function of UNC-84 was disrupted. Therefore these signals function, in part, redundantly to ensure the efficient trafficking of proteins to the INM. We also tested whether the second predicted cNLS or the SUN-NELS alone were sufficient to drive a GFP::LacZ reporter to the nucleoplasm. Neither signal was sufficient, suggesting that the signals we identified as necessary for targeting might be conformational or more extended than the primary sequence.
A model for trafficking UNC-84 to the INM
Localization of proteins to the INM requires at least three distinct steps: 1) ER-to-NE trafficking, 2) transport across the nuclear pore, and 3) retention by associating with the nuclear lamina (Figure 10). Whereas the diffusion-retention model postulates that Steps 1 and 2 of this process are passive, other models favor that ER-to-NE trafficking and translocation across the pore are active processes. The possibility that multiple steps are required to localize proteins to the INM, as well as the observation that cNLSs often overlap with nuclear retention domains (LaCasse and Lefebvre, 1995; Cokol et al., 2000), makes examining the relative roles of multiple targeting mechanisms difficult. Furthermore, experimental systems that rely on binary readouts of NE accumulation and stable association of INM proteins with the nuclear lamina have two limitations: an inability to observe or distinguish earlier trafficking events and a failure to assay in vivo functions of the INM protein. Using C. elegans embryos, we developed an assay that allowed us to observe differences in the timing of NE accumulation. Our system also allowed us to measure the function of UNC-84 during nuclear migration as an indirect readout of retention at the nuclear lamina. Using this system, we identified four extralumenal signals that function to target UNC-84 to the INM. Some of these signals may be involved in retention at the nuclear lamina, active import into the nucleus, or both.
FIGURE 10:
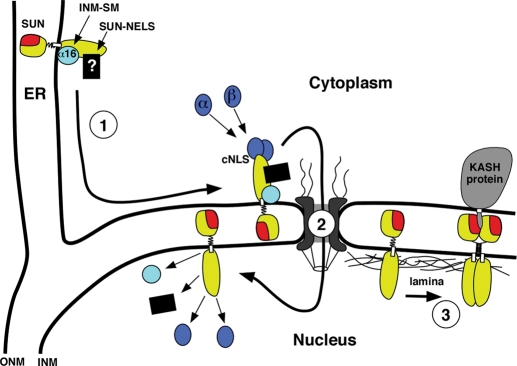
A model for trafficking of UNC-84 to the INM. See the text for full details. (Step 1) UNC-84 (yellow with a red SUN domain) is integrated into the ER membrane with its N-terminal nucleoplasmic domain facing the cytoplasm and its C-terminal SUN domain (red) in the perinuclear space. Membrane-associated importin-α-16 (light blue) associates with the INM-SM of UNC-84, and an unidentified factor (black box) binds to the SUN-NELS. UNC-84 is then actively transported from the peripheral ER to the NE. (Step 2) UNC-84 translocates across the nuclear pore complex. Some combination of importin-α/importin-β (dark blue) at the cNLS(s), the unknown SUN-NELS factors, and/or the small importin-α-16 isoform at the INM-SM facilitate trafficking across the nuclear pore. (Step 3) UNC-84 is released into the INM, and the import factors are recycled. Finally, UNC-84 binds to the nuclear lamina and recruits KASH proteins (gray) to the ONM to establish an NE bridge.
In Step 1 of our model, the INM-SM and SUN-NELS function in trafficking from the peripheral ER toward the NE (Figures 7 and 10). Mutations in either the SUN-NELS or the INM-SM resulted in a delay of targeting UNC-84 to the NE. Early in embryogenesis, mutant UNC-84 only partially localized to the NE as compared with wild type (Figure 6C or compare Figure 7, E and G, to C), but later in development, during hyp7 nuclear migration, normal levels of UNC-84 (INM-SM or SUN-NELS) mutants localized to the NE (compare Figure 8, B and C, to A). In insect cells, the INM-SM becomes associated with a divergent importin-α-16 at the translocon, which remains associated with the INM-SM after release from the translocon (Saksena et al., 2006). A 16 kDa isoform of human importin-α, KPNA-4–16, also cross-links to INM-SMs, suggesting that this ER-to-NE shuttling mechanism is conserved (Braunagel et al., 2007). In C. elegans, IMA-3 is the only importin-α expressed in the embryo (Geles and Adam, 2001). IMA-3 also has a high level of identity to the last 16 kDa of human KPNA-4–16. We hypothesize that a short isoform of IMA-3 functions as the worm importin-α-16 (light blue circle in Figure 10) and predict that it associates with the INM-SM of UNC-84 at the translocon in the ER. The mechanisms of how the SUN-NELS functions remain unknown. A 21-residue region of mouse Sun1 that contains the SUN-NELS, however, was recently found to participate in the localization of Sun1 to the INM (Haque et al., 2010).
