Abstract
Purpose
A common genetic variant (rs3812718) in a splice donor consensus sequence within the neuronal sodium channel gene SCN1A (encoding NaV1.1) modulates the proportion of transcripts incorporating either the canonical (5A) or alternative (5N) exon 5. A pharmacogenetic association has been reported whereby increased expression of exon 5N containing NaV1.1 transcripts correlated with lower required doses of phenytoin in epileptics. We tested the hypothesis that SCN1A alternative splicing affects the pharmacology of NaV1.1 channels.
Methods
To directly examine biophysical and pharmacological differences between the exon 5 splice variants, we performed whole-cell patch clamp recording of tsA201 cells transiently co-expressing either NaV1.1-5A or NaV1.1-5N with the β1 and β2 accessory subunits. We examined tonic inhibition and use-dependent inhibition of NaV1.1 splice isoforms by phenytoin, carbamazepine, and lamotrigine. We also examined the effects of phenytoin and lamotrigine on channel biophysical properties and determined concentration-response relationships for both splice variants.
Key Findings
We observed no significant differences in voltage-dependence of activation, steady-state inactivation, and recovery from inactivation between splice variants. However, NaV1.1-5N channels exhibited enhanced tonic block by phenytoin and lamotrigine compared to NaV1.1-5A. Additionally, NaV1.1-5N exhibited enhanced use-dependent block by phenytoin and lamotrigine across a range of stimulation frequencies and concentrations. Phenytoin and lamotrigine induced shifts in steady-state inactivation and recovery from fast inactivation for both splice isoforms. No splice isoform differences were observed for channel inhibition by carbamazepine.
Significance
These results suggest NaV1.1 channels containing exon 5N are more sensitive to the commonly used antiepileptic drugs phenytoin and lamotrigine.
Keywords: antiepileptic drugs, ion channel gene defects, alternative splicing
Introduction
Voltage-gated sodium (NaV) channels are critically important for initiating and propagating action potentials in most excitable cells (Hille, 1993). Sodium channels exist as heteromultimeric complexes comprised of a large (~260 kDa) pore-forming α-subunit and smaller accessory β-subunits (Yu and Catterall, 2003, George, 2005). Nine genes (SCN1A, SCN2A, etc.) encode distinct sodium channel α-subunits (NaV1.1, NaV1.2, etc.) and four genes encode the accessory β-subunits (George, 2005, Isom, 2001). Mutations in NaV channel genes are associated with various genetic epilepsies and more than 500 mutations have been identified in SCN1A (encoding NaV1.1) (Claes et al., 2009, Lossin, 2009). The central role of NaV channels in neuronal excitability is further highlighted by the numerous clinically utilized antiepileptic drugs (AEDs) including phenytoin, carbamazepine, and lamotrigine, which exhibit anticonvulsant effects by inhibiting NaV channel activity (Ragsdale et al., 1991, Kuo and Lu, 1997, Kuo, 1998).
Sodium channel transcripts undergo alternative mRNA splicing. One splice variation in SCN1A leads to the incorporation of an alternate exon 5 encoding a portion of the domain 1 S3/S4 helices (Copley, 2004). A corresponding splicing event is conserved among other neuronal (NaV1.2, NaV1.3, NaV1.6 and NaV1.7) as well as the cardiac (NaV1.5) NaV channel isoforms (Copley, 2004, Diss et al., 2004, Onkal et al., 2008). The resultant splice isoforms differ by 1 to 6 amino acids within the voltage-sensing portion of domain 1 and functional studies have revealed splice isoform specific channel gating (Diss et al., 2004). Interestingly, the biophysical effects of disease causing NaV channel mutations can be modified by the splice isoform background, as demonstrated for NaV1.7 (Jarecki et al., 2009).
Previous pharmacogenetic studies have demonstrated that a common SCN1A genetic variant (rs3812718) located within an intron splice donor site alters the proportion of human brain SCN1A transcripts incorporating the canonical (exon 5A) or alternative (exon 5N) exon 5 (Tate et al., 2005). The 5N/5A terminology arose due to initial reports suggested that exon 5N was expressed primarily during the neonatal period, while the exon 5A was the adult exon (Sarao et al., 1991, Gustafson et al., 1993). However, strict developmental regulation of this splicing event may be relaxed as evidenced by the detection of NaV1.1 transcripts (up to 50% of transcripts) incorporating exon 5N in adult human brain (Tate et al., 2005, Heinzen et al., 2007). In population studies, the two alleles generated by rs3812718 (A or G) occur with approximately equal frequency producing genotype frequencies of 25% AA, 50% AG and 25% GG. Importantly, the A allele disrupts the consensus splice donor sequence immediately following exon 5N, thereby completely blocking Nova2-mediated incorporation of exon 5N into mature SCN1A mRNA transcripts. Heinzen et al. reported that inclusion of exon 5N ranged from 1% to 50% for the AA and GG haplotypes, respectively (Heinzen et al., 2007).
Genetic variation in SCN1A or other NaV channel genes might also influence susceptibility to or treatment responses in non-Mendelian forms of epilepsy. The aforementioned pharmacogenetic study demonstrated that epileptic patients expressing the GG genotype (expressing NaV1.1-5N) required lower doses of phenytoin and carbamazepine for treatment (Tate et al., 2006, Tate et al., 2005), but subsequent analyses indicated that this common variant was not associated with carbamazepine dosage levels in other populations, suggesting other genetic factors may contribute to carbamazepine sensitivity, and illustrating the variability associated with pharmacogenetic studies (Abe et al., 2008, Zimprich et al., 2008). Nonetheless, these findings prompted the hypothesis that NaV1.1 channels translated from transcripts containing exon 5N may respond differently to AEDs.
