Akt activation sequesters FOXO1a away from its target genes and serves as an endpoint of a complex signaling network. A cell-based RNAi screen reveals an extensive network of genes, including UCP5, which directs nuclear localization of FOXO1a. Silencing of UCP5 disrupts the mitochondria and induces JNK1, creating a link to the Akt signaling network.
Abstract
Forkhead transcription factors (FOXOs) alter a diverse array of cellular processes including the cell cycle, oxidative stress resistance, and aging. Insulin/Akt activation directs phosphorylation and cytoplasmic sequestration of FOXO away from its target genes and serves as an endpoint of a complex signaling network. Using a human genome small interfering RNA (siRNA) library in a cell-based assay, we identified an extensive network of proteins involved in nuclear export, focal adhesion, and mitochondrial respiration not previously implicated in FOXO localization. Furthermore, a detailed examination of mitochondrial factors revealed that loss of uncoupling protein 5 (UCP5) modifies the energy balance and increases free radicals through up-regulation of uncoupling protein 3 (UCP3). The increased superoxide content induces c-Jun N-terminal kinase 1 (JNK1) kinase activity, which in turn affects FOXO localization through a compensatory dephosphorylation of Akt. The resulting nuclear FOXO increases expression of target genes, including mitochondrial superoxide dismutase. By connecting free radical defense and mitochondrial uncoupling to Akt/FOXO signaling, these results have implications in obesity and type 2 diabetes development and the potential for therapeutic intervention.
INTRODUCTION
The three Akt isoforms in mammalian cells function in cell growth, development, cell cycle, cell attachment, and migration by integrating multiple signals in normal and cancerous cells (Los et al., 2009). For example, Akt activation is central for repression of tuberous sclerosis complex and mammalian target of rapamycin complex 1 (mTORC1) in order to up-regulate ribosomal S6 kinase and protein translation (Tee et al., 2003). During glycolysis, Akt activity is vital for glucose uptake and is an essential part of the skeletal muscle response to calorie restriction (McCurdy and Cartee, 2005; Sakamoto et al., 2006). Additionally, anomalous Akt signaling is implicated in the development and progression of diabetes (Kim and Chung, 2002). Therefore extensive understanding of the Akt signaling network will lead to improved clarification of diabetes and cancer development.
The Forkhead transcription factor (FOXO) is a key downstream effector of Akt action. In many cases, FOXO continuously shuttles between the nucleus and the cytoplasm. In response to direct Akt phosphorylation, FOXO is rapidly escorted out of the nucleus through binding to the nuclear export receptor exportin-1 (XPO1). In the cytoplasm, FOXO is sequestered away from its target genes that are involved in stress response and apoptosis (Burgering and Medema, 2003; Greer and Brunet, 2005; Carter and Brunet, 2007). Inhibition of Akt signaling leads to nuclear FOXO and an increase in its gene regulatory function. As a result, FOXO localization serves as an informative endpoint of the Akt signaling network.
Among the more than 100 members of the forkhead/winged helix transcription factor family, FOXOs are the only members under control of Akt signaling (Barthel et al., 2005; Carter and Brunet, 2007). While there are four FOXO paralogues in mammals (FOXO1a, -3a, -4 and -6), FOXO1a is the only embryonically lethal member in knockout mice (Greer and Brunet, 2005). Upon Akt signal repression, nuclear FOXO binds to promoters containing insulin response elements and directs the transcription of genes involved in metabolism (G6P, PEPCK), detoxification (SOD2, catalase), cell cycle arrest (p21Cip1, p27Kip1, cyclin G2), and apoptosis (FasL, BIM-1, Chop; Guo et al., 1999; Ramaswamy et al., 2002; Greer and Brunet, 2005; Martinez et al., 2006; Oliveira et al., 2009; Onuma et al., 2006;). Ultimately, the precise nature of FOXO gene regulation depends on the cell or tissue type and growth conditions.
In this study, we utilized the cytoplasmic-to-nuclear enhanced green fluorescent protein (EGFP)-FOXO1a reporter-based system to elucidate the hierarchical network centered on Akt activity. Through siRNA-mediated silencing, we identified components of cell adhesion, the nuclear pore, and the mitochondria that were not previously implicated in FOXO1a nuclear localization. Categorization of these complexes revealed that silencing of a subset of translation factors was indicative of general nuclear export aberrations, whereas loss of focal adhesion and mitochondrial factors was specific to FOXO1a localization. Furthermore, we demonstrated that reduction of the mitochondrial uncoupling protein 5 (UCP5) leads to mitochondrial dysfunction, superoxide increase, and FOXO1a activation through the induction of c-Jun N-terminal kinase 1 (JNK1). Taken together, these results revealed a cellular network encompassing FOXO localization and activity that is paramount to understanding Akt's function in adhesion, metabolism, and stress response.
RESULTS
Establishment of a system for high-throughput analysis of FOXO1a localization
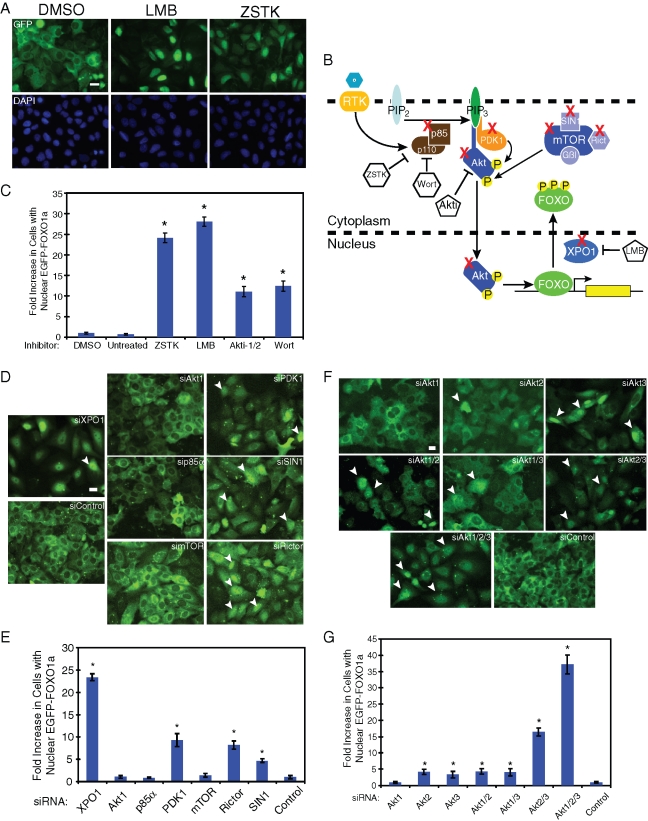
Since nuclear localization is essential for FOXO transcriptional activation, a visual assay evaluating nuclear inclusion of a GFP-tagged FOXO1a in U2OS cells was developed. We generated a cell line that stably expresses V5-tagged EGFP-FOXO1a (FOXO1a) confirming expression through Western blot analysis (Supplemental Figure S1A) and fluorescence microscopy (Figure 1A). These cells have normal expression of the insulin signaling pathway and respond to serum stimulation (Figure S1B). Under normal growth conditions, FOXO1a was phosphorylated and cytoplasmic (Figure 1, A and B). Initially, we blocked nuclear export using leptomycin B (LMB), an exportin-1 inhibitor, and found FOXO1a retained in the nucleus (Figure 1, A and B). By blocking the Akt signaling pathway with an Akt inhibitor (Akti-1/2) (DeFeo-Jones et al., 2005) or PI3K inhibitors (wortmannin and ZSTK474), we inhibited phosphorylation of FOXO1a, which led to its nuclear accumulation (Figure 1, A and B, and unpublished data). Through development of an automated nuclear translocation analysis (see Materials and Methods), we determined that all inhibitors caused a substantial fold increase in the number of cells with nuclear FOXO1a when compared to dimethyl sulfoxide (DMSO) -treated or -untreated cells (Figure 1C). With these results, we confirmed that FOXO1a stable expression in U2OS cells responded to changes in the Akt and nuclear export pathways.
FIGURE 1:
Knockdown of the Akt signaling pathway using small molecules and RNAi localizes FOXO1a to the nucleus in U2OS cells. (A) Microscope images of U2OS EGFP-FOXO1a cells taken after treatment with labeled small molecule for 24 h. GFP images represent EGFP-FOXO1a expression and DAPI represents nuclear DNA. Scale bar: 20 μm. (B) Simplified model showing specific proteins in Akt signaling pathway that were inhibited by small molecules or targeted for siRNA knockdown (X). p85α is a regulatory subunit of PI3K, which converts phosphatidylinositol 4,5-bisphosphate (PIP2) to phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 recruits Akt and phosphoinositide-dependent protein kinase (PDK1) to the plasma membrane, where PDK1 and mTORC2 (mTOR/SIN1/Rictor/Gβl) activate Akt. Akt localizes to the nucleus and phosphorylates FOXO, leading to its nuclear export by exportin-1 (XPO1). Wortmannin (Wort) and ZSTK474 (ZSTK) block the activation of PI3K, deactivating Akt. Akti-1/2 (Akti) blocks Akt activation directly. LMB alkylates and directly inhibits XPO1. (C) Automated counting of cells using nuclear translocation analysis (see Materials and Methods). About 1000 cells were counted for each well, with eight wells (of 96 wells) counted for each treatment. GFP nuclei/total number of cells (%) was used to calculate fold increase of cells with GFP nuclei when compared to DMSO treatment. Student's t test was used to compare test to control siRNAs (*p < 0.05). (D) Microscope images of U2OS EGFP-FOXO1a after siRNA knockdown. Arrows are representative cells with nuclear EGFP-FOXO1a. Scale bar: 20 μm. (E) The nuclear translocation analysis was used on four wells of each siRNA pool in a 96-well plate to determine fold increase in cells with nuclear EGFP-FOXO1a when compared to nontargeting siRNA cells (siControl). (F) Microscope images of EGFP-FOXO1a after siRNA knockdown. Arrows represent nuclear EGFP-FOXO1a cells. Scale bar: 20 μm. (F) Nuclear translocation analysis used in six wells of each siRNA pool in a 96-well plate to determine fold increase in cells with nuclear EGFP-FOXO1a when compared to nontargeting siRNA cells (siControl). Student's t test was used to compare test to control siRNAs (*p < 0.05) (see also Figures S1–S3).