In Step 2 of our model, UNC-84 is transported across the nuclear pore (Figure 10). The yeast INM protein Heh2 has a functional cNLS that associates with importin-α and importin-β and uses the Ran GTPase cycle to mediate translocation across the nuclear pore (King et al., 2006). In contrast, mutating both predicted cNLSs of UNC-84 had no effect on the localization of UNC-84. A construct, however, with both predicted cNLSs but not the SUN-NELS or the INM-SM localized UNC-84 to the INM, but a construct mutant for all four signals did not. Therefore at least one of the predicted cNLSs is sufficient, but not required, for localization of UNC-84 to the INM. Additionally, the UNC-84(1–370 2x cNLS) soluble mutant protein was efficiently targeted to the NE, suggesting that the SUN-NELS may function as an active nuclear import signal (Figure 5F). Together these data support a model in which at least one predicted cNLS functions partially redundantly to the SUN-NELS and the INM-SM to move UNC-84 across the nuclear pore.
After UNC-84 enters the INM, the factors used for import are released and UNC-84 associates with the nuclear lamina (Figure 10). The C. elegans B-type lamin, LMN-1, is required for UNC-84 retention at the INM (Lee et al., 2002), but it is unknown whether the association is direct. Finally, the lumenal SUN domain of UNC-84 recruits the KASH protein UNC-83 to the ONM, creating a bridge across the NE to mediate microtubule-motor-driven nuclear migration (McGee et al., 2006; Meyerzon et al., 2009; Fridolfsson et al., 2010).
The trafficking mechanisms used by INM proteins to gain access into the nucleus have remained elusive. Data presented here suggest that this is most likely due to functionally redundant signals spread across the cargo protein. The identification of a novel SUN-NELS suggests that other INM proteins may contain unidentified signals or targeting mechanisms. Complementary to our data, Turgay et al. (2010) recently reported that human Sun2 also uses multiple signals to target to the INM: a cNLS, a Golgi retrieval signal, and the SUN domain. They were unable to test the contribution of the predicted Sun2 INM-SM in their system (Turgay et al., 2010), and Sun2 does not have an extralumenal SUN-NELS. The Golgi retrieval signal in Sun2 consists of four arginine residues (Turgay et al., 2010), suggesting that the second predicted cNLS in UNC-84 (170HRRR173) might instead be a Golgi retrieval signal. Thus different INM proteins use various combinations of INM-localization signals. We propose that the novel SUN-NELS and the INM-SM function in ER-to-NE transport, reducing the amount of time INM proteins spend in ER membranes with their nucleoplasmic domains exposed to the cytoplasm. A more complete understanding of how nuclear import receptors interact with targeting signals to ensure the timely localization of proteins to the INM, as well as the identity of cytoplasmic/nucleoplasmic binding partners, should lead to a better understanding of the assembly of the nuclear lamina, establishment of NE bridges, and the formation or development of laminopathies.
MATERIALS AND METHODS
C. elegans strains, maintenance, and phenotypic characterization
C. elegans were cultured using standard conditions, and the N2 strain was used for wild type (Brenner, 1974). The null allele unc-84(n369) was previously described (Malone et al., 1999). Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources.
Transgenic lines were created by standard DNA microinjection techniques using 2–4 ng/μl of the construct of interest with 100 ng/μl of podr1::rfp or sur-5::gfp constructs as transformation markers (Mello et al., 1991; Yochem et al., 1998; Sagasti et al., 2001). Constructs encoding UNC-84ΔN::GFP under the control of the unc-84 promoter, and SV40 NLS::GFP::LacZ and GFP::LacZ both under the control of the unc-54 promoter, were injected into wild type. All other transgenes were expressed in an unc-84(n369) background.