We investigated the biophysical and pharmacological properties of NaV1.1 splice variants containing the different exon 5 sequences. Although these NaV1.1 splice variants differ by three amino acids near the domain 1 voltage sensor, no significant biophysical differences were apparent. In addition, the two splice variants exhibited similar levels of inhibition by carbamazepine. However, NaV1.1-5N channels were more sensitive to phenytoin compared to NaV1.1-5A, which correlates with findings from previous pharmacogenetic studies. Further, NaV1.1-5N channels were also more sensitive to inhibition by lamotrigine, a commonly used AED that has not yet been investigated for pharmacogenetic association with SCN1A.
Materials and Methods
Mutagenesis and heterologous expression of NaV1.1
Mutagenesis of recombinant NaV1.1 was performed as described previously (Lossin et al., 2002, Rhodes et al., 2004, Kahlig et al., 2008). Three mutations (Y202F, D208N, and V212F) were introduced into full length NaV1.1 (NaV1.1-5A) to generate NaV1.1-5N. To minimize spontaneous mutagenesis of NaV1.1 cDNA in bacterial culture, recombinants were always propagated in Stbl2 cells (Invitrogen, Carlsbad, CA) at 30°C.
Heterologous expression of NaV1.1 splice variants was performed in tsA201 cells (HEK293 cell line expressing SV40 large T antigen). Cells were grown in Dulbecco modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 50 units/ml penicillin and 50 μg/ml streptomycin in a humidified 5% CO2 atmosphere at 37°C. Expression of NaV1.1 and the accessory β1 and β2 subunits was achieved using transient transfection with Qiagen Superfect reagent (2 μg total plasmid DNA was transfected with a cDNA ratio of 10:1:1 for α1:β1:β2 subunits). Human β1 and β2 cDNAs were cloned into plasmids also encoding the CD8 receptor (CD8-IRES-hβ1) or EGFP (EFGP-IRES-hβ2), respectively, as transfection markers (Lossin et al., 2002). Unless otherwise noted, all chemicals were from Sigma-Aldrich (St. Louis, MO).
Electrophysiology and data analysis
Whole-cell voltage-clamp recordings of tsA201 cells were performed as described previously (Lossin et al., 2002, Rhodes et al., 2004, Kahlig et al., 2008). Sodium channel currents were recorded at room temperature (20-22°C) 24 – 72 hours after transfection. Patch pipettes were fabricated from borosilicate glass (Warner Instruments) with a P-97 multistage Flaming-Brown micropipette puller (Sutter Instruments) and fire polished using a microforge (MF 830, Narishige, Japan). Final pipette resistance was between 1.0 and 2.0 MΩ. The pipette solution consisted of (in mM) 110 CsF, 10 NaF, 20 CsCl, 2 EGTA, 10 HEPES, with pH 7.35 and osmolarity 300 mOsmol/kg. Bath solution contained (in mM) 145 NaCl, 4 KCl, 1.8 CaCl2, 1 MgCl2, 10 HEPES with pH 7.35 and osmolarity 310 mOsmol/kg. Bath solution was continually exchanged by gravity driven superfusion. Cells were allowed to stabilize for 10 min after establishment of the whole-cell configuration before currents were measured. Cells with peak current less than −0.6 pA were excluded from analysis to avoid data contamination by endogenous currents. Cells with currents larger than −6 nA were excluded to ensure accurate voltage control. Whole-cell capacitance and access resistance were determined by integrating capacitive transients in response to a +10 mV voltage step from −120 mV filtered at 50 kHz. Series resistance was compensated 90%, with 70% prediction, to assure that the command potential was reached within microseconds with a voltage error <2 mV. Leak currents were subtracted by using an online P/4 procedure. All whole-cell currents were low-pass Bessel filtered at 5 kHz and digitized at 50 kHz.
Specific voltage-clamp protocols assessing channel activation and inactivation are depicted in figure insets. Whole-cell conductance was calculated from the peak current amplitude by GNa = INa / (V − ENa) and normalized to the maximum conductance between −80 and +20 mV. Conductance-voltage and steady state channel availability curves were fit with Boltzmann functions to determine the voltage for half-maximal channel activation or inactivation (V½) and slope factor (k). Time-dependent recovery from inactivation was evaluated by fitting peak current recovery with a two-exponential equation in the form of I/Imax = Af × [1 − exp(−t/τf)] + As × [1 − exp(−t/τs)], where τf and τs denote time constants (fast and slow components, respectively), Af and As represent the fast and slow fractional amplitudes.
Pharmacology
Phenytoin, carbamazepine, and lamotrigine stock solutions (50 mM) were prepared in DMSO. All drugs were diluted in bath solution to the indicated concentrations the day of the experiment, and pH was readjusted to pH 7.35. The concentration of DMSO did not exceed 0.6% and control experiments with 0.6% DMSO showed no change in current density, with very small changes in voltage-dependence of activation, steady-state inactivation, recovery from inactivation, and use dependent rundown at 10 Hz. DMSO induced a small (~1.5 mV) shift in V1/2 of steady state inactivation, a small (~2.5 ms) increase in τf of recovery from fast inactivation, and ~10% use-dependent block, but these effects were not different between the two splice variants. Cells were continuously superfused with either bath solution or solution containing drug using a single cell perfusion system (Perfusion Pencil system; AutoMate Scientific, Berkeley, CA). Cells were superfused with drug containing solution for 3 minutes prior to measuring the effect of drugs.
For concentration response experiments, cells were exposed to a sequence of incrementing drug concentrations ranging from 3 to 100 μM (for phenytoin) or 300 μM (for lamotrigine) for 3 minutes at each concentration. Currents were elicited with a 20 ms pulse to 0 mV from a holding potential of −90 mV at a frequency of 0.2 Hz. The final five traces recorded for each condition were averaged, then normalized to the drug free control condition. Approximately 7% current run-up was observed at 25 minutes compared to 10 minutes after achieving whole cell configuration (the end of the stabilization period) for both splice variants. Concentration-response curves were fit with the Hill equation: I/Imax = 1/[1 + 10ˆ(logIC50 − I) × k], where IC50 is the concentration that produces half inhibition and k is the Hillslope factor.
