Abstract
The surface marker PROM1 is considered one of the most important markers of tumor-initiating cells, and its expression is believed to be an adverse prognostic factor in gliomas and in other malignancies. To date, to our knowledge, no specific studies of its expression in medulloblastoma series have been performed. The aims of our study were to evaluate the expression profile of the PROM1 gene in medulloblastoma and to assess its possible role as a prognostic factor. The PROM1 gene expression was evaluated by quantitative– polymerase chain reaction on 45 medulloblastoma samples by using specific dye-labeled probe systems. A significantly higher expression of PROM1 was found both in patients with poorer prognosis (P= .007) and in those with metastasis (P= .03). Kaplan–Meier analysis showed that both overall survival (OS) and progression-free survival (PFS) were shorter in patients with higher PROM1 mRNA levels than in patients with lower expression, even when the desmoplastic cases were excluded (P= .0004 and P= .002, for OS and PFS for all cases, respectively; P= .002 and P= .008 for OS and PFS for nondesmoplastic cases, respectively). Cox regression model demonstrated that PROM1 expression is an independent prognostic factor (hazard ratio, 4.56; P= .008). The result was validated on an independent cohort of 42 cases by microarray-based analysis (P= .019). This work suggests that high mRNA levels of PROM1 are associated with poor outcome in pediatric medulloblastoma. Furthermore, high PROM1 expression levels seem to increase the likelihood of metastases. Such results need to be confirmed in larger prospective series to possibly incorporate PROM1 gene expression into risk classification systems to be used in the clinical setting.
Keywords: cancer stem cells, medulloblastoma, PROM1, q-PCR
Medulloblastoma (MBL) is an embryonal tumor arising in the cerebellum, and is the most common brain malignancy in childhood.1 The current World Health Organization (WHO) classification distinguishes 4 main variants of MBL: classic, large cell/anaplastic (lumped into a “non desmoplastic group”), desmoplastic/nodular, and with extensive nodularity (lumped into a “desmoplastic” group).2 Because of morphologic and clinical factors, this classification has recently been confirmed on the basis of the histogenesis,3 suggesting the existence of stem or progenitor cell populations, as assumed in the first description of this neoplasm.4 Indeed, it has been possible to identify a cell subpopulation from MBL that exhibits cancer stem cell (CSC) properties.5–7 Recently, we succeeded both in obtaining a new permanent cell line with stem cell–like features from an anaplastic medulloblastoma and in isolating neurospheres from classic MBLs.8 CSCs possess properties that may confirm a latent stem cell program of 1 small subpopulation of cells in the tumor mass, also called tumor-initiating cells (TICs), that may be the cause of the aggressiveness of the tumors.9,10 One of the features that identifies brain TICs is the expression of the PROM1 (CD133) gene encoding a 5 transmembrane domain protein,11,12 which is considered one of the most important markers in both normal and tumoral neuronal progenitor cells.5,10,13 Thus, in animal models and in cell culture studies, CD133-bearing TICs demonstrate the ability to initiate and drive tumor progression,3,5 displaying strong tumor resistance to chemotherapy (CT) and/or radiotherapy (RT).14–16 Although the correlation of PROM1 with patient survival has already been demonstrated in various human tumors, including gliomas,17 to date, no specific studies of its expression as related to patient outcome in medulloblastoma have been performed. Thus, detection of CD133 should contribute to better stratification of patients with MBL.10 Our study aimed to evaluate the expression profiles of the PROM1 gene in MBL with use of quantitative–reverse-transcription (qRT)–polymerase chain reaction (PCR) and to determine its possible prognostic significance.
Methods
Tumor Specimens and Patients
Forty-five patients who underwent surgery at the Neurosurgery Unit of the Giannina Gaslini Children's Hospital (Genoa, Italy) during the period 1991–2007 were enrolled in this study. The only inclusion criteria were the availability of their complete clinical data and the availability of fresh tissue specimens. Each sample was required to have a tumor cell content of at least 80%. Histopathological findings for the selected cases were reviewed in accordance with the latest WHO classification.2 Thirty-five tumors (77.8%) fulfilled the criteria for classic MBL, 6 tumors (13.3%) had the desmoplastic variant, and 4 (8.9%) had the large-cell/anaplastic variant. Only tumors with severe, widespread anaplasia were classified as anaplastic. Nuclear expression of INI1 was retained in all cases. Immediately after surgical resection, tissue specimens surplus to diagnostic needs were snap-frozen into 2-mL cryogenic vials (Nalge Nunc International), stored in liquid nitrogen, and collected in the Neuro-Oncology Bio-Bank of the Giannina Gaslini Childrens' Hospital. The selected cases were described in a study published elsewhere.18 Cases were staged on the basis of current national cooperative protocols, and patients were treated at a single institution. Four children (8.9%) were ≤3 years of age at the time of surgery and were treated with up-front CT using 2 main protocols (UK920419 and AIEOP SNC 950120); to avoid or delay irradiation, one of them (whose condition was diagnosed after 1997) also received a more intensive protocol (high-dose chemotherapy [HDCT]). Forty-one children (91.1%) were >3 years of age and were treated with the Packer regimen.21 After 1997, patients aged >3 years who were considered at high risk (12 patients [26.7%]) were treated with an institutional protocol using HDCT followed by craniospinal irradiation.22 To categorize the selected cases, postsurgical treatment ranks were pooled in 3 groups: the NOR group, CT or RT only; HD, HDCT plus RT; and SD, standard dose CT plus RT.
