Abstract
Genetic methods of manipulating or eradicating disease vector populations have long been discussed as an attractive alternative to existing control measures because of their potential advantages in terms of effectiveness and species specificity1–3. The development of genetically engineered malaria-resistant mosquitoes has shown, as a proof-of-principle, the possibility of targeting the mosquito’s ability to serve as a disease vector4–7. The translation of these achievements into control measures requires an effective technology to spread a genetic modification from laboratory mosquitoes to field populations8. We have previously suggested that homing endonuclease genes (HEGs), a class of simple selfish genetic elements, could be exploited for this purpose9. Here we demonstrate that a synthetic genetic element, consisting of mosquito regulatory regions10 and the homing endonuclease gene I-SceI11–13, can substantially increase its transmission to the progeny in transgenic mosquitoes of the human malaria vector Anopheles gambiae. We show that the I-SceI element is able to rapidly invade receptive mosquito cage populations, validating mathematical models for the transmission dynamics of HEGs. Molecular analyses confirm that expression of I-SceI in the male germline induces high rates of site-specific chromosomal cleavage and gene conversion, which results in the gain of the I-SceI gene, and underlies the observed genetic drive. These findings demonstrate a new mechanism by which genetic control measures can be implemented. Our results also show in principle how sequence-specific genetic drive elements like HEGs could be used to take the step from the genetic engineering of individuals to the genetic engineering of populations.
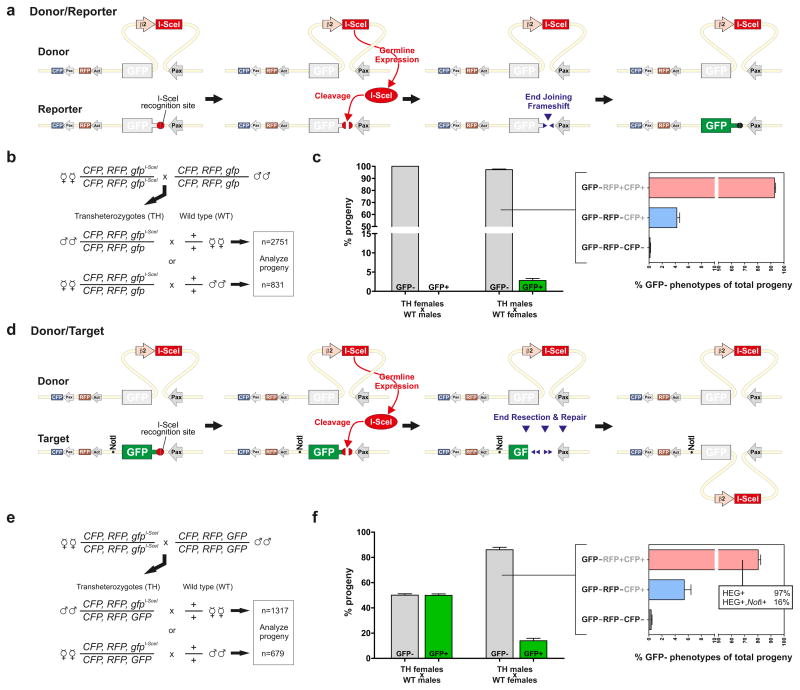
HEGs encode highly specific endonucleases with recognition sequences that typically occur only once per host genome, and have been identified in unicellular organisms in all three biological domains14. HEG-induced DNA double strand breaks (DSB) activate the recombinational repair system of the cell, which uses the homologous chromosome carrying the HEG as a template for repair. As a result the HEG is copied to the broken chromosome in a process referred to as ‘homing’. HEGs use this transmission distortion mechanism to spread through populations15. To investigate I-SceI activity in vivo we have developed an experimental system consisting of three distinct transgenic mosquito lines, the Donor, the Reporter and the Target, carrying either the I-SceI gene or its recognition site at identical positions on homologous chromosomes (Supplementary Fig. 1). For this purpose we used an A. gambiae docking line16 that allowed the site-specific integration of 3 different plasmids carrying the RFP (red fluorescent protein) transformation marker on chromosome 3R (Supplementary Fig. 2). The Donor line was generated using the construct pHome-D, containing a 3xP3-GFP (green fluorescent protein) transcription unit interrupted by a synthetic HEG element consisting of the I-SceI gene and the regulatory regions of the male testis-specific A. gambiae β2-tubulin gene10. The Reporter line was developed using the construct pHome-R, containing an I-SceI cleavage site that shifts out of frame the coding sequence of the GFP gene. The Reporter locus allows the scoring