Pulse train experiments with varying pulse frequencies were performed sequentially under control conditions, in the presence of drug, then in the presence of 0.5 μM TTX to enable determination of TTX-sensitive current by offline digital subtraction.
Results
Biophysical characterization of NaV1.1 splice variants
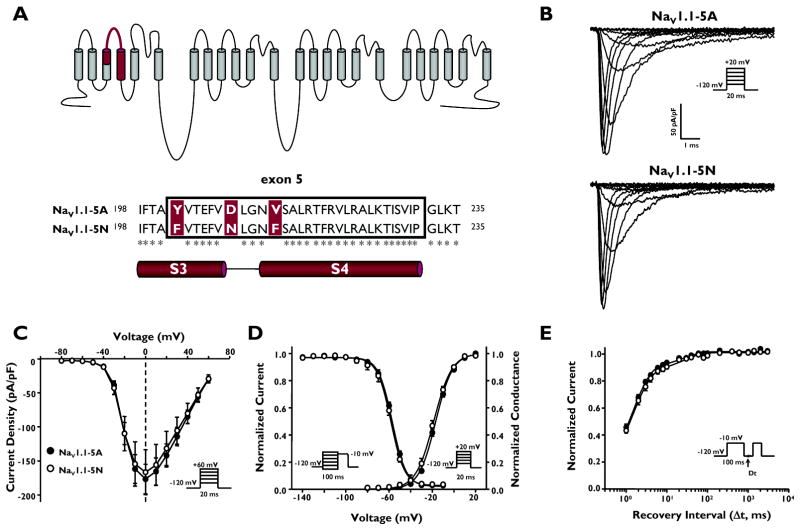
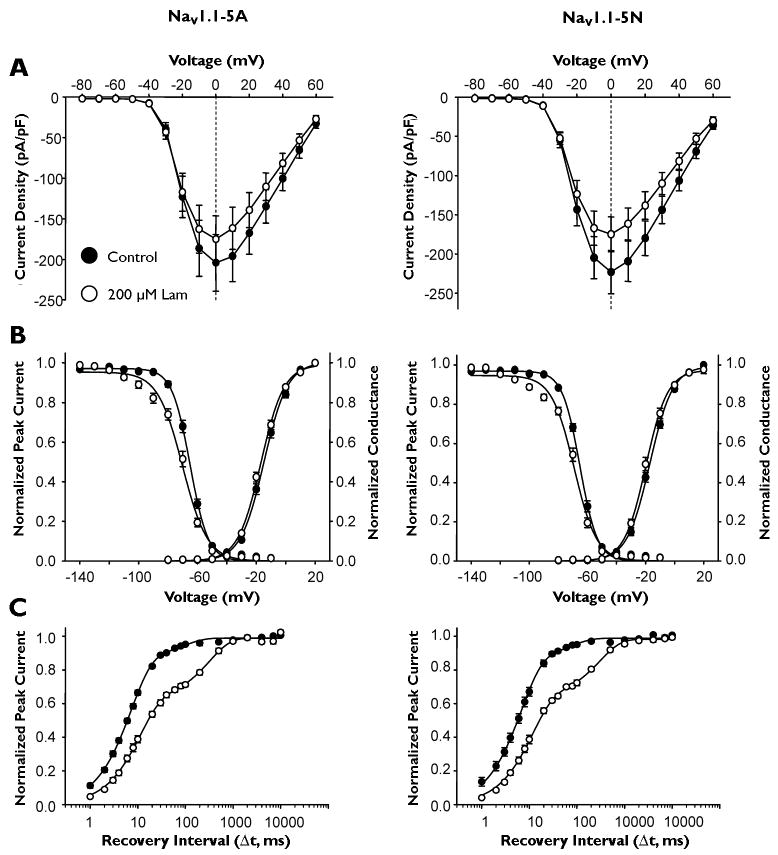
The two alternatively spliced SCN1A transcripts, incorporating either exon 5A or 5N, differ by three amino acid residues (5A to 5N: Y202F, D208N, and V212F) within a 30 amino acid region encoding the S3 and S4 segments of domain 1 (Fig. 1A). To examine the biophysical properties of the two NaV1.1 splice variants, we performed whole-cell recordings of human NaV1.1-5A or NaV1.1-5N co-expressed with human β1 and β2 in cultured human tsA201 cells. Figure 1B illustrates that both NaV1.1 splice variants express whole-cell sodium currents with similar peak current density and general gating behavior.
Figure 1.
Alternative splicing of SCN1A exon 5 results in two splice variants, NaV1.1-5A and NaV1.1-5N. (A) Predicted transmembrane topology of NaV.1.1 showing the location of the exon 5 coding region and amino acid alignment of NaV1.1-5A and NaV1.1-5N. Asterisks indicate identical amino acids. (B) Whole-cell sodium currents recorded from tsA201 cells co-expressing either NaV1.1-5A or NaV1.1-5N with both β1 and β2 accessory subunits. Channels were activated by voltage steps to between −80 and +60 mV from a holding potential of −120 mV. (C) Peak current density elicited by test pulses to various membrane potentials and normalized to cell capacitance. (D) Voltage dependence of channel activation measured between −80 to +20mV and voltage dependence of inactivation measured following a 100 ms inactivating prepulse to between −140 and −10 mV. (E) Time-dependent recovery from inactivation assessed with a 100 ms inactivating prepulse to −10 mV. Closed symbols represent NaV1.1-5A and open symbols represent NaV1.1-5N.
To determine if the splice isoforms exhibit divergent biophysical properties, we examined the voltage-dependence of activation and inactivation gating as well as recovery from inactivation. Current-voltage relationships for NaV1.1-5A and NaV1.1-5N were not significantly different with regard to current density and voltage at peak current (Fig. 1C). NaV1.1-5N channels exhibited a non-significant trend toward activation at more hyperpolarized membrane potentials (Fig. 1D, Supplementary Table S1). There was no difference in the voltage dependence of steady-state inactivation following a 100 ms depolarizing prepulse (Fig. 1D, Supplementary Table S1), and we observed no significant difference in the recovery from inactivation time course between the splice variants (Fig. 1E, Supplementary Table S1). These data demonstrated that although NaV1.1-5A and NaV1.1-5N differ by three amino-acid residues within a critical portion of a voltage-sensing domain, the biophysical properties of the two channel isoforms were indistinguishable.