To show efficacy of small interfering RNA (siRNA) knockdown in the FOXO1a nuclear translocation assay, we used interfering RNA (RNAi) to silence candidate genes from the Akt and nuclear export pathways (Figure 1B). We confirmed that these target proteins were depleted by RNAi (Figure S2). Using automated nuclear translocation analysis, knockdown of Akt activators PDK1, Rictor, and SIN1, as well as XPO1, led to an increase in nuclear localization of FOXO1a (Figure 1, B, D, and E). Surprisingly, reduction of Akt1, p85α (phosphatidylinositol-3 kinase [PI3K] regulatory subunit), and mTOR did not significantly change FOXO1a localization.
Since Akt1 silencing had no effect on FOXO1a localization, we asked whether Akt2 and/or Akt3 could regulate FOXO1a and thereby compensate for the loss of Akt1 function. Previous studies have shown that Akt2 directs FOXO1a phosphorylation and transcriptional activity in cardiomyocytes (DeBosch et al., 2006), yet the functional contribution of all three Akt isoforms to FOXO1a localization has not been examined. We depleted Akt gene expression by RNAi individually and in combination. Using real-time PCR, we validated that Akt siRNA knockdowns were specific for each targeted isoform (Figure S3). Akt2 and Akt3 knockdowns had a small but statistically significant effect on FOXO1a nuclear localization as compared to Akt1 knockdown (Figure 1, F and G). Although knockdown of different isoform combinations demonstrated that Akt2/3 silencing had a substantial effect on FOXO1a, reduction of all three isoforms was the strongest inducer of FOXO1a nuclear localization.
High-throughput screening reveals new effectors of FOXO1a localization
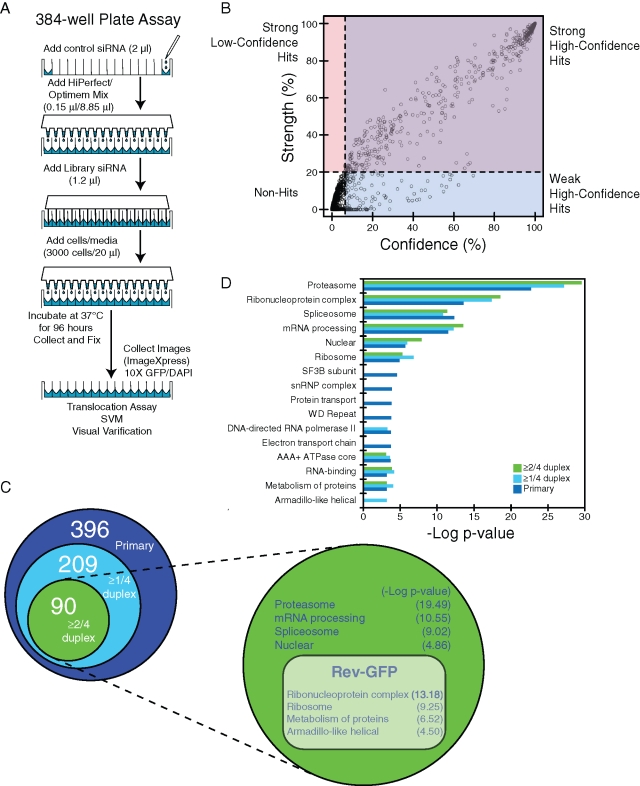
Using FOXO1a nuclear localization as a readout, we examined how silencing genes from the entire human genome influenced FOXO1a localization. Figure 2A schematizes our screen of a human genome siRNA library to determine a more complete set of genes that regulate FOXO1a nuclear localization. The siRNA library contained a nonrandomly arrayed set of 21,121 SMARTpools (four unique duplexes; EMD Millipore, Billerica, MA) corresponding to genes and open reading frames from the entire human genome. We performed the assay in 384-well plates in triplicate. After 4 d of siRNA incubation, cells were collected, fixed, and stained with 4′,6-diamidino-2-phenylindole (DAPI). GFP and DAPI images were examined by automated nuclear translocation analysis.
FIGURE 2:
High-throughput screening determines regulators of FOXO1 nuclear localization. (A) Schematic of the 384-well plate format screened in U2OS EGFP-FOXO1a and Rev-GFP cells. (B) False discovery thresholds for each model (horizontal and vertical dashed lines) were used to identify strong hits (purple) and weak hits (blue), as well as potential hits that would have been misclassified due to confidence model overfitting (pink). (C) A Venn diagram of hits from primary (dark blue) and validation screens (≥ 1/4 duplexes, light blue; ≥ 2/4 duplexes, green). Projection is functional analysis of two or more out of four duplexes from the validation screen tested in Rev-GFP cells. The DAVID database (http://david.abcc.ncifcrf.gov/) was used to group related categories and eliminate redundancy. This group's categories are specific to EGFP-FOXO1a nuclear localization (dark green) and those present in both screens (light green). Negative log p-values are present in parentheses. (D) Functional analysis using the primary and validation list from Tables S1 and S2 (see also Figure S4).
A support vector machine (SVM) model was developed to determine a list of siRNAs for follow-up validation (see Materials and Methods) (Birmingham et al., 2009). The SVM method of assigning hits took into account the positive (XPO1) and negative (scrambled siRNA) controls and determined a probability strength (magnitude) and confidence (reproducibility) score for each control (Figure S4A). The individual screen siRNAs were then assigned strength and confidence probability values based on the controls. Using an empirically determined false discovery rate of 5%, the strength and confidence cutoff probabilities (%) were set at 20% and 6.4%, respectively (Figure 2B, purple area). High confidence and strength scores resulted in a list of 574 genes that, when siRNA-silenced, led to FOXO1a nuclear localization (Table S1). The individual images of positive wells were examined by eye to eliminate obvious false positives. This culled the list to 396 genes for validation screening (Figure 2C and Table S1). Systematic analysis of primary hits revealed functional enrichment of factors from the proteasome complex, spliceosome, electron transport chain (ETC), protein transport, RNA polymerase II, and the ribosome (Figure 2D).
Validation of hits was determined using the four individual siRNA duplexes from the deconvolved SMARTpools in the previously described 384-well assay. This validation screening resulted in confirmation of 209 siRNA-targeted genes from the primary screen, with at least one out of four duplexes being positive (Figure 2C and Table S2). A more stringent cutoff resulted in 90 siRNA-targeted genes confirming at least two out of four duplexes (Figure 2C). When the primary and validation screens were compared, there was a strong correlation between primary (both strength and confidence) and validation screen scores using SVM (Figure S4B). In addition, functional enrichments for the proteasome, splicesome, ribosome, and nuclear proteins remained after the validation screen removed weak primary hits (Figure 2D, green and light blue). Despite validation of some factors from the ETC and protein transport, these functional categories were no longer significant (Figure 2D).
Effectors of general nuclear export
To distinguish FOXO1a-specific regulation from general effects of nuclear export, we examined the FOXO1a-validated genes in U2OS cells expressing an HIV Rev-GFP fusion protein (Rev-GFP) using the same 384-well and SVM assays. HIV Rev has been shown to undergo XPO1-dependent nuclear export similar to FOXO1a (Wolff et al., 1997; Kau et al., 2003). We confirmed that 26 out of 90 (≥ 2/4 duplexes) hits influenced localization of both reporters (Figure 2C and Table S3). In addition, we found functional enrichment of ribosomes, ribonucleoprotein complexes, and armadillo-like helical domain proteins influenced both reporters, while the proteasome, spliceosome, and nuclear proteins remained specific to FOXO1a localization (Figure 2C and Table 1).
TABLE 1:
Categorized list of genes when siRNA-silencing leads to nuclear localization of FOXOIa.