Metastasis stage (M stage) distribution, as defined by Chang et al,23 was performed, but owing to the small number of cases, analysis was limited to 2 stage groups: 29 patients (64.4%) were classified as M0, 15 patients (33.4%) were classified as M+, and the remaining patient (2.2%) was not evaluable. The mean age at the time of surgery of our cohort was 85.2 months (range, 19.4–156 months). Twenty-two patients (48.9%) died, and the 23 survivors (51.1%) had a median duration follow-up of 124.4 months (range, 41.9–219 months) as of the time of this report. Recurrent disease was diagnosed in the presence of positive MRI. The clinical-pathological features of the 45 patients are summarized in Table 1. Written informed consent was obtained from all the patients’ parents or guardians, and the local ethics committee for human studies approved the research.
Table 1.
Relationship between the clinical-pathologic characteristics and PROM-1 expression levels of the 45 cases
| Characteristic |
No. (%) of patients | PROM1 expression | P | |
|---|---|---|---|---|
| Age, months | ||||
| <36 | 4 (8.9) | 4.39 (0.77–18.4) | .47 | |
| >36 | 41 (91.1) | 1.84 (0.06–257) | ||
| Sex | ||||
| Male | 30 (66.7) | 1.67 (0.06–257) | .74 | |
| Female | 15 (33.3) | 1.84 (0.26–38.3) | ||
| Period of diagnosis | ||||
| 1991–1996 | 18 (40) | 1.97 (0.16–257) | .74 | |
| 1997–2007 | 27 (60) | 1.45 (0.06–38.3) | ||
| Histological variant | ||||
| Classic | 35 (77.8) | 1.84 (0.06–257) | .05a | |
| LC/A | 4 (8.9) | 5.94 (2.52–23.8) | ||
| Desmoplastic | 6 (13.3) | 0.95 (0.26–3.99) | ||
| Nondesmoplastic (Classic + LC/A) | 39 (86.7) | 1.97 (0.16–257) | .09 | |
| Desmoplastic | 6 (13.3) | 0.95 (0.26–3.99) | ||
| M stage | ||||
| M0 | 29 (64.4) | 1.13 (0.06–38.3) | .03 | |
| M+ | 15 (33.4) | 2.56 (0.64–257) | ||
| NV | 1 (2.2) | |||
| Surgical resection | ||||
| Complete (<1.5 cm2 residual tumor) | 36 (80) | 2.04 (0.14–257) | .02 | |
| Partial (>1.5 cm2 residual tumor) | 9 (20) | 0.87 (0.06–2.56) | ||
| Relapse | ||||
| Yes | 23 (51.2) | 2.11 (0.06–257) | .10 | |
| No | 21 (46.6) | 1.07 (0.11–29.9) | ||
| NV | 1 (2.2) | |||
| Treatment | ||||
| CT or RT only | NOR | 7 (15.5) | 5.86 (0.77–23.8) | .15a |
| HDCT + RT | HD | 14 (31.2) | 1.09 (0.06–38.3) | |
| Standard CT + RT | SD | 24 (53.3) | 1.53 (0.16–257) | |
| Outcome | ||||
| Alive | 23 (51.1) | 1.05 (0.06–29.9) | .007 | |
| Died | 22 (48.9) | 3.46 (0.16–257) | ||
P values refer to Mann–Whitney U-test, unless otherwise specified. NV denotes that data not available; M0 denotes the absence of metastasis; M+ denotes the presence of metastasis; CT denotes chemotherapy; RT denotes radiotherapy; and HDCT denotes high-dose chemotherapy.
aKruskal–Wallis test
RNA Isolation
Total RNA was extracted using 10-μm frozen sections using Trizol reagent (Invitrogen Life Technologies), following the standard procedure. RNA was quantified using Nanodrop spectrophotometric analysis (Celbio), and its integrity was assessed qualitatively on the Agilent 2100 Bioanalyzer (Agilent). Double-stranded cDNA synthesis was performed using a Two-Step cDNA Synthesis kit (Invitrogen Life Technologies) with Oligo(dT)20priming.
qRT-PCR Analysis and Assay Conditions
The human PROM1 gene expression (NM_006017) was tested by real-time PCR using a specific double-labeled fluorescent probe (ABI PRISM 7500 HT Sequence Detection System; Applied Biosystems). Beta actin (ACTB; NM_001101), Pyruvate kinase (PMK2; NM_002654), and Beta-2-microglobulin (B2M; NM_004048) were used as the endogenous control genes for each tumor specimen, and 3 samples of normal cerebellum were used as tissue references.