Tonic and use-dependent block of NaV1.1-5A and NaV1.1-5N
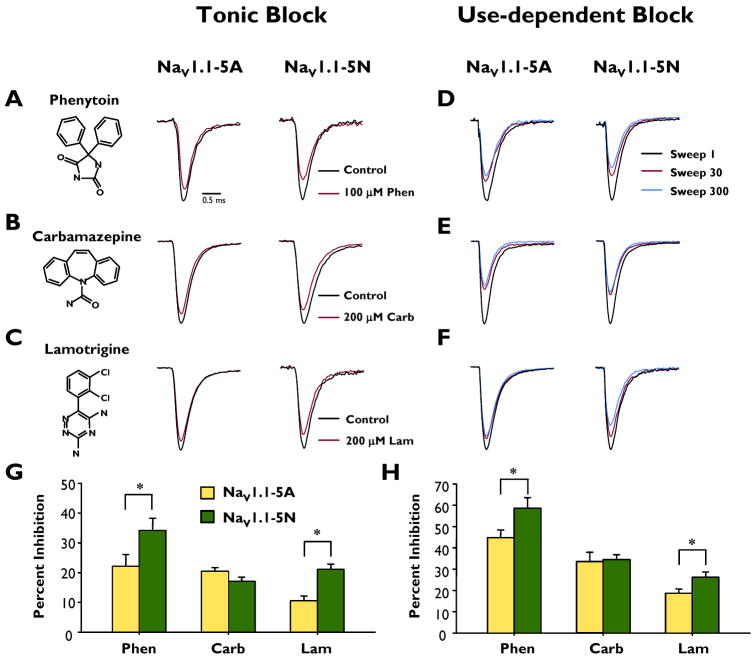
We next determined the ability of the antiepileptic drugs phenytoin, carbamazepine, and lamotrigine to exert tonic block of NaV1.1-5A and NaV1.1-5N. Tonic block of peak current was measured using a voltage-step to 0 mV from a holding potential of −90mV. Phenytoin (100 μM) modestly reduced the peak current generated by NaV1.1-5A (22.1 ± 3.9%, n = 9, Fig. 2A). By contrast, NaV1.1-5N channels were more sensitive to phenytoin block (34.2 ± 4.0%, n = 10) and the degree of inhibition was significantly greater as compared to NaV1.1-5A (p < 0.05). Carbamazepine (200 μM) blocked NaV1.1-5A and NaV1.1-5N to a similar degree (20.4 ± 1.2%, n = 8 and 17.0 ± 1.5%, n = 9, respectively, Fig.2B). Similar to phenytoin, lamotrigine (200 μM) exerted significantly greater tonic block of NaV1.1-5N compared to NaV1.1-5A (10.5 ± 1.7%, n = 13 and 21.1 ± 1.8%, n = 13, respectively; p < 0.05, Fig. 2C). Figure 2G illustrates that NaV1.1-5N is significantly more sensitive to tonic block by phenytoin and lamotrigine compared to NaV1.1-5A.
Figure 2.
Tonic block and use-dependent block of peak sodium current by phenytoin, carbamazepine and lamotrigine. Representative whole-cell recordings during superfusion with control (drug free, black trace) or drug (red trace) solutions: (A) phenytoin, (B) carbamazepine or (C) lamotrigine. Representative whole cell recordings showing the 1st (black trace), 30th (red trace) and 300th (blue) pulse illustrating use-dependent inhibition during superfusion of (D) phenytoin, (E) carbamazepine or (F) lamotrigine. Currents are normalized to peak current recorded in control solution. Average tonic block (G) and use-dependent block (H) of peak current of NaV1.1-5A (closed bars) and NaV1.1-5N (open bars) by phenytoin, carbamazepine and lamotrigine. An asterisk indicates p < 0.05. All points represent mean ± SEM for 9 - 14 experiments.
Antiepileptic drugs targeted to NaV channels exhibit use-dependent block during high frequency stimulation due to an accumulation of drug bound channels. We compared the use-dependent block of NaV1.1-5A and NaV1.1-5N by phenytoin, carbamazepine, and lamotrigine using a depolarizing pulse train (0 mV, 5 ms, 10 Hz, 300 steps) from a holding potential of −90mV. In drug-free control solution there was only a slight reduction in the peak current density over the course of the pulse train (pooled control data pulse 300 vs pulse 1: NaV1.1-5A, −4.3 ± 0.9%, n = 21, NaV1.1-5N, −4.9 ± 0.8%, n = 25). Phenytoin (100 μM) produced a use-dependent block of 44.7 ± 3.7% (n = 9) for NaV1.1-5A, which was significantly less than the block of 58.6 ± 4.9% for NaV1.1-5N (n = 10, p < 0.05, Fig. 2D). In contrast, carbamazepine (200 μM) induced use-dependent block of NaV1.1-5A and NaV1.1-5N to levels that were not significantly different (33.7 ± 4.3%, n = 8 and 34.6 ± 2.2%, n = 9, respectively; Fig. 2E). Lamotrigine (200 μM) caused significantly greater use-dependent block of NaV1.1-5N compared to NaV1.1-5A (18.7 ± 2.0%, n = 12, vs 26.2 ± 2.5%, n = 10, respectively; p < 0.05, Fig. 2F). Figure 2H illustrates that NaV1.1-5N is significantly more sensitive to use-dependent block by phenytoin and lamotrigine compared to NaV1.1-5A. In control experiments, vehicle (0.6% DMSO) treatment evoked modest effects on use-dependent rundown, but these effects were not different between the two splice variants, and therefore cannot explain the observed divergence in splice variant pharmacology.