Hyp7 dorsal cord nuclei were counted in L1 larvae using differential interference contrast (DIC) as described (Meyerzon et al., 2009). All counts were blind in that the animal was only identified as transgenic or not transgenic after counting.
Immunofluorescence and in C. elegans embryos
C. elegans embryos were extruded and stained for UNC-83 and UNC-84 as previously described (Starr et al., 2001; Lee et al., 2002; McGee et al., 2006). Embryos were fixed for 20 min in –20°C methanol and 10 min in –20°C acetone for UNC-84 staining and 10 min in –20°C methanol for UNC-83 staining. UNC-83 monoclonal 1209D7D5 was used undiluted (Starr et al., 2001). Affinity-purified anti–UNC-84 sera from rat CA2608 or CA2609 was used undiluted; sera from both rats behaved similarly (McGee et al., 2006). Rabbit anti-GFP (Novus Biologicals, Littleton, CO) was diluted 1:500 (vol/vol) in phosphate-buffered saline (PBS). Mouse monoclonal 9E10 anti-myc antibody (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) was used at a 1:500 dilution. Guinea pig anti–LMN-1 was provided by Jun Kelly Liu (Cornell University, Ithaca, NY) and used at a 1:1000 (vol/vol) dilution (McGee et al., 2009). Cy2-conjugated donkey anti–mouse immunoglobulin (Ig)G, Cy3-conjugated goat anti–mouse IgG, Cy3-conjugated donkey anti–rat IgG, and DyLight 649-conjugated donkey anti–guinea pig IgG (Jackson ImmunoResearch, West Grove, PA) diluted 1:200 (vol/vol) in PBS were used as secondary antibodies. DNA was stained with 1 μg/ml 4’,6-diamidino-2-phenylindole (DAPI) for 10 min. Images were captured with a 63× PLAN APO 1.40 objective on a Leica DM 6000 compound microscope with Leica AF4000 software or with a DeltaVision microscope and deconvolved using SoftWorx (Applied Precision, Issaquah, WA). Images were uniformly enhanced using the levels commands in Adobe Photoshop (Adobe Systems, Mountain View, CA). When comparing relative fluorescence intensities of anti–UNC-83 or UNC-84 staining, single focal planes were quantified using ImageJ.
Transgenic mutant UNC-84 expression constructs
Schematics of all the constructs used to make transgenic C. elegans for this study and their corresponding plasmid names are shown in Figure 1. In general, for the experiments shown in Figures 2, 7, 8, and 9, we engineered mutant versions of our unc-84 rescuing construct (pSL38; McGee et al., 2006) to contain either small deletions for our in vivo topology experiments or small deletions and mutations for our analysis of NLSs. Mutations, alanine substitutions, or small deletions were introduced by using PCR splicing by overlapping extension (SOEing) (Horton et al., 1990) or, in one case, by using the Quickchange XL system (Invitrogen, Carlsbad, CA). In all cases, the PCR products were TOPO cloned (Invitrogen) and confirmed by sequencing. Restriction enzyme fragments containing mutations were then moved out of the TOPO vectors and back into the pSL38, replacing the wild-type sequences. Double, triple, and quadruple mutations were introduced into pSL38 by multiple steps of such subcloning.
For the experiments in Figure 3, PCR products from pSL38, including 2.2 kb of the endogenous unc-84 promoter and genomic sequences encoding either residues 1–535 or 478–1111, were cloned into pPD95.77, a promoterless Fire Lab Vector with a C-terminal GFP (Addgene, Cambridge, MA). Constructs were confirmed by sequencing. The four UNC-84ΔC::GFP deletion constructs in Figure 4 were created by PCR SOEing as mentioned earlier in the text and subcloned back into the UNC-84ΔC::GFP construct.