of I-SceI cleavage activity by monitoring the frequency of GFP+ individuals in which the GFP reading frame was restored via non-homologous end joining (NHEJ) in the progeny of Donor/Reporter trans-heterozygous (TH) males (Fig. 1a, b). Finally, the Target line was developed using pHome-T, containing the I-SceI cleavage site within the coding sequence of a functional GFP gene. This construct contains a diagnostic NotI recognition site that facilitates the molecular genotyping of homing events. The Target locus allows the assessment of I-SceI homing activity in the progeny of Donor/Target TH males crossed with wild type females by measuring the increase in the frequency over a 1:1 ratio of GFP− to GFP+ individuals arising from the insertion of the HEG gene into the GFP open reading frame (Fig. 1d, e).
Figure 1. Analysis of HEG activity in transgenic mosquitoes.
a, d Anticipated molecular events unfolding in Donor/Reporter (a) or Donor/Target (d) trans-heterozygous (TH) males. The Donor locus expresses I-SceI under the control of the male germline promoter β2-tubulin. The Reporter and the Target loci contain an I-SceI recognition site within the GFP gene. a, In Donor/Reporter TH males I-SceI activity is detected by scoring events that restore the GFP reading frame upon cleavage of the I-SceI recognition site. d, In Donor/Target TH males cleavage of the Target locus is followed by end resection and homing of the I-SceI gene from the homologous chromosome. This leads to the inactivation of the GFP reporter gene and can also lead to co-conversion of the NotI molecular marker (Pax; 3xP3 promoter, Act; Actin5C promoter). c, f Phenotypic analysis of progeny from crosses of Donor/Reporter (b, c) or Donor/Target (e, f) trans-heterozygote with wild-type (WT) mosquitoes. The column graphs show the percentage of GFP− and GFP+ individuals. The bar graphs on the right show, as a percentage of the total progeny, the GFP−RFP+CFP+, GFP−RFP−CFP+ and GFP−RFP−CFP− individuals observed. The inset (f, right panel) shows the molecular genotype of GFP−RFP+CFP+ individuals analyzed for the presence of the HEG and the NotI molecular markers.
When Donor/Reporter TH females were crossed to WT males all progeny showed the expected GFP− phenotype, as the β2-tubulin promoter regulating I-SceI is not active in females. By contrast 3% of the progeny from Donor/Reporter TH males and WT females showed a GFP+ phenotype (Fig. 1c). Sequencing of PCR products from the region around the I-SceI site showed that in 5 out of 20 GFP+ individuals the correct reading frame had been restored by NHEJ repair events. The remaining 15 GFP+ mosquitoes showed in place of the I-SceI site a sequence that resembled the region joining the 3xP3 promoter and the CFP gene, which lacks a unique restriction site present in the 3xP3-GFP cassette (Supplementary Fig. 2). We established from one such GFP+ individual the HEG-resistant Control strain, containing all three fluorescent marker genes but lacking the I-SceI site within the GFP sequence (Supplementary Fig. 1). The remaining 97% of the progeny from Donor/Reporter TH males and WT females were GFP− and the majority of these mosquitoes (93%) showed a GFP−RFP+CFP+ phenotype expected to arise either from an intact GFP− parental locus, NHEJ events that did not restore GFP expression or I-SceI homing events (Fig. 1c).