Blocking efficacy of antiepileptic drugs for NaV1.1 splice variants
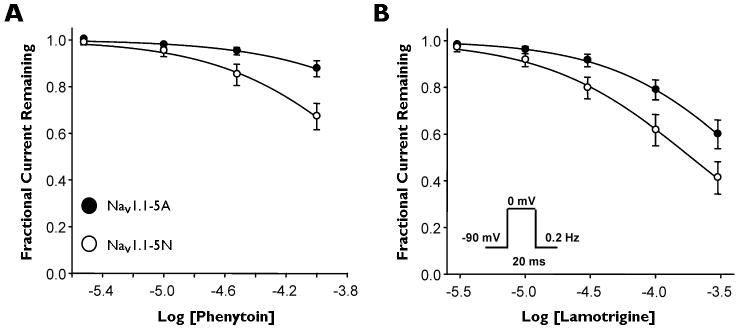
Due to the preferential activity shown by phenytoin and lamotrigine, we chose to examine blocking efficacy of these drugs for NaV1.1 splice variants. To examine blocking efficacy, we performed whole-cell patch clamp experiments in the absence and presence of either phenytoin or lamotrigine. Currents were stimulated by a 20 ms pulse to 0 mV from a holding potential of −90 mV, and then stimulated at a frequency of 0.2 Hz for the duration of the experiment. Using this protocol, we calculated an IC50 of 948 μM for phenytoin block of NaV1.1-5A compared with 213 μM for NaV1.1-5N (Fig. 3A), indicating that phenytoin has a 4-fold greater efficacy for NaV1.1-5N than NaV1.1-5A. Similarly, lamotrigine showed an IC50 of 485 μM for NaV1.1-5A compared with 187 μM for NaV1.1-5N (Fig. 3B), a 2.5-fold difference (Supplementary Table S2). These data illustrate significant pharmacological differences between the two splice variants.
Figure 3.
Phenytoin and lamotrigine exhibit greater blocking efficacy for NaV1.1-5N. (A and B) Concentration-response curves for tonic block of NaV1.1-5A (closed circles) or NaV1.1-5N (open circles) by phenytoin and lamotrigine. Currents were elicited by a 20 ms pulse to 0 mV from a holding potential of −90 mV and a frequency of 0.2 Hz. All points represent mean ± SEM for 9 - 10 experiments.
Biophysical properties of NaV1.1 splice variants in the presence of antiepileptic drugs
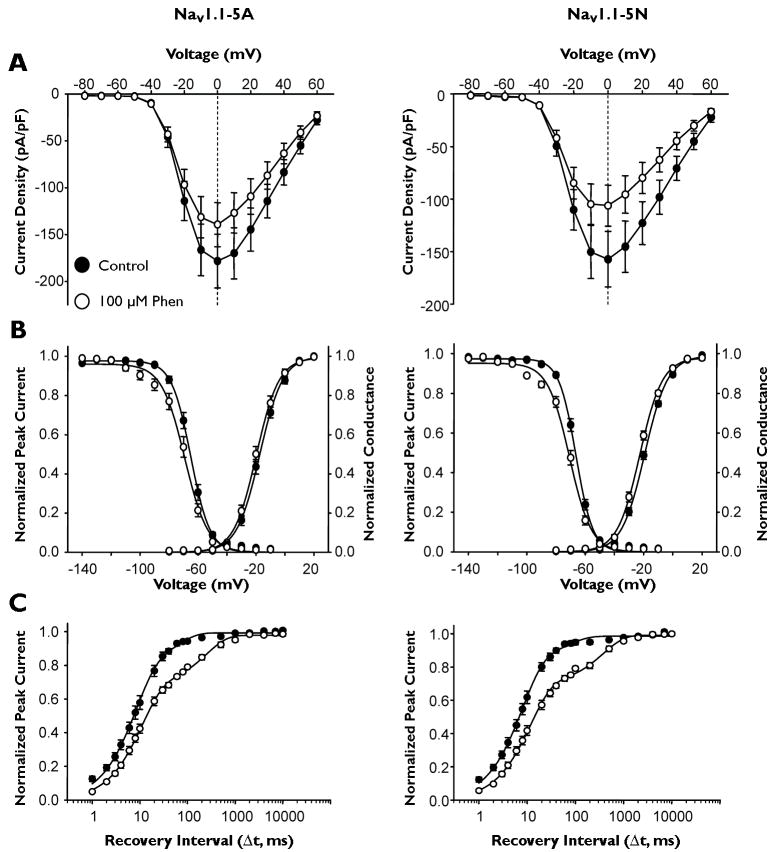
We examined the effects of phenytoin and lamotrigine on biophysical properties of NaV1.1 splice variants. We examined voltage-dependence of activation, steady-state inactivation, and recovery from fast inactivation in the absence and presence of phenytoin or lamotrigine. In the presence of phenytoin, current density was reduced, as expected for both splice isoforms, with NaV1.1-5N showing a greater reduction (Fig. 4A, Table 1). Both splice isoforms exhibited the expected shift in steady-state inactivation (~4 mV) in the presence of drug, but neither drug caused a significant change in voltage-dependence of activation (Figure 4B, Table 1). The most dramatic effect of phenytoin for both splice isoforms was a large shift in the relative contributions of fast and slow components of recovery from inactivation. Under control conditions for both splice isoforms, the fast component contributed ~90% to total recovery, while in the presence of phenytoin, this contribution was reduced to 70% (Table 1), suggesting a stabilization of the inactivated state in the presence of drug.
Figure 4.
Effect of phenytoin on NaV1.1 splice variant biophysical properties. (A) Peak current density elicited by test pulses to various membrane potentials and normalized to cell capacitance. (B) Voltage dependence of channel activation measured between −80 to +20mV and voltage dependence of inactivation measured following a 100 ms inactivating prepulse to between −140 and −10 mV. (C) Time-dependent recovery from inactivation assessed with a 100 ms inactivating prepulse to −10 mV. All recordings were performed with a holding potential of −90 mV. Closed symbols represent currents recorded under control condition and open symbols currents in the presence of 100 μM phenytoin. All points represent mean ± SEM for 9 - 10 experiments.