| Gene IDa | Entreza | Descriptionb | Functional categoryc |
|---|---|---|---|
| HAS1 | 3036 | Hyaluronan synthase 1 | Cell adhesion |
| ISLR | 3671 | Immunoglobulin superfamily containing leucine-rich repeat protein precursor | Cell adhesion |
| ITGAV | 3685 | Integrin alpha-V | Cell adhesion |
| KRT82 | 3888 | Keratin, type II articular Hb2 | Cell adhesion |
| MFAP1 | 4236 | Microfibrillar-associated protein 1 | Cell adhesion |
| MFAP2 | 4237 | Microfibrillar-associated protein 2 | Cell adhesion |
| NLGN2 | 57555 | Neuroligin-2 precursor | Cell adhesion |
| NUDT3 | 11165 | Diphosphoinositol polyphosphate phosphohydrolase 1 | Cell adhesion |
| SON | 6651 | SON protein | Cell adhesion |
| TLN1 | 7094 | Talin-1 | Cell adhesion |
| FSPAN9 | 83441 | Tetraspanin 9 | Cell adhesion |
| CDK1 | 983 | Cyclin-dependent kinase 1 | Cell cycle |
| TTK | 7272 | Dual specificity protein kinase TTK | Cell cycle |
| KERA | 11081 | Keratan sulfate proteoglycan keratocan | Cytoskeleton |
| FACC3 | 10460 | Transforming acidic coiled-coil-containing protein 3 | Cytoskeleton |
| TNP2 | 7142 | Nuclear transition protein 2 | Differentiation |
| THUMPD2 | 80745 | THUMP domain-containing protein 2 | DNA |
| XAB2 | 56949 | Pre-mRNA–splicing factor SYF1; XPA-binding protein 2 | DNA |
| SNAT3 | 10991 | Solute carrier family 38, member 3 | Glutamate metabolism |
| CHMP2A | 27243 | Charged multivesicular body protein 2a | Golgi |
| COPA | 1314 | Coatomer subunit alpha | Golgi |
| GGA1 | 26088 | ADP-ribosylation factor-binding protein | Golgi |
| SACM1L | 22908 | Phosphatidylinositide phosphatase SAC1 (suppressor of actin mutations 1-like protein) | Golgi |
| AGFG2 | 3268 | Arf-GAP domain and FG repeats-containing protein 2 | G-proteins |
| TBC1D29 | 26083 | Putative TBC1 domain family member 29 | G-proteins |
| AKR1CL1 | 340811 | Aldo-keto reductase family 1, member C-like 1 | Mitochondria |
| UCP5 | 9016 | Mitochondrial uncoupling protein 5 (SLC25A14) | Mitochondria |
| NUP205 | 23165 | Nuclear pore complex protein 205 | Nuclear export |
| PSMA2 | 5683 | Proteasome subunit alpha type-2 | Proteasome |
| PSMA3 | 5684 | Proteasome subunit alpha type-3 | Proteasome |
| PSMA4 | 5685 | Proteasome subunit alpha type-4 | Proteasome |
| PSMA5 | 5686 | Proteasome subunit alpha type-5 | Proteasome |
| PSMB1 | 5689 | Proteasome subunit beta type-1 precursor | Proteasome |
| PSMB2 | 5690 | Proteasome subunit beta type-2 | Proteasome |
| PSMC2 | 5701 | 26S protease regulatory subunit 7 | Proteasome |
| PSMC4 | 5704 | 26S protease regulatory subunit 6B | Proteasome |
| PSMC6 | 5706 | 26S protease regulatory subunit S10B | Proteasome |
| PSMD12 | 5718 | 26S proteasome non-ATPase regulatory subunit 12 | Proteasome |
| PSMD14 | 10213 | 26S proteasome non-ATPase regulatory subunit 14 | Proteasome |
| PSMD2 | 5708 | 26S proteasome non-ATPase regulatory subunit 2 | Proteasome |
| PSMD6 | 9861 | 26S proteasome non-ATPase regulatory subunit 6 | Proteasome |
| PSMD7 | 5713 | 26S proteasome non-ATPase regulatory subunit 7 | Proteasome |
| PSMD8 | 5714 | 26S proteasome non-ATPase regulatory subunit 8 | Proteasome |
| UBC | 7316 | Ubiquitin | Proteasome |
| HSP90AA2 | 3324 | Heat shock protein 90-kDa alpha, class A member 2 | Protein folding |
| RPL11 | 6135 | 60S ribosomal protein L1 1 | Ribosome |
| AQR | 9716 | Intron-binding protein aquarius | Splicing |
| CWC22 | 57703 | Pre-mRNA–splicing factor CWC22 homologue | Splicing |
| EFTUD2 | 9343 | 116 kDa U5 small nuclear ribonucleoprotein component | Splicing |
| HNRNPC | 3183 | Heterogeneous nuclear ribonucleoproteins C1/C2 | Splicing |
| POP1 | 10940 | Ribonucleases P/MRP protein subunit POP1 | Splicing |
| RBM8A | 9939 | RNA-binding protein 8A | Splicing |
| SF3B2 | 10992 | Splicing factor 3B subunit 2 | Splicing |
| SF3B5 | 83443 | Splicing factor 3B subunit 5 | Splicing |
| SFRS3 | 6428 | Splicing factor, arginine/serine-rich 3 | Splicing |
| SMU1 | 55234 | WD40 repeat-containing protein | Splicing |
| SNRNP200 | 23020 | US small nuclear ribonucleoprotein 200-kDa helicase | Splicing |
| SNRPD2 | 6633 | Small nuclear ribonucleoprotein Sm D2 | Splicing |
| U2AF2 | 11338 | Splicing factor U2AF 65-kDa subunit | Splicing |
| USP39 | 10713 | U4/U6.U5 tri-snRNP-associated protein 2 | Splicing |
| POLR2E | 5434 | DNA-directed RNA polymerases I, II, and III subunit RPABC1 | Transcription |
| POLR2I | 5438 | DNA-directed RNA polymerase II subunit RPB9 | Transcription |
| KIAA0907 | 22889 | UPF0469 protein KIAA0907 | Unknown |
| KIAA1466 | 57612 | KIAA1 466 gene | Unknown |
aEntrez identification and number.
bGeneral gene description from NCBI.
cThe major functional category associated with each gene.
Factors that affect the Akt signaling network
Further examination of the siRNA screen results revealed an interesting set of uncharacterized factors that regulate FOXO1a localization (Table 1) that included tetraspanin 9 (TSPAN9), a novel RNA/DNA-binding protein (SON), uncoupling protein 5 (UCP5), and a glutamine transporter (SNAT3). Tetraspanins and integrin signaling have been connected to focal adhesion, Akt signaling, and FOXO localization (Protty et al., 2009; Zhang et al., 2009; Zoller, 2009; Aldaye et al., 2010). TSPAN9, together with other genes from the siRNA screen such as integrin αV (ITGA) and talin 1 (TLN1) revealed a network of genes linked to focal adhesions that specifically affect FOXO1a localization (Figure S5).
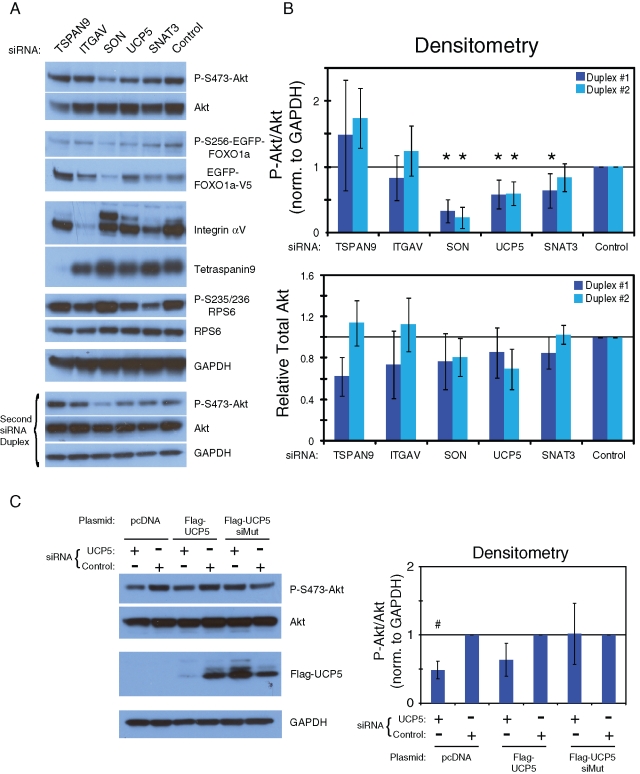
To determine whether the factors that regulate FOXO1a localization played a direct role in Akt signaling, we examined the phosphorylation state of Akt. Using two different siRNA duplexes per gene, we found that treatment of cells with all of the corresponding siRNAs, with the exception of TSPAN9, reduced the phosphorylation of Akt (Figure 3, A and B). Very few functional data are available on SON or SNAT3. SON has been associated with acute myeloid leukemia (AML) through antiapoptotic activity and may be a target of Akt activation (Ahn et al., 2008). We found that loss of the RNA/DNA-binding protein SON reduces phosphorylation of Akt while increasing ribosomal protein S6 (RPS6) phosphorylation (Figure 3A). These results suggest that the depletion of SON activity may induce growth arrest and apoptosis through reduction of Akt phosphorylation and nuclear localization of FOXO. SNAT3 is a glutamine transporter that has been shown to be phosphorylated by Akt (Boehmer et al., 2003). Its expression in islets of Langerhans indicates a possible coregulation of glutamine and insulin release (Gammelsaeter et al., 2009). Glutamine regulation is also essential for uptake of other amino acids and has been shown to be an important energy source (Bode, 2001). We showed that reduction of SNAT3 decreases Akt and RPS6 phosphorylation. Considering that mTOR is important for amino acid sensing (Kim, 2009), loss of SNAT3 and glutamine transport could influence Akt phosphorylation through the mTOR feedback loops.
FIGURE 3:
Knockdown of novel components influence the Akt signaling network. (A) U2OS cells expressing EGFP-FOXO1a were treated with 40 nM of the indicated siRNA duplex (TSPAN9, 5′-GCAAAAGCCUAGUGCAUUG-3′ and 5′-UGGGUUGGGUGGCGAUUAU-3′; ITGAV, 5′-CCAUGUAGAUCACAAGAUA-3′ and 5′- CGACAAAGCUGAAUGGAUU-3′; SON, 5′- CAAUGUCAGUGGAGUAUCA-3′ and 5′-GAUACAGAACUACGAUAUA-3′; UCP5, 5′-UGCCAUCGUUGUAGGAGUA-3′ and 5′-GGAAUGAUGGGCGAUACAA-3′; SNAT3, 5′-AGAAGGAGCCUGCAAGAUC-3′ and 5′-GGUCAUCGGUGCCACAUCU-3′). The lysates were then subjected to SDS–PAGE and Western blot analysis. The antibodies used are labeled to right of each blot. (B) Densitometry analysis of P-S473-Akt over total Akt from part (A). Statistics were calculated for multiple experiments (n = 4). Student's t test was performed for comparison of test to control siRNAs (*p < 0.05). (C) RNAi for UCP5 and scrambled control were performed in cells stably expressing pcDNA, Flag-UCP5, or Flag-UCP5-siRNA–insensitive mutant (Flag-UCP5-siMut). Cell lysates were blotted with the antibodies labeled next to each blot. Densitometry for P-S473-Akt was normalized to GAPDH and total Akt. Student's t test was used to compare test to control siRNAs for each transfection set (* P < 0.01; see also Figure S5).
Reduction of UCP5 affects mitochondrial membrane potential and ATP production
Uncoupling protein 5 (UCP5) was the most intriguing gene that we discovered to diminish Akt phosphorylation when reduced through RNAi (Figure 3, A and B). Because much of our focus is on UCP5, we demonstrated that stably expressed flag-tagged siRNA duplex-insensitive but not wild-type UCP5 rescued Akt phosphorylation after knockdown of endogenous UCP5 demonstrating RNAi specificity in U2OS EGFP-FOXO1a cells (Figure 3C).