To test for the occurrence of homing we analysed the progeny of crosses between Donor/Target TH and WT mosquitoes (Fig. 1f). As expected, the ratio of GFP+:GFP− phenotypes in the offspring of Donor/Target TH females crossed to WT males was about 50:50. By contrast, in the reciprocal cross of TH males and WT females the ratio was 14:86. The excess of GFP− progeny, the majority of which were RFP+CFP+, could originate either from NHEJ events or as a result of homologous repair involving the HEG+ chromosome (i.e. homing). To investigate the molecular nature of GFP inactivation we performed a PCR analysis of the region spanning the GFP locus and encompassing the I-SceI gene or its recognition site. The results showed that 97% of GFP−RFP+CFP+ individuals contained the HEG cassette (Fig. 1f). The estimated cleavage rate for I-SceI was therefore about 95%, and the overall homing rate 56%. Importantly, the diagnostic NotI marker present only on the Target locus allowed the identification of recombinant GFP− HEG+ NotI+ chromosomes that were generated as a result of homing events (Supplementary Fig. 3). We were able to detect the NotI site, located ~0.7kb from the I-SceI cleavage site, in 16% of HEG+ chromosomes analyzed, indicating that this marker was retained in 45% of all homing events (and lost, due to co-conversion in the remaining 55%). In both sets of male TH to WT crosses about 4–5% of the progeny were GFP−RFP−CFP+, and a small number of larvae lacked all 3 visible markers (Fig. 1c, f). These phenotypes were not observed in crosses of TH females, suggesting that they were the result of I-SceI activity accompanied by deletions encompassing parts of the RFP gene or the entire locus. These experiments are summarized in Supplementary Table 1.
Another independent transgenic line, referred to as Ectopic Target, was generated by transposase-mediated integration of the pHome-T plasmid on chromosome 2 (Supplementary Fig. 1). When Donor/Ectopic Target TH males were crossed to WT females the frequency of the GFP− phenotype in the progeny was 88%, compared to approximately 50% in the female TH control cross (Supplementary Fig. 4). However none of 94 GFP−RFP+CFP− individuals, the phenotypic class expected to contain non-parental HEGs, carried the HEG sequence. This experiment suggests that in the absence of a repair template on the homologous chromosome I-SceI cleavage activity does not induce detectable homing. Finally we observed no significant deviation from a 1:1 ratio of GFP− and GFP+ progeny from crosses of TH mosquitoes in which the Donor locus was combined with the Control locus (data not shown).
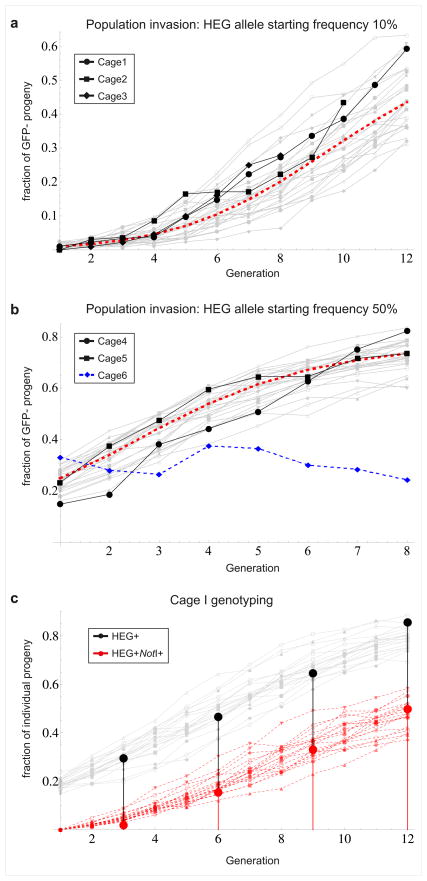
To test whether the observed transmission ratio distortion allows for efficient genetic drive of I-SceI in receptive A. gambiae populations we monitored its transmission dynamics in 5 cage populations of 600 individuals over 8 to 12 generations. Cage populations containing the I-SceI Target allele at initial frequencies of 90% or 50% were seeded with the I-SceI Donor allele at a frequency of 10% or 50%, respectively. GFP dominance results in an initial frequency of GFP− individuals of 1% or 25% in the two experimental conditions. In subsequent generations GFP+ individuals are expected to carry at least one allele of the original GFP+ target gene or a misrepaired GFP+ allele, whereas GFP− individuals contain two alleles in which GFP has been inactivated either by insertion of the HEG or NHEJ. At each generation a random sample of the progeny was visually analysed for the GFP marker at the larval stage. In all populations the frequency of GFP− individuals