Table 1.
Biophysical properties of NaV1.1 splice variants in the absence and presence of antiepileptic drugs
| NaV1.1-5A | Voltage Dependence of Activation | Voltage Dependence of Fast Inactivation | Recovery from Fast Inactivation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| V1/2(mV) | k (mV) | n | V1/2(mV) | k (mV) | n | τf (ms) (Amplitude) | τs (ms) (Amplitude) | n | |
| Control | −15.8 ± 0.8 | 7.7 ± 0.1 | 22 | −64.0 ± 0.6 | −5.9 ± 0.2 | 19 | 8.5 ± 0.6 (0.89 ± 0.01) | 997.9 ± 308.7 (0.11 ± 0.01) | 18 |
| Phenytoin | −19.5 ± 1.3 | 7.7 ± 0.3 | 9 | −67.6 ± 1.5 | −6.9 ± 0.4 | 9 | 12.6 ± 1.8 (0.69 ± 0.01) | 371.9 ± 89.8 (0.29 ± 0.01) | 9 |
| Δ Phenytoin | −2.1 ± 0.7 | −0.04 ± 0.4 | 9 | −3.8 ± 0.5 | −0.8 ± 0.4 | 9 | 5.8 ± 0.3 (−0.19 ± 0.03) | −696.5 ± 611.5 (0.16 ± 0.02) | 9 |
| Lamotrigine | −16.6 ± 0.8 | 7.7 ± 0.2 | 13 | −68.9 ± 0.9 | −7.6 ± 0.3 | 10 | 11.6 ± 0.9 (0.62 ± 0.01) | 352.0 ± 29.9 (0.37 ± 0.01) | 9 |
| Δ Lamotrigine | −1.7 ± 0.6 | −0.02 ± 0.1 | 13 | −4.6 ± 0.6 | −1.9 ± 0.3 | 10 | 3.8 ± 0.6 (−0.28 ± 0.01) | −639.8 ± 301.4 (0.26 ± 0.01) | 9 |
| NaV1.1-5N | V1/2(mV) | k (mV) | n | V1/2(mV) | k (mV) | n | τf (ms) (Amplitude) | τs (ms) (Amplitude) | n |
| Control | −17.8 ± 0.6 | 7.8 ± 0.1 | 24 | −65.7 ± 0.8 | −5.9 ± 0.2 | 21 | 7.7 ± 0.8 (0.87 ± 0.02) | 1038.6 ± 321.8 (0.13 ± 0.02) | 17 |
| Phenytoin | −22.2 ± 0.8 | 7.9 ± 0.3 | 10 | −70.9 ± 1.2 | −7.7 ± 0.3 | 9 | 12.3 ± 1.6 (0.69 ± 0.01) | 410.7 ± 74.4 (0.30 ± 0.01) | 8 |
| Δ Phenytoin | −3.1 ± 0.6 | −0.02 ± 0.2 | 10 | −4.7 ± 0.5 | −1.9 ± 0.3* | 9 | 3.9 ± 1.9 (−0.17 ± 0.03) | −317.2 ± 322.7 (0.17 ± 0.05) | 8 |
| Lamotrigine | −19.1 ± 0.9 | 7.5 ± 0.3 | 14 | −69.2 ± 1.1 | −7.5 ± 0.3 | 12 | 11.6 ± 0.8 (0.64 ± 0.01) | 333.5 ± 28.5 (0.34 ± 0.01) | 9 |
| Δ Lamotrigine | −2.3 ± 0.5 | −0. 3 ± 0.2 | 14 | −4.2 ± 0.5 | −1.6 ± 0.4 | 12 | 4.6 ± 0.7 (−0.24 ± 0.03) | −900.4 ± 559.2 (0.22 ± 0.03) | 9 |
Δ, AED induced change in paired experiments;
p < 0.05 between splice variants
Lamotrigine altered NaV1.1 splice variant biophysical behavior in a similar manner as phenytoin. NaV1.1-5N current density was reduced to a greater extent by 200 μM lamotrigine compared to NaV1.1-5A (Fig. 5A), and no significant shift in voltage-dependence of activation was observed for either splice isoform (Fig 5B, Table 1). Like phenytoin, lamotrigine exerted significant effects on steady-state inactivation, although the shifts were similar between splice isoforms (Fig. 5C, Table 1). Likewise, lamotrigine caused a reduction in the contribution of the fast component and a small increase of fast time constant of recovery from fast inactivation for both splice isoforms, although the degree of shift was similar between splice isoforms (Fig 5D, Table 1).
Figure 5.
Effect of lamotrigine on NaV1.1 splice variant biophysical properties. (A) Peak current density elicited by test pulses to various membrane potentials and normalized to cell capacitance. (B) Voltage dependence of channel activation measured between −80 to +20mV and voltage dependence of inactivation measured following a 100 ms inactivating prepulse to between −140 and −10 mV. (C) Time-dependent recovery from inactivation assessed with a 100 ms inactivating prepulse to −10 mV. All recordings were performed with a holding potential of −90 mV. Closed symbols represent currents recorded under control condition and open symbols currents in the presence of 200 μM lamotrigine. All points represent mean ± SEM for 10 - 14 experiments.