In Figure 5, pPD96.02 (Addgene), which encodes an SV40 NLS::GFP::LacZ fusion protein under control of the muscle-specific promoter unc-54, was used for a nucleoplasmic control. For a cytoplasmic control, pPD96.02 was digested with Kpn1 and ligated back together to remove the NLS. Various fragments of the unc-84 cDNA yk402g1 (Kohara, 1996; McGee et al., 2006) were amplified by PCR and cloned into the KpnI and AgeI sites of pPD96.02 to replace the SV40 cNLS and create a construct that uses the muscle-specific unc-54 promoter to drive an UNC-84 fragment::GFP::LacZ fusion protein. The UNC-84(1–370 2x cNLS)::GFP::LacZ mutant construct was cloned by amplifying a 1.6 kb fragment from the UNC-84(2x cNLS) mutant in the rescuing construct just described and subcloned into pPD96.02 using Kpn1 and AgeI restriction sites. To create the UNC-84 ΔH2::6x myc construct PCR SOEing was used to delete the stop codon and add a unique Pac1 site in pSL359. In addition, 6x myc with Pac1 overhangs was amplified from pCS2+MT (Rupp et al., 1994) and cloned into the Pac1 site to create psL550. All fragments were confirmed by sequencing.
To create the UNC-84::6x myc constructs shown in Figure 9, pSL550 was digested with Kpn1 and Sal1. A 3.2 kB fragment containing the C-terminal 519 residues of UNC-84 in frame with a 6x myc tag was subcloned into all UNC-84 NLS mutant constructs used in Figures 6–8. Mutants were confirmed by sequencing.
UNC-84 immunoblotting
Whole protein extracts from transgenic animals were isolated and blotted as previously described (Lee et al., 2002). Blots were probed with mouse monoclonal 9E10 anti-myc antibody at 1:1000 (vol/vol) or mouse monoclonal anti-GFP (Covance, Princeton, NJ) at 1:2000 (vol/vol). For the secondary antibody, anti-mouse conjugated to horseradish peroxidase (Jackson ImmunoResearch) was used at 1:10,000 (vol/vol) and developed with the enhanced chemiluminescence kit (GE Healthcare, Piscataway, NJ).
Acknowledgments
We thank S. Braunagel and M. Summers (Texas A&M, College Station, TX) for many helpful discussions. We thank Jun Kelly Liu (Cornell) for the LMN-1 antibodies. We thank L. Rose, J. Engebreht, D. Elnatan, Daniel Chu, M. Paddy (UC Davis), and our colleagues in the Starr lab and Molecular and Cellular Biology for many helpful comments on the manuscript and research. This research was supported by grant 5R01GM073874 from the NIH. E.C.T. was supported by NIH training grant 5T32GM008799 in biomolecular technology.
Abbreviations used:
- cNLS
classical nuclear localization signal
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- ER
endoplasmic reticulum
- GTP
guanosine 5′-triphosphate
- INM
inner nuclear membrane
- IMN-SM
inner nuclear membrane–sorting motif
- KASH
Klarsicht, ANC-1, Syne homology
- NE
nuclear envelope
- NLS
nuclear localization signal
- ONM
outer nuclear membrane
- PBS
phosphate-buffered saline
- SOEing
splicing by overlapping extension
- SUN
Sad1–UNC-84
- SUN-NELS
SUN nuclear envelope localization signal
- SV40
simian virus 40
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0733) on March 16, 2011.