increased rapidly over time (Fig. 2). The frequency rose from about 1% to 60% in 12 generations (cage 1), and from about 1% to 40% in 10 generations (cage 2). In the two populations seeded with higher initial HEG frequencies GFP− individuals reached about 75–80% after 8 generations. By contrast the frequency of GFP− individuals did not change significantly in a population (cage 6) in which the HEG Donor line was used in combination with the non-receptive Control line (Fig. 2b) indicating that the absence of GFP expression in GFP−RFP+CFP+ mosquitoes did not result in a measurable fitness advantage over GPF+RFP+CFP+ mosquitoes. We generated deterministic and stochastic population genetic models using as parameters the experimentally derived rates of cleavage, homologous repair and NHEJ, assuming no fitness differences among genotypes (Supplementary Fig. 5a). The observed dynamics in the population cages fall well within the stochastic variation expected from the model (Fig. 2), indicating a quantitative match between the experimental data and our theoretical understanding of HEG transmission dynamics. If I-SceI had any effect on mosquito fitness, it was not large enough to significantly affect this concordance.
Figure 2. HEG invasion in mosquito cage populations.
a, b Temporal dynamics of GFP− mosquitoes in populations in which the HEG Donor allele was seeded at a frequency of 10% (a) or 50% (b) into a background of GFP+ mosquitoes carrying the HEG Target allele. The experimental points (Black) are overlaid onto predicted dynamics derived from a deterministic population genetic model (Dashed Red Line) and from 20 iterations of a stochastic model (Grey Lines). The dynamics of a cage population in which the HEG Donor and Control alleles were combined at a frequency of 50% is also shown (Dashed Blue Line). c, Molecular genotyping performed on individuals randomly collected from cage 1 at generations 3,6,9 and 12 using a set of PCR primers that specifically amplifies the HEG cassette (Primerset 1b, Supplementary Fig. 2). Presence of the NotI marker was determined by in vitro digestion of PCR products using NotI. The graph shows the fraction of mosquitoes carrying the HEG (Black) and the fraction carrying the HEG and the NotI marker on the same chromosome (Red) overlaid onto predictions from 20 stochastic simulations (Grey lines and Dashed Red Lines, respectively).
Detailed phenotypic and molecular analyses were carried out at different generations on individuals sampled from the mosquito population of cage 1. More than 90% of all GFP− mosquitoes were RFP+CFP+ for 12 generations (Supplementary Fig. 5b). To confirm that the rise of the GFP−RFP+CFP+ phenotype reflected a parallel increase in the HEG allele we performed a PCR assay on randomly chosen mosquitoes to determine the presence of the I-SceI gene. The frequency of individuals positive for the HEG cassette rose from about 19% to 86% by generation 12 (Fig. 2c). Moreover, NotI digests of the PCR products showed that the frequency of individuals with chromosomes carrying both the HEG and the NotI marker, a combination that was absent at the beginning of the experiment, increased to 50% by generation 9 (Fig. 2c). The dynamics of both HEG+ and NotI+ allele frequencies matched expectations from stochastic simulations (Fig. 2c). We conclude that the rise in the frequency of GFP− individuals reflected the corresponding increase in the frequency of the HEG allele. The increase in the frequency of the NotI marker in the Donor allele pool indicates that homing is the cause for the observed rise in the frequency of HEG+ individuals.