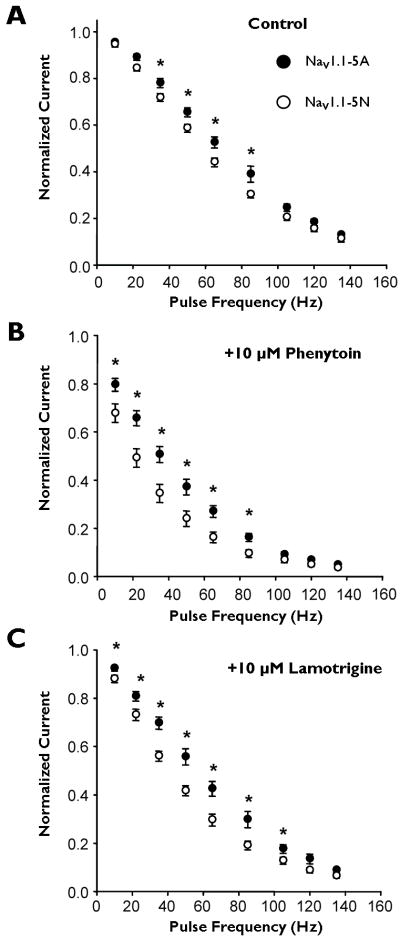
Finally, we examined use-dependent inhibition of both NaV1.1 splice variants over a range of stimulation frequencies from 10 to 135 Hz. Importantly, we used a therapeutically achievable concentration (10 μM) for both drugs to illustrate potential clinical relevance. In the absence of drug, we observed loss of channel availability as stimulation frequency increased (Fig. 6A) with NaV1.1-5N exhibiting a greater loss of channel availability at 35, 50, 65, and 85 Hz compared to NaV1.1-5A. In the presence of either 10 μM phenytoin (Fig. 6B), or 10 μM lamotrigine (Fig. 6C), NaV1.1-5N channels exhibited greater degrees of use-dependent channel inhibition by these drugs than NaV1.1-5A. Thus, at therapeutically relevant concentrations, NaV1.1-5N is more sensitive to phenytoin and lamotrigine.
Figure 6.
Use-dependent inhibition of NaV1.1 splice variants. Use-dependent inhibition of NaV1.1-5A (closed symbols) and NaV1.1-5N (open symbols) were assessed in the absence (A) or presence of 10 μM phenytoin (B) or 10 μM lamotrigine (C). Currents were elicited by a pulse to 0 mV from a holding potential of −90 mV at frequencies ranging from 10 to 135 Hz. Peak current at the 300th pulse was normalized to that of the 1st pulse. An asterisk indicates p < 0.05. All points represent mean ± SEM for 9 - 16 experiments.
Discussion
Alternative mRNA splicing occurs in up to 60% of human genes and is an important mechanism providing protein diversity (Copley, 2004, Matlin et al., 2005). Alternative splicing of NaV genes has been shown to generate channels with unique functional and pharmacological properties (Copley, 2004, Onkal et al., 2008, Diss et al., 2004, Tan et al., 2002). Our study examined the biophysical and pharmacological properties of NaV1.1 splice variants incorporating either exon 5A or 5N. This work was motivated by pharmacogenetic studies that identified a common genetic variant in an SCN1A intron splice donor site that influences the proportion of transcripts incorporating either exon 5A or exon 5N (Tate et al., 2006, Tate et al., 2005). Control and epileptic patients carrying the G allele had a larger proportion of NaV1.1-5N transcript in brain (~50% of channels) and required lower therapeutic doses of phenytoin, respectively. We found that NaV1.1-5N channels were more sensitive to both phenytoin and lamotrigine.
The appropriate therapeutic dosage for phenytoin, carbamazepine, and lamotrigine must be empirically determined to account for the unique pharmacodynamic and pharmacokinetic profile of each patient. Our study provides the first direct evidence that the variation in therapeutic phenytoin dosage observed by Tate, et al. (Tate et al., 2005) arises in part due to differences in how these drugs interact with an alternatively spliced brain sodium channel. Previous work has demonstrated that phenytoin, carbamazepine and lamotrigine preferentially interact with the inactivated states (Kuo, 1998, Kuo et al., 1997, Yang and Kuo, 2002, Kuo and Bean, 1994, Ragsdale et al., 1991) and this interaction can be described by the modulated receptor model (Hille, 1977). Therefore, altered binding site presentation and differences in drug block could result from isoform specific activation and/or inactivation gating. However, even though NaV1.1-5N trended toward activation at more hyperpolarized voltages, no statistically significant differences in biophysical properties between the splice variants were observed (Fig. 1). Only frequency dependent channel rundown differed between splice variants. Despite the absence of major biophysical differences between splice variants, even in the presence of antiepileptic drugs (Figs 4. and 5), NaV1.1-5N channels exhibited greater tonic and use-dependent inhibition by phenytoin and lamotrigine than NaV1.1-5A (Figs. 2 and 6), suggesting that binding sites for these drugs may be altered, and that the pharmacological differences may arise from slower inactivation processes. Because the binding sites for the studied drugs are predicted to lie near the DIV-S6 and exon 5 encodes D1-S3/S4, we predict that the divergent pharmacological properties observed for NaV1.1-5A and NaV1.1-5N result from allosteric modification of the binding sites through subtle structural rearrangements.
Our data suggests that phenytoin, carbamazepine, and lamotrigine interact with NaV1.1 with low blocking efficacy (IC50 >100 μM) that at first appears to conflict with the free serum concentration for these drugs at therapeutic levels (~10 μM) (Rambeck et al., 1987, Johannessen and Tomson, 2006, Dasgupta, 2007). However, the degree of channel inhibition is strongly dependent on the holding potential, which in turn determines binding site presentation (Ragsdale et al., 1991, Kuo, 1998). Thus, measurements of tonic block by these drugs only reveals binding to a specific set of states that are determined by the holding potential used during the experiment, and the available states change as the membrane potential of the cell changes. Therefore, relative tonic inhibition measurements made in vitro cannot be directly compared with values expected from therapeutic concentrations of these drugs. However, experiments with a therapeutic concentration of these drugs using repetitive stimulation over a range of frequencies, we demonstrated conclusively that phenytoin and lamotrigine both evoke greater use-dependent block of NaV1.1-5N (Fig. 6).