To obtain the highest amplification efficiency of the systems, primers and “dual-labeled” probes (Table 2) were designed using Primer Express (PE Applied-Biosystem), Oligo 4.1 (National-), and PrimerPy v0.97 (a GUI utility for quantitative-polymerase chain reaction (q-PCR) primer design software) to assess the best thermodynamically performing sequences. Primers were selected to hybridize on different exons, and the absence of region-containing single nucleotide polymorphisms was tested using the e!Ensembl website (http://www.ensembl.org). In addition, the folding of the amplified sequences and their flanking regions were checked using the DNA mfold suite on the M. Zuker web site,24 according to the thermodynamic parameters established by J. Santalucia.25 Finally, the specificity of primers and probes was tested using NCBI Blast, GeneWorks 2.5.1 (Oxford Molecular Group) and MacVector (MacVector) software suites. TIBMolbiol (Genoa, Italy) performed the synthesis.
Table 2.
Primers and probes used and calibration curve of the systems
| Primers and Probes 5′-3′ |
|||||||
|---|---|---|---|---|---|---|---|
| Gene | Slope | R2 | E = 10 [−1/slope] | Forward | Reverse | Probe (FAM/BHQ1) | Amplicons (bp) |
| PROM1 | −3,53 | 0,99 | 1.92 | ATTGGCATCTTCTATGGTTT | GCCTTGTCCTTGGTAGTGT | CAAATCACCAGGTAAGAACCCGG | 167 |
| ACTB | −3,489 | 0,994 | 1.93 | CAACTGGGACGACATGG | CTGGGGTGTTGAAGGTC | CTGGCACCACACCTTCTACAATGA | 161 |
| PMK2 | −3,43 | 0,991 | 1.95 | AGAGAAGGGAAAGAACATCAA | GCACCGTCCAATCATCAT | CAGCAAAATCGAGAATCATGAGGG | 139 |
| B2M | −3,47 | 0,996 | 1.94 | TCACAGCCCAAGATAGTTAAG | GAGGTTTGAAGATGCCG | TGCTGCTTACATGTCTCGATCCC | 67 |
Efficiencies values of each system. Efficiencies were calculated as E = 10 [−1/slope], and all differences were <0.1, indicating that the data could be compared. Amplifications were carried out on 25 μL using 20 ng of cDNA using Platinum Quantitative PCR SuperMix-UDG Master Mix (Invitrogen). Polymerase chain reaction was performed according to the following conditions: primers at 300 nM, probes at 200 nM, and ROX reference dye at 100 mM. The cycling conditions included 2 min of degradation of pre-amplified templates at 50°C, followed by 2 min of denaturation at 95°C, 40 cycles of denaturation at 95°C for 20 s, and annealing/extension at 54°C for 60 s.
Validation of each system was performed using standard curves on cDNA derived from the 1603-MED medulloblastoma cell line.8 The q-PCR efficiencies were calculated using the following equation: E = 10 [−1/slope]. Data were considered comparable when the difference between the efficiencies was <0.1.26 The normalized fluorescent signal (ΔRn) was automatically calculated by an algorithm that normalizes the reporter emission signal. The threshold value applied to the algorithm generating the threshold cycle (Ct) was set at 0.05 in all experiments. These assays did not generate any fluorescent signals when genomic DNA was used as a template, confirming that the assays only measured mRNA expression. The relative quantification of PROM1 transcript for each sample was performed according to the comparative method (2−ΔΔCt; Applied Biosystems User Bulletin no. 2P/N 4303859),27,28 using 3 endogenous control genes as the normalizer (Ctref) based on the geometric mean, as suggested by geNorm algorithm, established by Jo Vandesompele et al. (http://medgen.ugent.be/~jvdesomp/genorm/).29 An average of 3 values obtained from normal pediatric cerebellar tissues (9.53 ± 0.031) was used as tissue control (ΔCtref).
The minimum information for publication of quantitative real-time PCR experiments (MIQE) are provided elsewhere.30
Valition of the Prognostic Role of PROM1
In order to validate the prognostic role of PROM1 expression 42 medulloblastoma samples were used. Such additional series of cases were homogenously selected on the basis of similar clinical features to those analyzed by qPCR. The PROM1 expression profile was evaluated by micro-array-based data-analysis as reported elsewhere.31 Patients with medulloblastoma were dichotomized on the basis of expression of PROM1; overall survival (OS) curves were drawn up, and the best P value from the sequence was used to represent the final PROM1 expression cutoff.
Statistical Analysis
Descriptive analyses were performed reporting absolute frequencies and percentages for qualitative variables, whereas medians, 25th and 75th percentiles, and ranges were used for quantitative variables. Comparisons of the quantitative data of PROM1 expression between 2 groups of patients were performed using the Mann–Whitney U-test, because the normality and homoscedasticity assumptions were not fulfilled. Comparisons of >2 groups were performed by nonparametric analysis of variance (ie, the Kruskal–Wallis test).
The quantitative variable PROM1 expression was dichotomized according to the best cutoff value obtained by the receiver operating characteristics (ROC) analysis using the life status (dead vs alive) or the event of relapse (presence vs absence of relapse) as the outcome variable, respectively.32
Progression-free survival (PFS) and OS curves were calculated for all patients (n = 45) as well as for patients with nondesmoplastic variants (n = 39) on the basis of high or low levels of PROM1 gene expression. All curves were compared using the log-rank test.