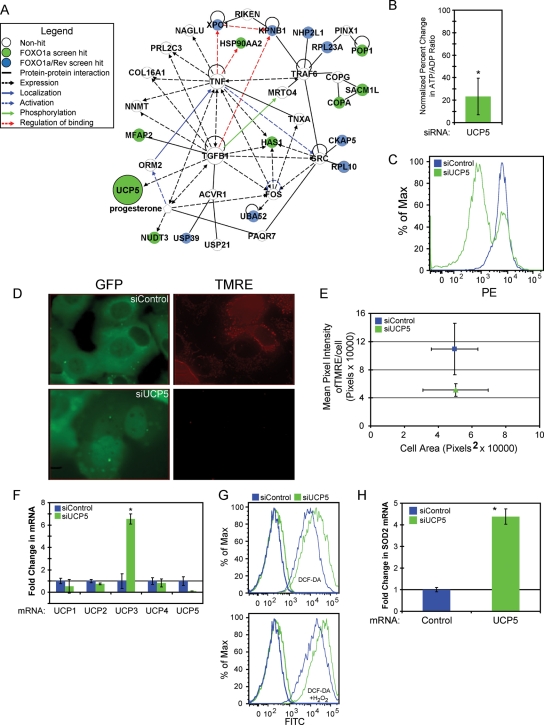
Uncoupling proteins are involved in disengagement of the (ETC) from ATP synthase, allowing for proton transport across the inner mitochondrial membrane without ATP production (Krauss et al., 2005). Despite UCP5 studies showing mammalian expression is influenced by H2O2 and insulin, which uncouple mitochondria with some neuroprotective properties, UCP5's functional importance is still largely unknown (Ho et al., 2006; Santandreu et al., 2009; Yonezawa et al., 2009; Kwok et al., 2010). By using Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA), which utilized published data to create a network interaction map, we highlighted that UCP5 mRNA expression was connected to transforming growth factor-β (TGF-β) and tangentially to nuclear import/export proteins KPNB1 and XPO1 (Figure 4A). Considering TGF-β involvement in apoptosis induction and differentiation (Moses et al., 1990; Moses and Serra, 1996), this network suggested that increased UCP5 mRNA expression may be an important effector of these processes.
FIGURE 4:
Knockdown of UCP5 causes ROS production and changes in the mitochondria membrane potential (MMP). (A) Network map of validated screen hits associated with UCP5 were created using Ingenuity Pathway Analysis software (Ingenuity Systems); ○ genes not affecting FOXO1a localization;  siRNA targets affecting FOXO1a localization;
siRNA targets affecting FOXO1a localization;  siRNA targets affecting FOXO1a/Rev localization. Edge relationships:
siRNA targets affecting FOXO1a/Rev localization. Edge relationships:  protein–protein interaction;
protein–protein interaction;  expression;
expression;  localization;
localization;  activation;
activation;  phosphorylation;
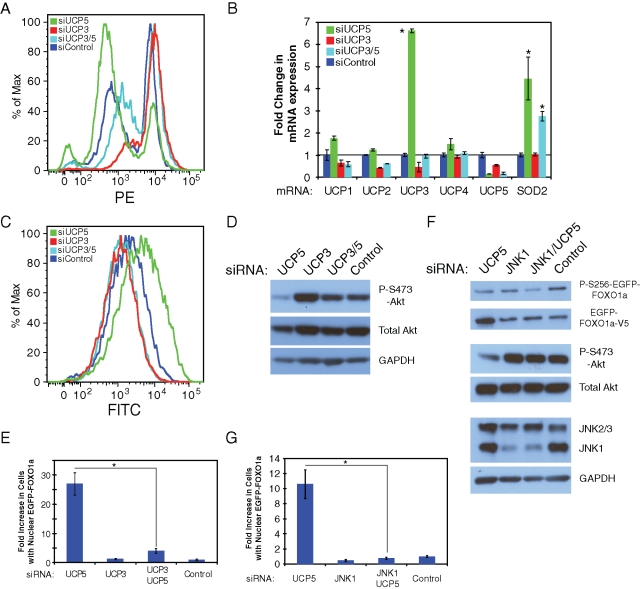
phosphorylation;  regulation of binding. (B) HPLC analysis of metabolites extract from U2OS EGFP-FOXO1a cells. ATP:ADP ratios were normalized to siRNA control (n = 6). Student's t test was used to compare UCP5 to control siRNA (*p < 0.05). (C) Knockdown cells are stained with TMRE-labeled active mitochondria. TMRE was detected by flow cytometry. Log PE was recorded versus histogram percentages of 10,000 cells. Representative results of at least three independent experiments were used. (D) Images of U2OS EGFP-FOXO1a cells treated with labeled siRNA and then stained with TMRE. Knockdown of UCP5 increases uncoupling, which decreases TMRE incorporation. Representative results of at least three independent experiments were used. (E) Quantization of cells stained with TMRE from part (D). (F) Real-time PCR of UCP mRNAs from cells treated with labeled knockdown. Representative results of at least three independent experiments were used. Student's t test was used to compare test to control siRNA (*p < 0.05). (G) U2OS cells were treated with labeled siRNA, then DCF-DA (light lines), and used for flow cytometry. Log FITC was recorded versus histogram percentages of 10,000 cells. Cells were additionally treated with H2O2 in the lower panel (light lines). Untreated cells were represented as darker lines in both panels. Representative results of at least three independent experiments were used. (H) Real-time PCR SOD2 mRNA from cells treated with labeled knockdown. Representative results of at least three independent experiments were used. Student's t test was used to compare test to control siRNAs (*p < 0.05; see also Table S3).
regulation of binding. (B) HPLC analysis of metabolites extract from U2OS EGFP-FOXO1a cells. ATP:ADP ratios were normalized to siRNA control (n = 6). Student's t test was used to compare UCP5 to control siRNA (*p < 0.05). (C) Knockdown cells are stained with TMRE-labeled active mitochondria. TMRE was detected by flow cytometry. Log PE was recorded versus histogram percentages of 10,000 cells. Representative results of at least three independent experiments were used. (D) Images of U2OS EGFP-FOXO1a cells treated with labeled siRNA and then stained with TMRE. Knockdown of UCP5 increases uncoupling, which decreases TMRE incorporation. Representative results of at least three independent experiments were used. (E) Quantization of cells stained with TMRE from part (D). (F) Real-time PCR of UCP mRNAs from cells treated with labeled knockdown. Representative results of at least three independent experiments were used. Student's t test was used to compare test to control siRNA (*p < 0.05). (G) U2OS cells were treated with labeled siRNA, then DCF-DA (light lines), and used for flow cytometry. Log FITC was recorded versus histogram percentages of 10,000 cells. Cells were additionally treated with H2O2 in the lower panel (light lines). Untreated cells were represented as darker lines in both panels. Representative results of at least three independent experiments were used. (H) Real-time PCR SOD2 mRNA from cells treated with labeled knockdown. Representative results of at least three independent experiments were used. Student's t test was used to compare test to control siRNAs (*p < 0.05; see also Table S3).
Previous work has shown that uncoupling proteins influence the ATP:ADP ratios through uncoupling of the ETC from ATP synthesis (Krauss et al., 2005; Dietrich and Horvath, 2010). We found reduction of UCP5 led to a 23% increase in the ATP:ADP ratio (Figure 4B). There are two possibilities that could lead to a change in this ratio: 1) the loss of mitochondrial membrane uncoupling increases ATP production in order to prevent backup of the ETC, or 2) ADP levels are reduced by adenylate kinase activity due to a loss of ATP production. Both of these conditions are directly linked to proton transport across the mitochondrial inner membrane (mitochondrial membrane potential [MMP]). To examine MMP in cells treated with siRNA, we used tetramethylrhodamine, ethyl ester perchlorate (TMRE). TMRE stains active mitochondria with polarized inner membranes. With the depletion of UCP5 and the increase in the ATP:ADP ratio, we expected the membrane potential to increase. Instead, we found that MMP decreased in the majority of cells treated with UCP5 siRNA (Figure 4C). Microscopy confirmed the results seen with flow cytometry (Figure 4, D and E).
We explored ways in which reduced UCP5 may be connected to decreased membrane potential. The simplest explanation could be a change in one of the other uncoupling proteins, which would influence MMP. Examination of real-time PCR of all UCP genes revealed that UCP3 expression increased following UCP5 reduction (Figure 4F). This result would explain the decreased TMRE incorporation.
There are previous studies showing that UCPs change the MMP and influence reactive oxygen species (ROS) production (Dietrich and Horvath, 2010). We measured ROS production using 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (DCF-DA) in flow cytometry. We showed that loss of UCP5 increased DCF-DA oxidation corresponding to increased ROS (Figure 4G). Treatment of the cells with H2O2 increased DCF-DA oxidation in all cases, which accentuated the effect seen with siRNA to UCP5. We observed an increase in SOD2 mRNA as a result of ROS production (Figure 4H). Nuclear FOXO has been shown to control the expression of mitochondria superoxide dismutase (SOD2), indicating a possible connection to FOXO (Nakae et al., 2008; Rodriguez et al., 2010).
Induction of UCP3 expression after UCP5 reduction alters activity of Akt and localization of FOXO1a through JNK1
We investigated whether UCP3 expression is responsible for MMP reduction and the ROS increase observed after UCP5 reduction. We reduced UCP3 using siRNA SMARTpool in combination with UCP5 siRNA duplex and measured TMRE and DCF-DA. Reduction of UCP3 resulted in a slight increase in TMRE incorporation when compared to control siRNA (Figure 5A, dark blue vs. red lines). Knockdown of both UCP3 and UCP5 brought the membrane potential back to control levels (Figure 5A, light blue vs. dark blue lines). These data were confirmed by real-time PCR expression analysis of UCP3 (Figure 5B). There was also a reduction in SOD2 expression, suggesting a reduction in ROS production and nuclear FOXO1a. Examination of DCF-DA confirmed that reduction of UCP3 alone and in combination with UCP5 reduced ROS production to control levels well below siRNA silencing of UCP5 alone (Figure 5C). These data confirm that induced expression of UCP3 after UCP5 loss leads to a decrease in membrane potential and increased ROS production.
FIGURE 5:
UCP5 loss up-regulates UCP3 and is connected to Akt/FOXO1a through JNK1 activity. (A) U2OS EGFP-FOXO1a cells are treated with UCP5 duplex alone, UCP3 SMARTpool, UCP3 plus UCP5, or control siRNAs for 96 h, then treated with TMRE for flow cytometry analysis of MMP. Log PE was recorded vs. histogram percentages of 10,000 cells. Representative results of at least three independent experiments were used. (B) Real-time PCR of UCP isoforms and SOD2 mRNAs from cells treated with labeled knockdown. Representative results of at least three independent experiments were used. Student's t test was used to compare test to control siRNA (*p < 0.01). (C) U2OS cells were treated with labeled siRNA and DCF-DA, and used for flow cytometry. Log FITC was recorded versus histogram percentages of 10,000 cells. Representative results of at least three independent experiments were used. (D) Western blot of RNAi knockdown of UCP5 and/or UCP3 compared to control siRNA. Antibody labels appear on the right. Representative results of at least three independent experiments were used. (E) Nuclear translocation analysis of EGFP-FOXO1a after siRNA knockdown of UCP3 and/or UCP5. Representative results of at least three independent experiments were used. Student's t test was used to compare UCP5 siRNA to UCP3/UCP5 siRNA (*p < 0.01). (F) Western blot of RNAi knockdown of UCP5 and/or JNK1 compared to nontargeting control siRNA. Antibody labels appear on the right. Representative results of at least three independent experiments were used. (G) Nuclear translocation analysis of EGFP-FOXO1a after siRNA knockdown of UCP5 and/or JNK1 compared to nontargeting control siRNA. Representative results of at least three independent experiments were used. Student's t test was used to compare UCP5 siRNA to JNK1/UCP5 siRNA (*p < 0.01).