REFERENCES
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Bolender RP. Stereological analysis of the guinea pig pancreas. I. Analytical model and quantitative description of nonstimulated pancreatic exocrine cells. J Cell Biol. 1974;61:269–287. doi: 10.1083/jcb.61.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunagel SC, Williamson ST, Ding Q, Wu X, Summers MD. Early sorting of inner nuclear membrane proteins is conserved. Proc Natl Acad Sci USA. 2007;104:9307–9312. doi: 10.1073/pnas.0703186104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunagel SC, Williamson ST, Saksena S, Zhong Z, Russell WK, Russell DH, Summers MD. Trafficking of ODV-E66 is mediated via a sorting motif and other viral proteins: facilitated trafficking to the inner nuclear membrane. Proc Natl Acad Sci USA. 2004;101:8372–8377. doi: 10.1073/pnas.0402727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Epstein HF, Casey DL, Ortiz I. Myosin and paramyosin of Caenorhabditis elegans embryos assemble into nascent structures distinct from thick filaments and multi-filament assemblages. J Cell Biol. 1993;122:845–858. doi: 10.1083/jcb.122.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol. 2010;338:237–250. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geles KG, Adam SA. Germline and developmental roles of the nuclear transport factor importin alpha3 in C. elegans. Development. 2001;128:1817–1830. doi: 10.1242/dev.128.10.1817. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shannahan CM, Shackleton S. Mammalian SUN protein networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S, Guttinger S, Muhlhausser P, Anderegg F, Burgler S, Kutay U. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 2006;580:1263–1268. doi: 10.1016/j.febslet.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD. Sun2 is a novel mammalian inner nuclear membrane protein. J Biol Chem. 2004;279:25805–25812. doi: 10.1074/jbc.M313157200. [DOI] [PubMed] [Google Scholar]
- Holmer L, Worman HJ. Inner nuclear membrane proteins: functions and targeting. Cell Mol Life Sci. 2001;58:1741–1747. doi: 10.1007/PL00000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Kohara Y. [Large scale analysis of C. elegans cDNA] Tanpakushitsu Kakusan Koso. 1996;41:715–720. [PubMed] [Google Scholar]
- LaCasse EC, Lefebvre YA. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Starr DA, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in C. elegans. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Wu X, Summers MD, Lee A, Ryan KJ, Braunagel SC. Truncated isoforms of Kap60 facilitate trafficking of Heh2 to the nuclear envelope. Traffic. 2010;11:1506–1518. doi: 10.1111/j.1600-0854.2010.01119.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, Burke B, Roux KJ. Functional association of Sun1 with nuclear pore complexes. J Cell Biol. 2007;178:785–798. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Shi Y, Lu Q, Ma Y, Luo J, Wang Q, Ji J, Jiang Q, Zhang C. Requirement for lamin B receptor and its regulation by importin β and phosphorylation in nuclear envelope assembly during mitotic exit. J Biol Chem. 2010;285:33281–33293. doi: 10.1074/jbc.M110.102368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk CP, Blobel G, King MC. Highway to the inner nuclear membrane: rules for the road. Nat Rev. 2007;8:414–420. doi: 10.1038/nrm2165. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee MD, Stagljar I, Starr DA. KDP-1 is a nuclear envelope KASH protein required for cell-cycle progression. J Cell Sci. 2009;122:2895–2905. doi: 10.1242/jcs.051607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009;136:2725–2733. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Ohba T, Schirmer EC, Nishimoto T, Gerace L. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol. 2004;167:1051–1062. doi: 10.1083/jcb.200409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund C, Sullivan T, Stewart CL, Worman HJ. Dependence of diffusional mobility of integral inner nuclear membrane proteins on A-type lamins. Biochemistry. 2006;45:1374–1382. doi: 10.1021/bi052156n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L, Burke B. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the interaction of p55 with the nuclear lamina. J Cell Biol. 1990;111:2225–2234. doi: 10.1083/jcb.111.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hisamoto N, Hyodo J, Tanaka-Hino M, Matsumoto K, Bargmann CI. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell. 2001;105:221–232. doi: 10.1016/s0092-8674(01)00313-0. [DOI] [PubMed] [Google Scholar]
- Saksena S, Summers MD, Burks JK, Johnson AE, Braunagel SC. Importin-alpha-16 is a translocon-associated protein involved in sorting membrane proteins to the nuclear envelope. Nat Struct Mol Biol. 2006;13:500–508. doi: 10.1038/nsmb1098. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- Stewart CL, Roux KJ, Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, Kutay U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;29:2262–2275. doi: 10.1038/emboj.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure predictionHydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Weibel ER, Staubli W, Gnagi HR, Hess FA. Correlated morphometric and biochemical studies on the liver cellIMorphometric model, stereologic methods, and normal morphometric data for rat liver. J Cell Biol. 1969;42:68–91. doi: 10.1083/jcb.42.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, et al. Several novel nuclear envelope transmembrane proteins identified in skeletal muscle have cytoskeletal associations. Mol Cell Proteomics. 2011;10:M110 003129. doi: 10.1074/mcp.M110.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-beta proteins. Curr Opin Struct Biol. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]