Finally, a Cox regression model was fitted to evaluate the role of some variables in influencing the negative outcome. Variables that reached statistical significance in the univariate analysis, as well as clinically important variables, were included into the model. A step-down strategy was used for modelling, and the likelihood ratio test was used for comparisons. Hazard ratios (HR) and 95% confidence intervals (95% CIs) were calculated and reported.
All statistical tests were 2-sided, and P values <.05 were considered to be statistically significant. The Statistica software package, release 8.0) (StatSoft), was used for descriptive and bivariate analyses, and Stata software, version 7 (Stata), was used for survival and multivariate analysis.
Results
PROM1 Expression in Pediatric Medulloblastoma
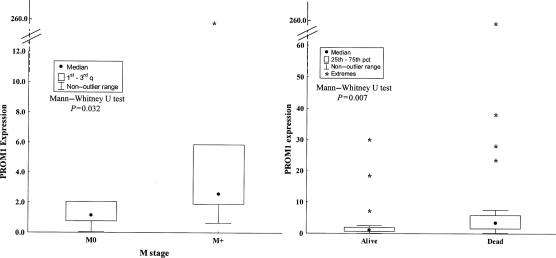
The reproducibility of the calibration curve of each system was analyzed and the q-PCR efficiencies of the all systems displayed slope differences <0.1, indicating that the data could be compared (Table 2). To determine the most stable reference genes, we also performed a preliminary analysis using GeNorm. Indeed, the Ct values were transformed to quantities by using the comparative Ct method, and the relative expression level for each reference gene was calculated on all MBL samples. PMK2 and B2M genes showed the same value of ratio between their lowest and highest values; however, ACTB had a different ratio (a 5-fold increase), indicating less gene stability. On the basis of the analysis, we excluded ATCB as a reference gene for this tissue, and the following PROM1 expression level was calculated for each tumor sample by using 2 endogenous control genes as normalizers (PMK2 and B2M). The median level of gene expression for PROM1 was 1.84-fold greater than that of normal cerebellar tissue and ranged from 0.06 to 257. Desmoplastic tumors (6 of 45) showed slightly lower levels of PROM1 transcript (median , 0.95; range, 0.26–3.99) than did nondesmoplastic tumors (39 of 45; median, 1.97; range, 0.06–257; P= .09) (Table 1). On the basis of the outcome of the whole group (desmoplastic plus nondesmoplastic cases), significantly higher levels of PROM1 were observed in patients with poorer prognosis: 22 (48.9%) of 45 died of disease, and 23 (51.1%) were alive at the end of the study (median level of PROM1 expression, 3.46 [range, 0.15–257] vs 1.05 [range, 0.06–29.9]; P= .007) (Table 1 and Fig. 1). PROM1 expression was higher in patients who underwent complete surgical resection (P= .02). PROM1 expression levels were significantly higher in the 15 M+ patients (median, 2.56; range, 0.64–257), compared with the 29 M0 patients (median, 1.13; range, 0.06–38.3; P= .03) (Table 1 and Fig. 1). The gene expression of PROM1 was not statistically different in the 3 groups of treatment (P= .15) (Table 1). None of the other clinical-pathologic features, such as age at surgery, sex, or recurrence, had any statistically significant relationship with PROM1 expression (Table 1).
Fig. 1.
PROM1 expression in pediatric medulloblastoma. The boxplots show the median of values: M+ patients have an increased level of PROM1 gene expression (P= .032, by Mann–Whitney test), and the survivors express significantly lower levels than did those who did not survive (P= .007, by Mann–Whitney test).
Association of PROM1 Expression with Patient Outcome
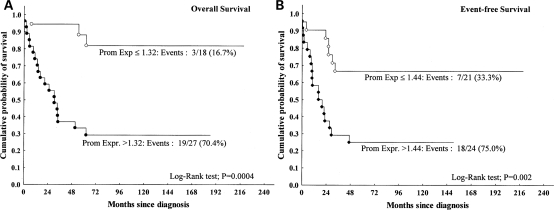
To perform survival analysis, the quantitative variable “PROM1 expression” was dichotomized by the ROC. The cutoff value between children who died of disease and those who survived was 1.32 (n = 45); it was 1.97 when patients with nondesmoplastic tumors alone were included in the analysis (39/45). The discriminating value between children who had relapse and those who did not was 1.44 and did not change even when nondesmoplastic cases alone were analyzed. Kaplan–Meier curves of estimated OS revealed a significantly shorter duration of survival for patients with PROM1 expression levels >1.32 (P= .0004) (Fig. 2A). PROM1 expression levels ≤1.32 were observed in 18 (40%) of 45 patients, 3 of whom died (16.7%). On the contrary, PROM1 expression levels >1.32 were observed in 27 (60%) of 45 patients, 19 (70.4%) of whom died. Kaplan–Meier curves of estimated PFS revealed a significantly shorter PFS among patients with PROM1 expression levels >1.44 than in those with PROM1 expression levels ≤1.44 (P= .002) (Fig. 2B).
Fig. 2.