We examined the phosphorylation status of Akt and FOXO1a localization after RNAi knockdown of UCP3 and UCP5. We confirmed a reduction in Akt phosphorylation after UCP5 knockdown that was recovered with the coablation of UCP5 and UCP3 by RNAi (Figure 5D). These data corresponded with localization of FOXO1a; knockdown of UCP5 and UCP3 together rescued the localization phenotype observed with siRNA of UCP5 alone (Figure 5E).
Changes in the ATP:ADP ratio, ROS production, and MMP after UCP5 reduction all suggested aberrations of the ETC . Perturbation of ETC function and ROS production pointed to a stress-induced response through JNK. JNK has been connected to Akt and FOXO in multiple instances (Lee et al., 2003; Sunayama et al., 2005; Vogt et al., 2005; Shao et al., 2006). To evaluate the level of JNK involvement, we treated cells with JNK1 SMARTpool siRNA alone and in combination with UCP5 knockdown. We observed a recovery of Akt phosphorylation after the combinatorial reduction of both UCP5 and JNK1 when compared to UCP5 knockdown alone (Figure 5F). There was only a slight change in FOXO1a phosphorylation after JNK1 RNAi. However, we identified cytoplasmic FOXO1a localization post-JNK1 and UCP5 coreduction consistent with an increase in Akt phosphorylation (Figure 5, F and G).
DISCUSSION
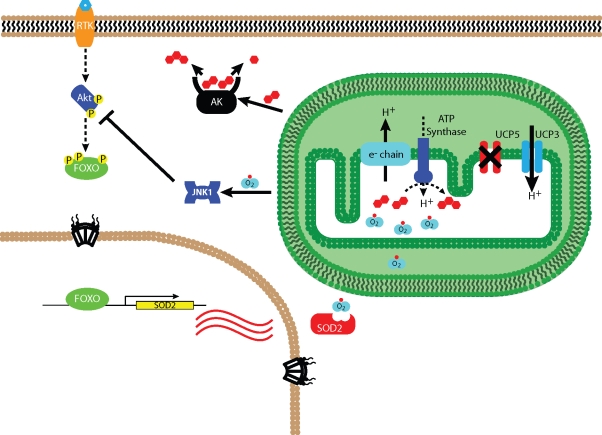
By exploiting FOXO1a's distinct localization in a high-throughput siRNA screen of the human genome, we discovered that knocking down a subset of components of focal adhesions, the nuclear pore complex, translation machinery, and the mitochondria resulted in nuclear accumulation of FOXO1a. Further analysis revealed that silencing a subset of translation factors affected general nuclear export, whereas losses of focal adhesion and mitochondrial components were specific FOXO1a effectors (Table 1). The most intriguing result was that the loss of UCP5 connects mitochondria to the Akt/FOXO1a signaling network through ROS and JNK1 (Figure 6).
FIGURE 6:
Mitochondrial function is connected to Akt through UCP5. Under normal growth conditions, Akt is actively sequestering phosphorylated FOXO in the cytoplasm. Mitochondria (green box) are operating properly to provide ATP for cellular energy consumption. After RNAi of UCP5, cells increase UCP3 expression, decreasing the MMP, which leads to aberrant ATP synthase production and increased stress on the ETC (e− chain) elevating free radical concentration (DCF-DA increase) and oxidative stress (O2*). ATP:ADP ratio decreases due to the activity of adenylate kinase activity (AK), which produces ATP and AMP () in order to maintain high cellular ATP levels. Increased ROS production activates JNK1 and promotes nuclear FOXO1a localization via dephosphorylation of Akt. FOXO's transcriptional activity controls expression of target genes such as mitochondrial SOD2 in order to battle ROS.
Depletion of UCP5 affects Akt signaling and the mitochondria through UCP3 and JNK1
UCP1 was discovered to be important in thermal regulation in brown fat (Krauss et al., 2005). In contrast, the other UCPs are more widely expressed and at present are not implicated in thermal regulation. Along with uncoupling mitochondrial oxidative phosphorylation, they are thought to be involved in β-oxidation through direct transport of fatty acids across the mitochondrial membrane (Krauss et al., 2005). Accordingly, UCP2 expression has been shown to be stimulated by free fatty acids (Rodriguez et al., 2010). Additional data show that UCP2 expression influences the release of insulin and glucose metabolism in β-islets (Krauss et al., 2005). In Drosophila, forced expression of human UCP2 attenuated insulin signaling, leading to a mild hyperglycemia and lifespan extension (Fridell et al., 2009). UCP3 is controlled by fasting, free fatty acids, and glucocorticoids (Amat et al., 2007; Rodriguez et al., 2010). Overexpression of UCP3 in skeletal muscle results in an increase in the ATP:ADP ratio and a decrease in mitochondrial membrane potential (Garcia-Martinez et al., 2001). This leads to a switch from glucose metabolism to β-oxidation and protection against fat-induced insulin resistance (Choi et al., 2007; Schrauwen et al., 2004). This is an important fact to consider, since insulin resistance is a hallmark of obesity and type 2 diabetes.
Here we demonstrate control of mitochondria inner membrane uncoupling through UCP5 reduction and a connection to Akt/FOXO signaling (Figure 6). Under normal growth conditions, Akt-phosphorylated FOXO is in the cytoplasm and the mitochondria operate properly. Stressing MMP through RNAi of UCP5 causes a compensatory increase in UCP3 (Figure 4F). This leads to a depletion of the MMP and an increase in ROS production through the stressed ETC (Figure 4, C and G). The lack of ATP synthase activity elevates adenylate kinase activity to maintain ATP levels (Noma, 2005). This action depletes ADP, which results in an increase in the ATP:ADP ratio (Figure 4B). The stressed ETC and ROS production induce activation of JNK1 (Figure 5F). In turn, JNK1 controls FOXO1a localization through dephosphorylation of Akt (Figure 5F) (Lee et al., 2003; Vogt et al., 2005). As a result, nuclear FOXO1a activates the transcription of genes involved in the stress response, including superoxide dismutase (SOD2) in order to battle mitochondrial ROS (Figure 5B).
Our model (Figure 6) proposes that Akt signaling provides a crucial response to changes in mitochondrial ETC alterations and energy output caused by UCP5 reduction. As such, the JNK1 link between mitochondria and Akt has implications in obesity, diabetes treatment, and lifespan (Sanchez-Blanco et al., 2006; Jia et al., 2009; Sabio and Davis, 2010). JNK has been shown to phosphorylate and alter the localization of FOXO3a directly and through the FOXO repressor, 14-3-3 (Sunayama et al., 2005; Vogt et al., 2005). In mouse fibroblasts, JNK negatively influences insulin signaling through repressive phosphorylation of IRS1 (Lee et al., 2003). In addition, chronic activation of JNK has been linked to insulin resistance through inflammation in muscle cells (Hirosumi et al., 2002; Tuncman et al., 2006). However, acute activation has recently been demonstrated to reverse insulin resistance in mouse myoblasts (Berdichevsky et al., 2010). Additionally, a study in ischemic-induced hypoxic cardiomyocytes showed that JNK phosphorylates Akt, priming it for full activation by PI3K signaling (Shao et al., 2006). Previous studies, together with our new data, suggest that targeting of uncoupling proteins could potentially influence insulin signaling and glycolytic balance, making it a potential therapeutic target for obesity and diabetes.
All three isoforms of Akt control FOXO1a localization
Additionally in this study, we verified that knockdown of Akt activators (PDK1, Rictor, and SIN1) and XPO1 localize FOXO1a to the nucleus. Interestingly, silencing of Akt1, p85α, and mTOR failed to result in accumulation of nuclear FOXO1a. The reason for this lack of effect may be that there are multiple isoforms or complexes of these proteins that affect FOXO1a localization (Jacinto et al., 2004; Chaussade et al., 2007). Indeed, previous reports have confirmed involvement of Akt2 in FOXO1a phosphorylation and activation (DeBosch et al., 2006). However, the participation of all three isoforms in FOXO1a localization has not been extensively studied. We systematically looked at the participation of all three Akt genes in FOXO1a localization. All isoforms were represented in U2OS cells, with the majority of mRNA being composed of Akt1 and Akt2. We demonstrated that all three Akt isoforms influence the localization of FOXO1a, with Akt2 and Akt3 making up half of the activity. This result is important to consider when designing drugs that target specific Akt isoforms.
Loss of critical complexes influences FOXO1a localization
Since multiple functions are associated with each gene found in the siRNA screen, there are inherent limitations of gene ontological analysis. Despite this fact, functional categorization from the FOXO1a and Rev screens suggest knockdown of some critical factors involved in transcription, splicing, and protein degradation influence FOXO1a localization. On the other hand, we found that reduction of a subset of translation factors is important for nuclear import and export. Additional studies will need to be undertaken to determine whether these factors are important hubs of both splicing control and Akt signaling, for example, or whether their function is tangential to the Akt signaling network, with a coincidental effect on FOXO1a localization.
Previous studies have shown that RNA splicing has been linked to mTOR signaling through the SKAR protein that recruits active ribosomal S6 kinase (mTOR substrate) to newly spliced mRNA for enhanced translation efficiency (Ma et al., 2008). Perhaps a loss of spliceosome and related components causes a loss of growth signaling to mTOR and Akt, thereby leading to nuclear accumulation and activation of FOXO1a.