Our results demonstrate that homing can occur at appreciable frequencies in the germline of A. gambiae and therefore address a fundamental uncertainty that previously had been associated with proposals to use HEGs for pest control, namely whether HEGs would function in animals as they do in microbes. HEGs do not occur naturally in the nuclear genomes of metazoans; our results indicate that this absence is not because homing cannot occur, and instead supports alternative explanations such as that the segregated germline of animals prevents the horizontal transmission amongst species that these selfish genes need to persist over long evolutionary timespans17. The transmission dynamics of HEGs in cage populations provide the first evidence of the potential of these genetic elements to serve as synthetic gene drive systems in insect pests and add a promising candidate to those under development18–19.
The sequence-specific activity of HEGs could be exploited to develop vector control strategies aimed at either disrupting the mosquito genes that contribute to its vectorial capacity or introducing at selected chromosomal locations novel genes that impair the mosquito’s ability to function as vector for malaria9. Any use of HEGs in natural A. gambiae populations will depend on the ability to re-engineer their specificity20–23 towards native mosquito sequences. We identified in the A. gambiae genome within intergenic regions of the left (2L) and right arms (2R) of chromosome 2 two sequences that show similarities to the recognition sites of the two HEGs I-AniI and I-CreI that have previously been shown to be amenable to re-engineering to target novel human and plant sequences23–29. A previously described HEG engineering strategy was then used to generate an I-AniI variant to selectively cleave the 2L site, and a variant of monomerized I-CreI (termed mCre30) to selectively cleave the 2R site (Supplementary Fig. 6). The change in specificity of these enzymes demonstrates that HEGs can be designed to recognise novel mosquito sequences and opens the possibility to investigate the biology of HEGs in wild type mosquito populations. Though technical hurdles in HEG engineering technology must still be addressed to reach the flexibility required to target specific mosquito genes essential for viability or disease transmission, our results suggest how these genetic elements could overcome a major scientific roadblock in developing genetic control measures targeting species like the main vector of human malaria: the genetic manipulation of entire field populations starting from a few laboratory individuals.
Methods Summary The generation of transgenic lines and population cage experiments are described in Methods. To monitor homing mosquito larvae were subjected to fluorescent microscopy at the larval stage to detect the presence of the marker genes or subjected to molecular PCR to detect the presence of the HEG gene at the adult stage.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
We thank Michael Ashburner, Steve Russell, David Huen and Sang Chan from the Department of Genetics, University of Cambridge for comments, assistance and for plasmids. We thank Michele P. Calos, Department of Genetics, Stanford University for providing the pET11phiC31polyA plasmid. We thank Malcolm J. Fraser, Jr., University of Notre Dame for providing the pBSII-IFP2-orf plasmid. We thank Janet Meredith and Paul Eggleston from Keele University for providing the docking strain. We thank Ann Hall, Tony Nolan, Kalle Magnusson, David Rogers and Silke Fuchs for assistance. We thank S. Arshiya Quadri and Mindy Szeto for experimental support and the members of the Northwest Genome Engineering Consortium laboratories of David Baker, Ray Monnat, Andy Scharenberg and Barry Stoddard for their collective support of HEG engineering. Alden F.M. Hackmann provided graphics support. Funded by a grant from the Foundation for the National Institutes of Health through the Vector-Based Control of Transmission: Discovery Research (VCTR) program of the Grand Challenges in Global Health initiative and by NIH RL1 awards GM084433 to D.B. and CA133831 to R.J.M., Jr.
Footnotes
Author Contributions NW designed the experiments. NW, MM and PAP performed the experiments. NW and PAP generated the transgenic lines. MM maintained mosquito populations. NW analyzed the data. AB and NW generated the population dynamic models. AC and AB inspired the work and wrote the paper together with NW. HEG redesign and target site cleavage analyses were performed by ST, HL, UU and BH with guidance from DB and RJM, Jr. All authors read and approved the final manuscript.