Kaplan–Meier analysis revealing that (A) overall survival (OS) is shorter in patients with PROM1 mRNA levels 1.32-fold those of normal cerebellar tissue (all cases: No = 45, P = .0004) and (B) showing a significant relationship between PFS and PROM1 mRNA levels (all patients: No = 45, P = .002). The cutoff values were obtained by using the receiver operating characteristic (ROC) analysis: the quantitative variable PROM1 expression was dichotomized using the life status (dead vs alive) or the event of relapse (presence or absence of relapse) as the outcome variable for OS and progression-free survival (PFS), respectively.
Because patients with the desmoplastic variant are known to have better survival rates, we decided to analyze the OS and PFS of patients with nondesmoplastic tumors (39 of 45 patients) alone. OS and PFS, as calculated by the Kaplan–Meier curve, revealed that nondesmoplastic variants with high levels of PROM1 expression were significantly associated with poor prognosis (OS, P = .002; PFS, P = .008) (data not shown).
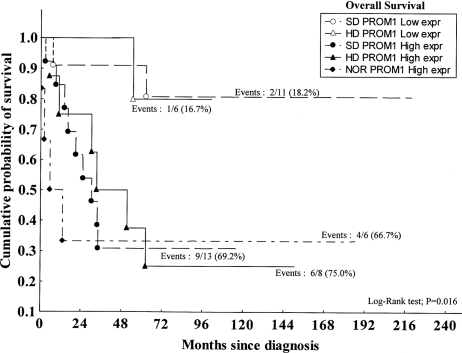
Furthermore, we analyzed the OS curve in the 3 treatment groups—that is, the NOR (for CT or RT only), HD (for HDCT plus RT), and SD (for standard dose CT plus RT) groups. The log-rank test revealed a significantly shorter duration survival among patients with high-level PROM1 expression than in patients with lower-level expression, independent of the treatment received (P= .016) (Fig. 3).
Fig. 3.
Kaplan–Meir survival plot showing prognostic significance of PROM1 expression pattern (≤1.32 [low expr] vs >1.32 [high expr]) and patient treatments (NOR, HD, and SD). To perform the statistical test (ie, the log-rank test), the group NOR/low PROM1 expression was excluded, because it was represent by only 1 patient. That patient is still alive after a follow-up of 13.6 years.
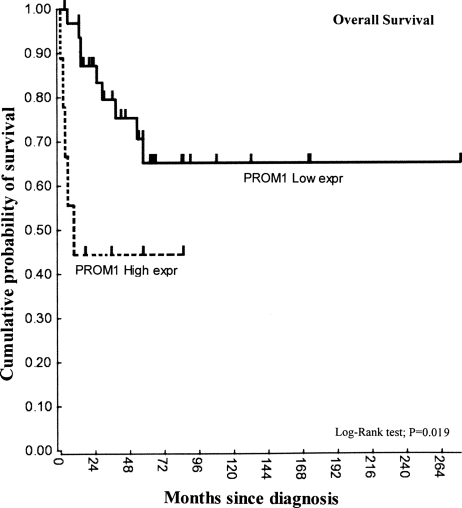
OS of an independent cohort of patients (n = 42), analyzed by microarray analysis, also confirmed that, in this cohort, high PROM1 expression levels were significantly associated with poor prognosis (P= .019) (Fig. 4).
Fig. 4.
Kaplan–Meir survival plot of independent series of cases (N = 42, P = .019). The duration of overall survival (OS) is shorter in patients with high PROM1 mRNA levels (n = 9). Medulloblastoma patients were sorted on the basis of expression of PROM1 and subsequently dichotomized on the basis of expression of PROM1. For each group separation, the log-rank significance was calculated. The best P value out of the sequence was used to represent the final PROM1 expression cutoff.
Prognostic Role of PROM1 Expression
In addition, we performed Cox regression model analysis to evaluate the possibly independent role played by the various prognostic factors, such as PROM1 expression level, M stage, extent of postoperative residual tumor, and treatment, on patient survival. As shown in Table 3, the analysis demonstrated that there were 2 independent prognostic factors statistically related to patient survival: PROM1 expression >1.32 (HR, 4.56; P= .008) and M+ status (HR, 3.20; P= .012). Postoperative residual tumor and treatment were not found to be statistically significant, but they were then forced into the model to obtain estimates of HRs adjusted for them. The role of PROM1 expression > 1.32 remained unmodified even when adjusted for the role of treatment and surgical intervention (HR, 4.93; P= .008). Moreover, the role of PROM1 expression > 1.32 (HR, 3.78; P= .02) and of M+ status (HR, 2.72; P= .028) did not change when only patients with nondesmoplastic variants were evaluated.
Table 3.