Additionally, other studies coupled with our data have linked FOXO and Akt signaling to protein degradation machinery activation. In cardiomyocytes, active FOXO promotes the transcription of atrogin-1, an E3 ligase that controls the activity and degradation of calcineurin and protein phosphatase 2A (PP2A) (Vogt et al., 2005; Ni et al., 2006, 2007). These and other phosphatases, such as protein phosphatase 1 (PP1) and PH domain and leucine-rich repeat protein phosphatases (PHLPP), have been shown to control the dephosphorylation of Akt (Miyamoto et al., 2010). This would connect the proteasome to the Akt pathway through a FOXO1a transcriptionally controlled negative feedback loop.
In addition to important complexes, our high-throughput siRNA screen identified individual genes that influence FOXO1a localization. These include proteins involved in cell adhesion and other novel genes, such as SON and SNAT3. Our data and the data of others have connected focal adhesion to FOXO localization and the Akt signaling network (Figure S5) (Gutierrez-Lopez et al., 2003; Lishner et al., 2008; Zoller, 2009; Aldaye et al., 2010). Considering tetraspanins have been linked to type 2 diabetes susceptibility (Zeggini et al., 2008), our evidence further confirms that link and extends the connection between Akt/FOXO regulation and cell attachment.
In conclusion, our list of RNAi-confirmed genes specific to FOXO1a localization (Table 1) presents an intriguing set of factors potentially linked to Akt signaling. Considering aberrant Akt signaling is a crucial step in diabetes and cancer progression (Manning, 2004), these genes, including UCP5, could be prospective targets for future drug development.
MATERIALS AND METHODS
Tissue culture and stable cell lines
U2OS cells were maintained in McCoy's 5A medium (Invitrogen, Life Technologies) supplemented with 10% fetal bovine serum (FBS; HyClone, Thermo Fisher, Lafayette, CO) and penicillin/streptomycin (Invitrogen, Carlsbad, CA). Stable clones were maintained in 200 μg/ml hygromycin. A U2OS cell line with a single FRT (5′-GAAGTTCCTATTCtctagaaaGTATAGGAACTTC) site of integration and the start codon for hygromycin was used for the expression of pEF5/FRT/V5 (Invitrogen) with EGFP, FOXO1a, or EGFP-FOXO1a. FOXO1a was PCR amplified and cloned from pcDNA3-Flag-FKHR plasmid (Ramaswamy et al., 2002). EGFP was PCR amplified and cloned from pEGFP-C1 (Clontech, Mountain View, CA). Stable integration was achieved by coexpressing flipase with pEF5/FRT/V5 constructs in FRT U2OS cells and selecting with 500 μg/ml hygromycin (Invitrogen). UCP5 long form was cloned from U2OS cDNA using the forward (5′-GGAATTCATGGGTATCTTTCCCGGAATAATCC-3′) and reverse FLAG (5′-CCGCTCGAGCGGttacttgtcatcgtcgtccttgtagtcGATTTGAAGCCTCTTTAGCTGCTC-3′) primers and cloned using EcoRI and XhoI into pcDNA3.1+ with puromycin mammalian resistance. An siRNA-insensitive mutant UCP5 was created using the following forward primer (5′-CTGCTCAGCGTGCTGCCATCGTgGTgGGAGTAGAGCTACCAGTCTA-3′) and the reverse complement. Both wild-type and mutant UCP5 were transfected into the EGFP-FOXO1a stable U2OS cell line, then selected with 1 μg/ml of puromycin/200 μg/ml hygromycin to establish double clones. Small molecules were obtained as follows: dimethyl sulfoxide (DMSO), 40 nM wortmannin, 4 nM leptomycin B (Sigma-Aldrich, St. Louis, MO); 5 μM ZSTK474 (LC Laboratories, Woburn, MA); 5 μM Akti-1/2 (DeFeo-Jones et al., 2005).
Western analysis
Cells were lysed on tissue culture plates in PTY buffer (50 mM HEPES, 50 mM NaCl, 5 mM EDTA, 1% TritonX-100, 50 mM NaF, 10 mM Na4P2O7, 1 mM Na3O4V, 10 μg/ml PMSF) for 30 min on ice. Cells were scraped with a cell lifter and passed through a 22-gauge needle at least five times. Lysates were incubated in 1.5 ml Eppendorf tubes for 15 min and then spun at 4ºC for 10 min at 15,000 rpm in a benchtop centrifuge. The concentration was determined by Bradford assay. Lysates were normalized and loaded into a 4–12% Bis-Tris NuPage gel (Invitrogen) and run with MOPS buffer. The lysates were then transferred to Immobilon-P membrane (Millipore) and probed with antibodies. Antibodies used were ITGA, Phospho-S473-Akt, Phospho-S256-FOXO, Pan-Akt, Akt1, p85α, mTOR, PDK1, ribosomal S6, Phospho-S235/236-S6, SAPK/JNK (Cell Signaling, Beverly, MA), V5, GAPDH-HRP conjugate (Sigma-Aldrich), Rictor (Bethyl Laboratories, Montgomery, TX), and TSPAN9 (Abcam, Cambridge, MA).
High-throughput screening of U2OS EGFP-FOXO1a cells using 384-well plates
A semi-automated protocol for high-throughput screening was developed to test the Dharmacon Human Genome Library in triplicate (Figure 3A). Control siRNAs were plated on Costar 384-well, clear, flat-bottomed plates (Corning Life Sciences, Lowell, MA). Two microliters of 600 nM siRNA (final concentration of 40 nM in 30 μl) was added to each plate by hand pipetting. A Matrix Wellmate (Thermo Scientific, Lafayette, CO) was used to pipette 9 μl of transfection mix (0.15 μl HiPerfect [Qiagen, Valencia, CA] and 8.85 μl Optimem [Invitrogen]) into each well. Velocity 11 Bravo liquid handler (Agilent Technologies, Santa Clara, CA) was used to pipette 1.2 μl of 1 μM siRNA from the human genome library (Dharmacon, Thermo Fisher, Lafayette, CO) into each well. The plates were incubated for at least 20 min before the cells were added. After trypsinization and counting, 3000 cells were added to each well using the Wellmate. The cells were gently spun to evenly spread them onto the plate surface. After 96 h of incubation, the cells were washed in phosphate-buffered saline (PBS), fixed in 3% paraformaldehyde (Sigma-Aldrich), and stained with DAPI (Sigma-Aldrich). The plates were taken to the ImageXpress high-throughput microscope for imaging (Molecular Devices, Sunnyvale, CA). To maximize the number of cells captured in one photograph, a 10× image of green fluorescent protein (GFP; 495 nm/519 nm) and DAPI (358 nm/461 nm) was taken of each well (200–1000 cells/image, depending on confluency).
Automated nuclear translocation analysis
GFP and DAPI images were used with an analysis program written for Metamorph software. In brief, a line was drawn around nuclei in the DAPI image to score the nuclear section, and a larger concentric circle was drawn outside the nuclei circle to score the cytoplasmic section. This image was then overlaid with the GFP image, and the nuclear-to-cytoplasmic ratio was determined. If this ratio was above a user-defined threshold, the cell was counted as positive for nuclear staining. The number of nuclear positive (GFP image) over total number of cells (DAPI image) indicates the percentage of positives. The nuclear/cytoplasmic threshold was determined by maximizing the difference in percentage between the positive (exportin-1) and negative (scrambled) siRNA controls.
Hit Identification
All data processing and analyses were performed in R (http://www.r-project.org/) using the BioConductor package cell HTS2 for structured data representation, sample annotation, and calculation of robust z-scores:
where X is the vector of measurements across samples, ![]() is the median, and MAΓ is the median absolute deviation. All data were normalized using median scaling (Boutros et al., 2006) and log transformed. Robust (platewise) z-scores were averaged across biological replicates. For the SVM models, each assayed value (sample i, replicate j) was stored in n-dimensional vectors Xi consisting of elements xij ∈ Xi : j ≤ n. To train the SVM models, we specified the set of control wells and their associated labels: X+p:P∈ {positive controls} and X−p: P∈ {negative controls}. Once the appropriate formulation was selected, the SVM algorithm defined the optimal boundary separating the positive and negative controls in n-dimensional replicate space. From here, the process of hit identification simply involved applying the trained SVM model to predict continuous class identifiers P(class = hit | Xr); for the remaining experimental data Xr∉{X+∪X−}. Using preexisting R libraries (Chang and Lin, 2001), we defined two separate classification SVM models using Gaussian radial basis (RBF) or linear kernel functions to predict hit confidence or hit strength, respectively. We used Platt's (Platt, 2000) method for probabilistic output, which allowed for false discovery detection calculated at arbitrary thresholds according to Zhang (Zhang et al., 2008). Using this method, we routinely analyzed genome-wide screening data in less than 5 min with minimal processing capacity (2.4 GHz Core 2 Duo laptop computers running Mac OS version 10.5.6). Complete coded methods are provided as supplementary R scripts.
is the median, and MAΓ is the median absolute deviation. All data were normalized using median scaling (Boutros et al., 2006) and log transformed. Robust (platewise) z-scores were averaged across biological replicates. For the SVM models, each assayed value (sample i, replicate j) was stored in n-dimensional vectors Xi consisting of elements xij ∈ Xi : j ≤ n. To train the SVM models, we specified the set of control wells and their associated labels: X+p:P∈ {positive controls} and X−p: P∈ {negative controls}. Once the appropriate formulation was selected, the SVM algorithm defined the optimal boundary separating the positive and negative controls in n-dimensional replicate space. From here, the process of hit identification simply involved applying the trained SVM model to predict continuous class identifiers P(class = hit | Xr); for the remaining experimental data Xr∉{X+∪X−}. Using preexisting R libraries (Chang and Lin, 2001), we defined two separate classification SVM models using Gaussian radial basis (RBF) or linear kernel functions to predict hit confidence or hit strength, respectively. We used Platt's (Platt, 2000) method for probabilistic output, which allowed for false discovery detection calculated at arbitrary thresholds according to Zhang (Zhang et al., 2008). Using this method, we routinely analyzed genome-wide screening data in less than 5 min with minimal processing capacity (2.4 GHz Core 2 Duo laptop computers running Mac OS version 10.5.6). Complete coded methods are provided as supplementary R scripts.
Secondary confirmatory screens
A select group of siRNAs was identified for reconfirmation. Generally, this group included genes that passed a strength score false discovery rate (FDR) threshold of 5%; however, hits that were associated with high inviability scores were eliminated, and discretion was used such that some siRNAs of particular biological interest with high z-scores (but low strength scores) were also rescreened. Individual duplexes (four per pool) were screened in biological triplicate using the same format described above. For both screens, reconfirmed duplexes were those ≥ 3 SD from the mean of the combined negative controls. Final results were reported as duplex validation efficiencies (0/4, 1/4, etc.).