Author Information The plasmids pHome-T and pHome-D have been deposited to Genbank under the accession numbers HQ159398 and HQ159399. The authors declare no competing financial interests. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Curtis CF. Possible use of translocations to fix desirable genes in insect pest populations. Nature. 1968;218:368–369. doi: 10.1038/218368a0. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton WD. Extraordinary sex ratios. A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science. 1967;156:477–488. doi: 10.1126/science.156.3774.477. [DOI] [PubMed] [Google Scholar]
- 3.Alphey L, et al. Malaria control with genetically manipulated insect vectors. Science. 2002;298:119–121. doi: 10.1126/science.1078278. [DOI] [PubMed] [Google Scholar]
- 4.Corby-Harris V, et al. Activation of Akt Signaling Reduces the Prevalence and Intensity of Malaria Parasite Infection and Lifespan in Anopheles stephensi Mosquitoes. PLoS Pathog. 2010;6:e1001003. doi: 10.1371/journal.ppat.1001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-Lorena M. Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature. 2002;417:452–455. doi: 10.1038/417452a417452a. [pii] [DOI] [PubMed] [Google Scholar]
- 6.Moreira LA, et al. Bee venom phospholipase inhibits malaria parasite development in transgenic mosquitoes. J Biol Chem. 2002;277:40839–40843. doi: 10.1074/jbc.M206647200M206647200. [pii] [DOI] [PubMed] [Google Scholar]
- 7.Li F, Patra KP, Vinetz JM. An anti-Chitinase malaria transmission-blocking single-chain antibody as an effector molecule for creating a Plasmodium falciparum-refractory mosquito. J Infect Dis. 2005;192:878–887. doi: 10.1086/432552. JID34510 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev Genet. 2006;7:427–435. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- 9.Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci. 2003;270:921–928. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catteruccia F, Benton JP, Crisanti A. An Anopheles transgenic sexing strain for vector control. Nat Biotechnol. 2005;23:1414–1417. doi: 10.1038/nbt1152. [DOI] [PubMed] [Google Scholar]
- 11.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 12.Bellaiche Y, Mogila V, Perrimon N. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics. 1999;152:1037–1044. doi: 10.1093/genetics/152.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Windbichler N, et al. Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos. Nucleic Acids Res. 2007;35:5922–5933. doi: 10.1093/nar/gkm632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 15.Goddard MR, Greig D, Burt A. Outcrossed sex allows a selfish gene to invade yeast populations. Proc Biol Sci. 2001;268:2537–2542. doi: 10.1098/rspb.2001.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meredith JM, et al. Site-Specific Integration and Expression of an Anti-Malarial Gene in Transgenic Anopheles gambiae Significantly Reduces Plasmodium Infections. PLoS One. 2011;6:e14587. doi: 10.1371/journal.pone.0014587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Chen CH, et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 19.McMeniman CJ, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323:141–144. doi: 10.1126/science.1165326. 323/5910/141 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Ashworth J, et al. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarjour J, et al. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009;37:6871–6880. doi: 10.1093/nar/gkp726. gkp726 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashworth J, et al. Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq283. gkq283 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thyme SB, et al. Exploitation of binding energy for catalysis and design. Nature. 2009;461:1300–1304. doi: 10.1038/nature08508. nature08508 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao H, et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010;61:176–187. doi: 10.1111/j.1365-313X.2009.04041.x. TPJ4041 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Grizot S, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. gkp548 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz IG, et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq801. gkq801 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redondo P, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. nature07343 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Arnould S, et al. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J Mol Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. S0022-2836(07)00579-7 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Rosen LE, et al. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. gkl645 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Pellenz S, Ulge U, Stoddard BL, Monnat RJ., Jr Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009;37:1650–1662. doi: 10.1093/nar/gkp004. gkp004 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.