Multivariate Cox model (overall survival)
| Hazard Ratio | 95% CI | P | |
|---|---|---|---|
| Best-fitted Cox model (n = 44a) | |||
| PROM expr > 1.32 (reference: PROM expr ≤1.32) | 4.56 | 1.27–16.40 | .008 |
| M stage M+ (reference: M0) | 3.20 | 1.27–8.10 | .012 |
| Saturated Cox model (n= 44a) | |||
| PROM expr > 1.32 (reference: PROM expr ≤1.32) | 4.93 | 1.32–18.38 | .008 |
| M stage, M+ (reference: M0) | 3.91 | 1.38–11.04 | .007 |
| Treatment HD (reference: SD) | 0.59 | 0.20–1.73 | .51 |
| Treatment NOR (reference: SD) | 1.25 | 0.32–4.88 | |
| Surgical resection, partial (reference: complete) | 1.44 | 0.46–4.50 | .54 |
| Best-fitted Cox model (nondesmoplastic; n = 38a) | |||
| PROM expr >1.32 (reference: PROM expr ≤1.32) | 3.78 | 1.07–13.37 | .020 |
| M stage, M+ (reference: M0) | 2.72 | 1.09–6.79 | .028 |
The Cox model demonstrates that PROM1 expression is an independent prognostic factor with regard to treatments, postsurgical tumor residue, and M stage. CI denotes confidence interval; PROM expr, PROM1 expression level; HD, high-dose chemotherapy (HDCT) followed by craniospinal irradiation; SD, standard dose chemotherapy plus radiotherapy; and NOR, chemotherapy or radiotherapy only.
aFor 1 patient, data were not available.
Discussion
This work suggests that there is a significant relationship between high mRNA levels of PROM1 and the prognosis of pediatric MBLs, finding that PROM1 expression is a potential predictor of survival independent to clinical parameters.
Therapeutic approaches for MBL have made a great deal of progress over the past 3 decades, leading to an improvement in the survival of affected children. Nonetheless, the permanent neurological deficits related to the treatment still occur, and the mortality rate exceeds 30%.33 To choose the best treatment for patients with MBL, it is crucial to stratify them into risk groups. In brief, current staging is based on histological classification, age at diagnosis, extent of tumor resection, and presence or absence of metastases. Because patients who are stratified as “average risk” have better prognosis and an average PFS rate of 80%, they undergo less aggressive therapy protocols. On the contrary, “high-risk” patients whose PFS is no greater than 40%–70% undergo multimodal therapy, including more intensive CT and RT regimens.34–37 Unfortunately, however, some patients who are currently classified as “average risk” experience treatment failure and thus require moreaggressive protocols. On the contrary, some selected high-risk patients might avoid undergoing intensive approaches, which imply possible treatment-related toxicities for long-term survivors.34–41 In other words, based on new biological markers, a refinement of staging is needed to optimize treatment for MBL.42,43 The presence of a subpopulation of cells with stem-cell like features has been demonstrated and could provide us with a suitable explanation for the biological behavior of such a tumor.44 TICs, like normal stem cells and progenitor cells, express the PROM1 gene (CD133).9 One of the reports regarding the role of PROM1 expression in cancer showed its correlation with patient survival in gliomas.17 Nonetheless, although this antigen is considered the “molecule of the moment,”45 its role has stirred up considerable controversy. To simply evaluate PROM1 expression as a prognostic factor, we evaluated its transcript levels by q-PCR in a cohort of 45 pediatric patients with MBL; to our knowledge, this represents the first study of this issue. We found that high levels of PROM1 expression were associated with adverse prognosis, and furthermore, a significant correlation was found with M+ patients that may support the assumption that high mRNA levels of PROM1 are associated with poor outcome. To define a cutoff value that could dichotomize the range of quantitative variables of PROM1 expression, a ROC curve was calculated, taking biological factors into account as well. As a result, we found that patients with higher PROM1 mRNA levels had shorter OS and PFS durations than did patients with lower PROM1 expression, and importantly, this observation was also confirmed when the desmoplastic cases, which bear a better prognosis, were excluded. Moreover, the prognostic role of PROM1 was confirmed in an independent cohort of 42 cases showing that OS is shorter when patients with medulloblastoma express high levels of PROM1.
PROM1 expression would appear to be more predictive than standard clinical factors, such as M-status, which is acknowledged as an important adverse prognostic factor.46 Indeed, it is worth making an additional comment—that is, that high levels of PROM1 seem to increase the metastatic phenotype and, as a prognostic factor, appear to have a higher degree of independence by treatment than the M-stage. Thus, keeping the TIC hypothesis in mind, this result might be supported by a recent review, which supposes that such cells may play a key role in the metastatic process.47 Thus, our findings suggest that there is a significant relationship between PROM1 expression and the prognosis of pediatric MBLs. Our data are complementary to and expand on previous brain tumor studies.
It is well known that the mechanisms regulating tumorigenesis are “multifactorial,” so it is unlikely that any single biological factor will be sufficiently robust to optimally stratify patients with MBL. On the other hand, without having addressed the problems related to the CSC hypothesis, we believe that PROM1 may play a potential role in determining the patient's prognosis and may thus be helpful in making treatment decisions.
Clearly, our results need to be confirmed in larger prospective studies, as do the prognostic cutoff levels of PROM1 mRNA expression. We therefore recommend prospectively assessing PROM1 expression levels in ongoing MBL clinical trials to validate the role of this gene for its possible future incorporation into risk classification systems to be used in the clinical setting.
Funding
This work was supported by grants from the Associazione Italiana per la Ricerca dei Tumori Cerebrali del Bambino and by the Italian Association for Cancer Research (AIRC).
Acknowledgments
We thank Valerie Perricone for her editorial assistance. We also thank Camusso Raffaella and Tedeschi Luca (Photographic Service, Giannina Gaslini Children's Research Hospital, Genoa, Italy) for their graphic assistance.
Conflict of interest statement. None declared.