Method validation and performance statistics
We tested three hit-identification methods: two SVM models (confidence and strength) and average z-scores. For comparative analysis of all three methods, average z-scores were normalized to resemble probabilities (i.e., zi,norm ∈ [0,1]) using the following transformation function:
where Z represents the vector of average z-scores calculated across all experimental wells. Performance statistics, including the true positive rate (sensitivity), false positive rate (1 − specificity), receiver operating characteristic (ROC), and area under the receiver operating characteristic (AUROC) were calculated using pre-existing R routines (Sing et al., 2005):
where TP is the number of true positives, FN is the number of false negatives, FP is the number of false positives, and TN is the number of true negatives.
Network pathway designer graphical representation
A network pathway is a graphical representation of the molecular relationships between molecules. Molecules are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base (Ingenuity Systems, Redwood, CA). Human, mouse, and rat orthologues of a gene are stored as separate objects in the Ingenuity Pathways Knowledge Base, but are represented as a single node in the network. Nodes are displayed using circles with various colors relating to the screen performed. Edges are displayed with various colors and styles that describe the nature of the relationship between the nodes.
High-performance liquid chromatography
A 60-mm plate of U2OS EGFP-FOXO1a cells were treated with siRNA and then trypsinized and lysed with methanol:chloroform:water (2:2:1.8) on ice. Lysates were vortexed three times for 15 s each to disrupt the cell membranes. Lysates were centrifuged for 10 min at 10,000 × g at 4ºC. The aqueous fraction was moved to a 15 ml centrifuge tube and frozen in liquid nitrogen. Supernatant was lyophilized at −40ºC overnight. Dried pellets were resuspended in 100 μl of anaerobic water and run immediately on Agilent 1100 series high-performance liquid chromatography (HPLC) instrument fitted with a Luna C18 5 μ column (250 × 4.5 mm; Phenomenex, Torrance, CA). Mobile phase A (10 mM tetrabutylammonium hydroxide, 10 mM KH2PO4, 0.25% methanol) and mobile phase B (2.8 mM tetrabutylammonium hydroxide, 100 mM KH2PO4, 30% methanol) were used for metabolite separation. Maximum separation was achieved at a flow rate of 0.75 ml/min from 55 to 57.7% mobile phase B, with a ramping rate of 0.073%B/min over 37 min. ATP, ADP, NAD+, and NADH were analyzed at 260 nm using a photodiode array detector (Waters 996; Meadows Instrumentation, Bristol, WI) comparing retention times to standards. The areas under the curve were used to calculate ratios.
TMRE and DCF-DA stain detection
Microscopy: U2OS EGFP-FOXO1a cells were treated with labeled siRNAs. Cells were grown on glass coverslips, then stained with 50 μM TMRE (Sigma-Aldrich) for 30 min in serum-free media at 37ºC. Live cells were used directly for microscopy analysis. Flow cytometry: U2OS EGFP-FOXO1a cells were treated with 50 μM of TMRE for 30 min in serum-free media at 37ºC. Normal U2OS cells were treated with DCF-DA (Invitrogen) or DCF-DA plus 2 mM H2O2 for 30 min in serum-free media at 37ºC. Cells were then trypsinized and washed in PBS with 1% serum. They were then stained with Sytox Blue (Molecular Probes, Invitrogen) to identify live and dead cells. Flow cytometry (LSR II; BD Biosciences, Billerica, MA) was used for TMRE and DCF-DA analysis.
Real-time PCR
RNA was harvested with the RNeasy kit from Qiagen using the standard protocol. First strand synthesis was performed using Superscript III kit with oligo dT primer from Invitrogen. Primers were determined at the exonic junction, whenever possible, by using the web-based Primer3 program (http://frodo.wi.mit.edu/primer3/). The cDNA plus primer sets (Table S3) were used with SYBR green mastermix (Applied Biosystems, Life Technologies, Carlsbad, CA) in a 96-well format qPCR machine (Eppendorf, Hauppauge, NY), and data were collected for 45 cycles. All mRNA was determined relative to GAPDH control primer sets.
Supplementary Material
Acknowledgments
We thank Zeev Waks for aid in setting up the HPLC assay and Michael Moore, Devin Burrill, Thomas Armel, Faisal Aldaye, Daniel Ducat, and Aristotelis Astrinidis for their helpful insight and critical review of the manuscript. RNAi screening capability was provided by the ICCB-Longwood screening facility at Harvard Medical School with assistance from Caroline Schmu, Sean Johnston, Stewart Rudnicki, Zachery Cooper, and David Wrobel. This work was supported by NIH grant R37GM36373 to P.A.S.
Abbreviations used:
- AK
adenylate kinase
- AML
acute myeloid leukemia
- AUROC
area under the receiver operating characteristic
- DAPI
4′,6-diamidino-2-phenylindole
- DCF-DA
5-(and-6)-chloromethyl-2′,7′ dichlorodihydrofluorescein diacetate, acetyl ester
- DMSO
dimethyl sulfoxide
- EGFP
enhanced green fluorescent protein
- ETC
electron transport chain
- FBS
fetal bovine serum
- FDR
false discovery rate
- FOXO
Forkhead transcription factor
- GFP
green fluorescent protein
- HPLC
high-performance liquid chromatography
- ITGA
integrin αV
- JNK1
c-Jun N-terminal kinase 1
- LMB
leptomycin B
- MMP
mitochondria membrane potential
- PDK1
phosphoinositide-dependent protein kinase
- PHLPP
PH domain and leucine-rich repeat protein phosphatase
- PI3K
phosphatidylinositol-3 kinase
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- PP1
protein phosphatase 1
- PP2A
protein phosphatase 2A
- RNAi
interfering RNA
- ROC
receiver operating characteristic
- ROS
reactive oxygen species
- RPS6
ribosomal protein S6
- siRNA
small interfering RNA
- SOD2
superoxide dismutase
- SVM
support vector machine
- TGF-β
transforming growth factor-β
- TLN1
talin 1
- TMRE
tetramethylrhodamine, ethyl ester perchlorate
- TSPAN9
tetraspanin 9
- UCP
uncoupling protein
- XPO1
exportin 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0854) on April 1, 2011.
REFERENCES
- Ahn EY, Yan M, Malakhova OA, Lo MC, Boyapati A, Ommen HB, Hines R, Hokland P, Zhang DE. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc Natl Acad Sci USA. 2008;105:17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldaye FA, Senapedis WT, Silver PA, Way JC. A structurally tunable DNA-based extracellular matrix. J Am Chem Soc. 2010;132:14727–14729. doi: 10.1021/ja105431h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amat R, Solanes G, Giralt M, Villarroya F. SIRT1 is involved in glucocorticoid-mediated control of uncoupling protein-3 gene transcription. J Biol Chem. 2007;282:34066–34076. doi: 10.1074/jbc.M707114200. [DOI] [PubMed] [Google Scholar]
- Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab. 2005;16:183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Guarente L, Bose A. Acute oxidative stress can reverse insulin resistance by inactivation of cytoplasmic JNK. J Biol Chem. 2010;285:21581–21589. doi: 10.1074/jbc.M109.093633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode BP. Recent molecular advances in mammalian glutamine transport. J Nutr. 2001;131:2475S–2485S;. doi: 10.1093/jn/131.9.2475S. discussion 2486S–2477S. [DOI] [PubMed] [Google Scholar]
- Boehmer C, Okur F, Setiawan I, Broer S, Lang F. Properties and regulation of glutamine transporter SN1 by protein kinases SGK and PKB. Biochem Biophys Res Commun. 2003;306:156–162. doi: 10.1016/s0006-291x(03)00921-5. [DOI] [PubMed] [Google Scholar]
- Boutros M, Bras LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7:R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgering BM, Medema RH. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. J Leukocyte Biol. 2003;73:689–701. doi: 10.1189/jlb.1202629. [DOI] [PubMed] [Google Scholar]
- Carter ME, Brunet A. FOXO transcription factors. Curr Biol. 2007;17:R113–R114. doi: 10.1016/j.cub.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin CJ. Training nu-support vector classifiers: theory and algorithms. Neural Comput. 2001;13:2119–2147. doi: 10.1162/089976601750399335. [DOI] [PubMed] [Google Scholar]
- Chaussade C, et al. Evidence for functional redundancy of class IA PI3K isoforms in insulin signalling. Biochem J. 2007;404:449–458. doi: 10.1042/BJ20070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CS, et al. Overexpression of uncoupling protein 3 in skeletal muscle protects against fat-induced insulin resistance. J Clin Invest. 2007;117:1995–2003. doi: 10.1172/JCI13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBosch B, Sambandam N, Weinheimer C, Courtois M, Muslin AJ. Akt2 regulates cardiac metabolism and cardiomyocyte survival. J Biol Chem. 2006;281:32841–32851. doi: 10.1074/jbc.M513087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFeo-Jones D, et al. Tumor cell sensitization to apoptotic stimuli by selective inhibition of specific Akt/PKB family members. Mol Cancer Ther. 2005;4:271–279. [PubMed] [Google Scholar]