References
- 1.Kuhl J, Doz F, Taylor RE. Embryonic tumors. In: Walker DA, Perilongo G, Punt JAG, Taylor RE, editors. Brain, and Spinal Tumors of Childhood. London: Arnold; 2004. pp. 314–330. [Google Scholar]
- 2.Giangaspero F, Eberhart CG, Haapasalo H, Pietsch T, Wiestler OD, Ellison DW. Medulloblastoma. In: Louis DN, Ohgaki H, Wiestler D, Cavenee WK, editors. WHO Classification of Tumours of the Central Nervous System. Lyon: IARC; 2007. pp. 132–140. [Google Scholar]
- 3.Eberhart CG. In search of the medulloblast: neural stem cells and embryonal brain tumors. Neurosurg Clin N Am. 2007;18:59–69. doi: 10.1016/j.nec.2006.10.005. doi:10.1016/j.nec.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Bailey P, Cushing H. Medulloblastoma cerebelli: a common type of mid-cerebellar glioma of chidhood. Arch Neurol Psychiatry. 1925;14:192–224. [Google Scholar]
- 5.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. doi:10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 6.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. doi:10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Research. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. doi:10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 8.Raso A, Negri F, Gregorio A, et al. Successful isolation and long-term establishment of a cell line with stem cell-like features from an anaplastic medulloblastoma. Neuropathol Appl Neurobiol. 2008;34:306–315. doi: 10.1111/j.1365-2990.2007.00896.x. doi:10.1111/j.1365-2990.2007.00896.x. [DOI] [PubMed] [Google Scholar]
- 9.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. doi:10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 10.Neuzil J, Stantic M, Zobalova R, et al. Tumour-initiating cells vs. cancer ‘stem’ cells and CD133: what's in the name? Biochem Biophys Res Commun. 2007;355:855–859. doi: 10.1016/j.bbrc.2007.01.159. doi:10.1016/j.bbrc.2007.01.159. [DOI] [PubMed] [Google Scholar]
- 11.Miraglia S, Godfrey W, Yin AH, et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90:5013–5021. [PubMed] [Google Scholar]
- 12.Corbeil D, Röper K, Hellwig A, et al. The human AC133 hematopoietic stem cell antigen is also expressed in epithelial cells and targeted to plasma membrane protrusions. J Biol Chem. 2000;275:5512–5520. doi: 10.1074/jbc.275.8.5512. doi:10.1074/jbc.275.8.5512. [DOI] [PubMed] [Google Scholar]
- 13.Zhang QB, Ji XY, Huang Q, et al. Differentiation profile of brain tumor stem cells: a comparative study with neural stem cells. Cell Res. 2006;16:909–915. doi: 10.1038/sj.cr.7310104. doi:10.1038/sj.cr.7310104. [DOI] [PubMed] [Google Scholar]
- 14.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. doi:10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hambardzumyan D, Squarto M, Holland EC. Radiation resistance and stem-like cells in brain tumors. Cancer Cell. 2006;10:454–456. doi: 10.1016/j.ccr.2006.11.008. doi:10.1016/j.ccr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. doi:10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 17.Zeppernick F, Ahmadi R, Campos B, et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res. 2008;14:123–129. doi: 10.1158/1078-0432.CCR-07-0932. doi:10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- 18.Garrè ML, Cama A, Bagnasco F, et al. Medulloblastoma variants: age-dependent occurrence and relation to Gorlin syndrome–a new clinical perspective. Clin Cancer Res. 2009;15:2463–2471. doi: 10.1158/1078-0432.CCR-08-2023. doi:10.1158/1078-0432.CCR-08-2023. [DOI] [PubMed] [Google Scholar]
- 19.Grundy RG, Wilne SA, Weston CL, et al. Primary postoperative chemotherapy without radiotherapy for intracranial ependymoma in children: the UKCCSG/SIOP prospective study. Lancet Oncol. 2007;8:696–705. doi: 10.1016/S1470-2045(07)70208-5. doi:10.1016/S1470-2045(07)70208-5. [DOI] [PubMed] [Google Scholar]
- 20.Garre’ ML, Massimino M, Cefalo G, et al. High-risk malignant CNS tumours in infants: standard vs myeloablative chemotherapy: the experience of the Italian cooperative study for children <3 years of age. Neurooncol. 2003;1:40. [Google Scholar]
- 21.Packer RJ, Gajjar A, Vezina G, et al. Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J Clin Oncol. 2006;24:4202–4208. doi: 10.1200/JCO.2006.06.4980. doi:10.1200/JCO.2006.06.4980. [DOI] [PubMed] [Google Scholar]
- 22.Dallorso S, Dini G, Ladenstein R, et al. Evolving role of myeloablative chemotherapy in the treatment of childhood brain tumours. Bone Marrow Transplant. 2005;35:S31–S34. doi: 10.1038/sj.bmt.1704841. doi:10.1038/sj.bmt.1704841. [DOI] [PubMed] [Google Scholar]
- 23.Chang CH, Housepian EM, Herbert C. An operative staging system and a megavoltage radiotherapeutic technic for cerebellar medulloblastomas. Radiology. 1969;93:1351–1359. doi: 10.1148/93.6.1351. [DOI] [PubMed] [Google Scholar]
- 24.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. doi:10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. doi:10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:45. doi: 10.1093/nar/29.9.e45. doi:10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. doi:10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 28.Fiermonte G, Palmieri L, Todisco S, et al. Identification of the mitochondrial glutamate transporter: bacterial expression, reconstitution, functional characterization and tissue distribution of two human isoforms. J Biol Chem. 2002;277:19289–19294. doi: 10.1074/jbc.M201572200. doi:10.1074/jbc.M201572200. [DOI] [PubMed] [Google Scholar]
- 29.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;8:3. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. doi:10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 31.Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3(8):e3088. doi: 10.1371/journal.pone.0003088. doi:10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker SG. The central role of receiver operating characteristic (ROC) curves in evaluating tests for the early detection of cancer. J Natl Cancer Inst. 2003;95:511–515. doi: 10.1093/jnci/95.7.511. doi:10.1093/jnci/95.7.511. [DOI] [PubMed] [Google Scholar]
- 33.Gajjar A, Hernan R, Kocak M, et al. Clinical, histopathologic, and molecular markers of prognosis: toward a new disease risk stratification system for medulloblastoma. J Clin Oncol. 2004;22:984–993. doi: 10.1200/JCO.2004.06.032. doi:10.1200/JCO.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 34.Jozwiak J, Grajkowska W, Wlodarski P. Pathogenesis of medulloblastoma and current treatment outlook. Med Res Rev. 2007;27:869–890. doi: 10.1002/med.20088. doi:10.1002/med.20088. [DOI] [PubMed] [Google Scholar]
- 35.Rossi A, Caracciolo V, Russo G, et al. Medulloblastoma: from molecular pathology to therapy. Clin Cancer Res. 2008;14:971–976. doi: 10.1158/1078-0432.CCR-07-2072. doi:10.1158/1078-0432.CCR-07-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Packer RJ, MacDonald T, Vezina G. Central nervous system tumors. Pediatr Clin North Am. 2008;55:121–145. doi: 10.1016/j.pcl.2007.10.010. doi:10.1016/j.pcl.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Gandola L, Massimino M, Cefalo G, et al. Hyperfractionated accelerated radiotherapy in the Milan strategy for metastatic medulloblastoma. J Clin Oncol. 2009;27:566–571. doi: 10.1200/JCO.2008.18.4176. doi:10.1200/JCO.2008.18.4176. [DOI] [PubMed] [Google Scholar]
- 38.Packer RJ, Rood BR, Macdonald TJ. Medulloblastoma: present concepts of stratification into risk groups. Pediatr Neurosurg. 2003;39:60–67. doi: 10.1159/000071316. doi:10.1159/000071316. [DOI] [PubMed] [Google Scholar]
- 39.Gilbertson RJ. Medulloblastoma: signaling a change in treatment. Lancet Oncol. 2004;5:209–218. doi: 10.1016/S1470-2045(04)01424-X. doi:10.1016/S1470-2045(04)01424-X. [DOI] [PubMed] [Google Scholar]
- 40.Mulhern RK, Palmer SL, Merchant TE, et al. Neurocognitive consequences of risk-adapted therapy for childhood medulloblastoma. Clin Oncol. 2005;23:5511–5519. doi: 10.1200/JCO.2005.00.703. doi:10.1200/JCO.2005.00.703. [DOI] [PubMed] [Google Scholar]
- 41.Gottardo NG, Gajjar A. Current therapy for medulloblastoma. Curr Treat Options Neurol. 2006;8:319–334. doi: 10.1007/s11940-006-0022-x. doi:10.1007/s11940-006-0022-x. [DOI] [PubMed] [Google Scholar]
- 42.Rutkowski S, von Bueren A, von Hoff K, et al. Prognostic relevance of clinical and biological risk factors in childhood medulloblastoma: results of patients treated in the prospective multicenter trial HIT’91. Clin Cancer Res. 2007;13:2651–2657. doi: 10.1158/1078-0432.CCR-06-1779. doi:10.1158/1078-0432.CCR-06-1779. [DOI] [PubMed] [Google Scholar]
- 43.Lamont JM, McManamy CS, Pearson AD, et al. Combined histopathological and molecular cytogenetic stratification of medulloblastoma patients. Clin Cancer Res. 2004;10:5482–5493. doi: 10.1158/1078-0432.CCR-03-0721. doi:10.1158/1078-0432.CCR-03-0721. [DOI] [PubMed] [Google Scholar]
- 44.Fan X, Eberhart CG. Medulloblastoma stem cells. J Clin Oncol. 2008;26:2821–2827. doi: 10.1200/JCO.2007.15.2264. doi:10.1200/JCO.2007.15.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. doi:10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 46.Albright AL, Wisoff JH, Zeltzer PM, et al. Effects of medulloblastoma resections on outcome in children: a report from the Children's Cancer Group. Neurosurgery. 1996;38:265–271. doi: 10.1097/00006123-199602000-00007. doi:10.1097/00006123-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 47.Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J Cell Mol Med. 2008;12:374–390. doi: 10.1111/j.1582-4934.2007.00211.x. doi:10.1111/j.1582-4934.2007.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]