- Dietrich MO, Horvath TL. The role of mitochondrial uncoupling proteins in lifespan. Pflug Arch. 2010;459:269–275. doi: 10.1007/s00424-009-0729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell YW, Hoh M, Kreneisz O, Hosier S, Chang C, Scantling D, Mulkey DK, Helfand SL. Increased uncoupling protein (UCP) activity in Drosophila insulin-producing neurons attenuates insulin signaling and extends lifespan. Aging (Albany NY) 2009;1:699–713. doi: 10.18632/aging.100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammelsaeter R, Jenstad M, Bredahl MK, Gundersen V, Chaudhry FA. Complementary expression of SN1 and SAT2 in the islets of Langerhans suggests concerted action of glutamine transport in the regulation of insulin secretion. Biochem Biophys Res Commun. 2009;381:378–382. doi: 10.1016/j.bbrc.2009.02.062. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez C, Sibille B, Solanes G, Darimont C, Mace K, Villarroya F, Gomez-Foix AM. Overexpression of UCP3 in cultured human muscle lowers mitochondrial membrane potential, raises ATP/ADP ratio, and favors fatty acid vs. glucose oxidation. FASEB J. 2001;15:2033–2035. doi: 10.1096/fj.00-0828fje. [DOI] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Lopez MD, Ovalle S, Yanez-Mo M, Sanchez-Sanchez N, Rubinstein E, Olmo N, Lizarbe MA, Sanchez-Madrid F, Cabanas C. A functionally relevant conformational epitope on the CD9 tetraspanin depends on the association with activated beta1 integrin. J Biol Chem. 2003;278:208–218. doi: 10.1074/jbc.M207805200. [DOI] [PubMed] [Google Scholar]
- Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- Ho PW, Chu AC, Kwok KH, Kung MH, Ramsden DB, Ho SL. Knockdown of uncoupling protein-5 in neuronal SH-SY5Y cells: effects on MPP+-induced mitochondrial membrane depolarization, ATP deficiency, and oxidative cytotoxicity. J Neurosci Res. 2006;84:1358–1366. doi: 10.1002/jnr.21034. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jia JJ, Zhang X, Ge CR, Jois M. The polymorphisms of UCP2 and UCP3 genes associated with fat metabolism, obesity and diabetes. Obes Rev. 2009;10:519–526. doi: 10.1111/j.1467-789X.2009.00569.x. [DOI] [PubMed] [Google Scholar]
- Kau TR, Schroeder F, Ramaswamy S, Wojciechowski CL, Zhao JJ, Roberts TM, Clardy J, Sellers WR, Silver PA. A chemical genetic screen identifies inhibitors of regulated nuclear export of a Forkhead transcription factor in PTEN-deficient tumor cells. Cancer Cell. 2003;4:463–476. doi: 10.1016/s1535-6108(03)00303-9. [DOI] [PubMed] [Google Scholar]
- Kim D, Chung J. Akt: versatile mediator of cell survival and beyond. J Biochem Mol Biol. 2002;35:106–115. doi: 10.5483/bmbrep.2002.35.1.106. [DOI] [PubMed] [Google Scholar]
- Kim E. Mechanisms of amino acid sensing in mTOR signaling pathway. Nutr Res Pract. 2009;3:64–71. doi: 10.4162/nrp.2009.3.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Zhang CY, Lowell BB. The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol. 2005;6:248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- Kwok KH, Ho PW, Chu AC, Ho JW, Liu HF, Yiu DC, Chan KH, Kung MH, Ramsden DB, Ho SL. Mitochondrial UCP5 is neuroprotective by preserving mitochondrial membrane potential, ATP levels, and reducing oxidative stress in MPP+ and dopamine toxicity. Free Radic Biol Med. 2010;49:1023–1035. doi: 10.1016/j.freeradbiomed.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Lee YH, Giraud J, Davis RJ, White MF. c-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J Biol Chem. 2003;278:2896–2902. doi: 10.1074/jbc.M208359200. [DOI] [PubMed] [Google Scholar]
- Lishner M, Zismanov V, Tohami T, Tartakover-Matalon S, Elis A, Drucker L. Tetraspanins affect myeloma cell fate via Akt signaling and FoxO activation. Cell Signal. 2008;20:2309–2316. doi: 10.1016/j.cellsig.2008.08.018. [DOI] [PubMed] [Google Scholar]
- Los M, Maddika S, Erb B, Schulze-Osthoff K. Switching Akt: from survival signaling to deadly response. Bioessays. 2009;31:492–495. doi: 10.1002/bies.200900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Manning BD. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SC, Cras-Meneur C, Bernal-Mizrachi E, Permutt MA. Glucose regulates Foxo1 through insulin receptor signaling in the pancreatic islet beta-cell. Diabetes. 2006;55:1581–1591. doi: 10.2337/db05-0678. [DOI] [PubMed] [Google Scholar]
- McCurdy CE, Cartee GD. Akt2 is essential for the full effect of calorie restriction on insulin-stimulated glucose uptake in skeletal muscle. Diabetes. 2005;54:1349–1356. doi: 10.2337/diabetes.54.5.1349. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, et al. PHLPP-1 negatively regulates Akt activity and survival in the heart. Circ Res. 2010;107:476–484. doi: 10.1161/CIRCRESAHA.109.215020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses H, Serra R. Regulation of differentiation by TGF-beta. Curr Opin Genet Dev. 1996;6:581–586. doi: 10.1016/s0959-437x(96)80087-6. [DOI] [PubMed] [Google Scholar]
- Moses H, Yang E, Pietenpol J. TGF-beta stimulation and inhibition of cell proliferation: new mechanistic insights. Cell. 1990;63:245–247. doi: 10.1016/0092-8674(90)90155-8. [DOI] [PubMed] [Google Scholar]
- Nakae J, Cao Y, Oki M, Orba Y, Sawa H, Kiyonari H, Iskandar K, Suga K, Lombes M, Hayashi Y. Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes. 2008;57:563–576. doi: 10.2337/db07-0698. [DOI] [PubMed] [Google Scholar]
- Ni YG, et al. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114:1159–1168. doi: 10.1161/CIRCULATIONAHA.106.637124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni YG, Wang N, Cao DJ, Sachan N, Morris DJ, Gerard RD, Kuro OM, Rothermel BA, Hill JA. FoxO transcription factors activate Akt and attenuate insulin signaling in heart by inhibiting protein phosphatases. Proc Natl Acad Sci USA. 2007;104:20517–20522. doi: 10.1073/pnas.0610290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma T. Dynamics of nucleotide metabolism as a supporter of life phenomena. J Med Invest. 2005;52:127–136. doi: 10.2152/jmi.52.127. [DOI] [PubMed] [Google Scholar]
- Oliveira SJ, Pinto JP, Picarote G, Costa VM, Carvalho F, Rangel M, de Sousa M, de Almeida SF. ER stress-inducible factor CHOP affects the expression of hepcidin by modulating C/EBPalpha activity. PLoS One. 2009;4:e6618. doi: 10.1371/journal.pone.0006618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuma H, Vander Kooi BT, Boustead JN, Oeser JK, O’Brien RM. Correlation between FOXO1a (FKHR) and FOXO3a (FKHRL1) binding and the inhibition of basal glucose-6-phosphatase catalytic subunit gene transcription by insulin. Mol Endocrinol. 2006;20:2831–2847. doi: 10.1210/me.2006-0085. [DOI] [PubMed] [Google Scholar]
- Platt J. Probabilistic outputs for support vector machines and comparison to regularized likelihood methods. In: Smola AJ, Bartlett P, Schölkopf B, Schuurmaus D, editors. In Advances in Large Margin Classifiers. Cambridge, MA: MIT Press; 2000. [Google Scholar]
- Protty MB, et al. Identification of Tspan9 as a novel platelet tetraspanin and the collagen receptor GPVI as a component of tetraspanin microdomains. Biochem J. 2009;417:391–400. doi: 10.1042/BJ20081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez AM, Sanchez J, Tobaruela A, Priego T, Pico C, Palou A. Time-course effects of increased fatty acid supply on the expression of genes involved in lipid/glucose metabolism in muscle cells. Cell Physiol Biochem. 2010;25:337–346. doi: 10.1159/000276566. [DOI] [PubMed] [Google Scholar]
- Sabio G, Davis RJ. cJun NH2-terminal kinase 1 (JNK1): roles in metabolic regulation of insulin resistance. Trends Biochem Sci. 2010;35:490–496. doi: 10.1016/j.tibs.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Arnolds DE, Fujii N, Kramer HF, Hirshman MF, Goodyear LJ. Role of Akt2 in contraction-stimulated cell signaling and glucose uptake in skeletal muscle. Am J Physiol Endocrinol Metab. 2006;291:E1031–E1037. doi: 10.1152/ajpendo.00204.2006. [DOI] [PubMed] [Google Scholar]
- Sanchez-Blanco A, Fridell YW, Helfand SL. Involvement of Drosophila uncoupling protein 5 in metabolism and aging. Genetics. 2006;172:1699–1710. doi: 10.1534/genetics.105.053389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santandreu FM, Valle A, Fernandez de Mattos S, Roca P, Oliver J. Hydrogen peroxide regulates the mitochondrial content of uncoupling protein 5 in colon cancer cells. Cell Physiol Biochem. 2009;24:379–390. doi: 10.1159/000257430. [DOI] [PubMed] [Google Scholar]
- Schrauwen P, Hardie DG, Roorda B, Clapham JC, Abuin A, Thomason-Hughes M, Green K, Frederik PM, Hesselink MK. Improved glucose homeostasis in mice overexpressing human UCP3: a role for AMP-kinase? Int J Obes Relat Metab Disord. 2004;28:824–828. doi: 10.1038/sj.ijo.0802629. [DOI] [PubMed] [Google Scholar]
- Shao Z, et al. c-Jun N-terminal kinases mediate reactivation of Akt and cardiomyocyte survival after hypoxic injury in vitro and in vivo. Circ Res. 2006;98:111–118. doi: 10.1161/01.RES.0000197781.20524.b9. [DOI] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Sunayama J, Tsuruta F, Masuyama N, Gotoh Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J Cell Biol. 2005;170:295–304. doi: 10.1083/jcb.200409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Tuncman G, Hirosumi J, Solinas G, Chang L, Karin M, Hotamisligil GS. Functional in vivo interactions between JNK1 and JNK2 isoforms in obesity and insulin resistance. Proc Natl Acad Sci USA. 2006;103:10741–10746. doi: 10.1073/pnas.0603509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt PK, Jiang H, Aoki M. Triple layer control: phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle. 2005;4:908–913. doi: 10.4161/cc.4.7.1796. [DOI] [PubMed] [Google Scholar]
- Wolff B, Sanglier JJ, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- Yonezawa T, Haga S, Kobayashi Y, Katoh K, Obara Y. Saturated fatty acids stimulate and insulin suppresses BMCP1 expression in bovine mammary epithelial cells. Biochem Biophys Res Commun. 2009;390:915–919. doi: 10.1016/j.bbrc.2009.10.077. [DOI] [PubMed] [Google Scholar]
- Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Kotha J, Jennings LK, Zhang XA. Tetraspanins and vascular functions. Cardiovasc Res. 2009;83:7–15. doi: 10.1093/cvr/cvp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, et al. Hit selection with false discovery rate control in genome-scale RNAi screens. Nucleic Acids Res. 2008;36:4667–4679. doi: 10.1093/nar/gkn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M, et al. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.