Our data demonstrate that NaV1.1 splice variants have divergent pharmacological properties, with NaV1.1-5N exhibiting increased sensitivity to the commonly prescribed AED phenytoin. An important additional finding was that lamotrigine, another widely prescribed drug for the treatment of epilepsy that was not considered in the previous pharmacogenetic studies, more effectively inhibits NaV1.1-5N than NaV1.1-5A. Additional studies are needed to determine whether genetic testing of patients for the SCN1A polymorphism enables more precise dosing of phenytoin, lamotrigine and possibly other AEDs that target this channel.
Supplementary Material
Acknowledgments
This work was supported by supported by NIH grant NS032387 (A.L.G.); C.H.T. was supported by institutional training grant MH065215.
Abbreviations
- IC50
drug concentration required for 50% inhibition
Footnotes
Author Disclosures:
Dr. Thompson received salary support from NIH training grant MH065215.
Dr. Kahlig received salary support from NIH grant NS032387.
Dr. George is funded by NIH grants NS032387, HL068880 and HL083374, and is a paid scientific consultant for Allergan, and received grant support from Gilead Sciences.
Ethical Publication Statement
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- Abe T, Seo T, Ishitsu T, Nakagawa T, Hori M, Nakagawa K. Association between SCN1A polymorphism and carbamazepine-resistant epilepsy. Br J Clin Pharmacol. 2008;66:304–307. doi: 10.1111/j.1365-2125.2008.03203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes LR, Deprez L, Suls A, Baets J, Smets K, Van DT, Deconinck T, Jordanova A, De JP. The SCN1A variant database: a novel research and diagnostic tool. Hum Mutat. 2009;30:E904–E920. doi: 10.1002/humu.21083. [DOI] [PubMed] [Google Scholar]
- Copley RR. Evolutionary convergence of alternative splicing in ion channels. Trends Genet. 2004;20:171–176. doi: 10.1016/j.tig.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Dasgupta A. Usefulness of monitoring free (unbound) concentrations of therapeutic drugs in patient management. Clin Chim Acta. 2007;377:1–13. doi: 10.1016/j.cca.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Diss JK, Fraser SP, Djamgoz MB. Voltage-gated Na+ channels: multiplicity of expression, plasticity, functional implications and pathophysiological aspects. Eur Biophys J. 2004;33:180–193. doi: 10.1007/s00249-004-0389-0. [DOI] [PubMed] [Google Scholar]
- George AL., Jr Inherited disorders of voltage-gated sodium channels. J Clin Invest. 2005;115:1990–1999. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson TA, Clevinger EC, O’Neill TJ, Yarowsky PJ, Krueger BK. Mutually exclusive exon splicing of type III brain sodium channel alpha subunit RNA generates developmentally regulated isoforms in rat brain. J Biol Chem. 1993;268:18648–18653. [PubMed] [Google Scholar]
- Heinzen EL, Yoon W, Tate SK, Sen A, Wood NW, Sisodiya SM, Goldstein DB. Nova2 interacts with a cis-acting polymorphism to influence the proportions of drug-responsive splice variants of SCN1A. Am J Hum Genet. 2007;80:876–883. doi: 10.1086/516650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Local anesthetics: Hydrophilic and hydrophobic pathways for the drug-receptor reaction. J Gen Physiol. 1977;69:497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. 3. Sinauer Associates Inc.; Sunderland, MA: 1993. [Google Scholar]
- Isom LL. Sodium channel beta subunits: anything but auxiliary. Neuroscientist. 2001;7:42–54. doi: 10.1177/107385840100700108. [DOI] [PubMed] [Google Scholar]
- Jarecki BW, Sheets PL, Xiao Y, Jackson JO, Cummins TR. Alternative splicing of Na(V)1.7 exon 5 increases the impact of the painful PEPD mutant channel I1461T. Channels (Austin) 2009;3:259–267. doi: 10.4161/chan.3.4.9341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen SI, Tomson T. Pharmacokinetic variability of newer antiepileptic drugs: when is monitoring needed? Clin Pharmacokinet. 2006;45:1061–1075. doi: 10.2165/00003088-200645110-00002. [DOI] [PubMed] [Google Scholar]
- Kahlig KM, Rhodes TH, Pusch M, Freilinger T, Pereira-Monteiro JM, Ferrari MD, van den Maagdenberg AM, Dichgans M, George AL., Jr Divergent sodium channel defects in familial hemiplegic migraine. Proc Natl Acad Sci U S A. 2008;105:9799–9804. doi: 10.1073/pnas.0711717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CC. A common anticonvulsant binding site for phenytoin, carbamazepine, and lamotrigine in neuronal Na+ channels. Mol Pharmacol. 1998;54:712–721. [PubMed] [Google Scholar]
- Kuo CC, Bean BP. Slow binding of phenytoin to inactivated sodium channels in rat hippocampal neurons. Mol Pharmacol. 1994;46:716–725. [PubMed] [Google Scholar]
- Kuo CC, Chen RS, Lu L, Chen RC. Carbamazepine inhibition of neuronal Na+ currents: quantitative distinction from phenytoin and possible therapeutic implications. Mol Pharmacol. 1997;51:1077–1083. doi: 10.1124/mol.51.6.1077. [DOI] [PubMed] [Google Scholar]
- Kuo CC, Lu L. Characterization of lamotrigine inhibition of Na+ channels in rat hippocampal neurones. Br J Pharmacol. 1997;121:1231–1238. doi: 10.1038/sj.bjp.0701221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossin C. A catalog of SCN1A variants. Brain Dev. 2009;31:114–130. doi: 10.1016/j.braindev.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Lossin C, Wang DW, Rhodes TH, Vanoye CG, George AL., Jr Molecular basis of an inherited epilepsy. Neuron. 2002;34:877–884. doi: 10.1016/s0896-6273(02)00714-6. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Onkal R, Mattis JH, Fraser SP, Diss JK, Shao D, Okuse K, Djamgoz MB. Alternative splicing of Nav1.5: an electrophysiological comparison of ‘neonatal’ and ‘adult’ isoforms and critical involvement of a lysine residue. J Cell Physiol. 2008;216:716–726. doi: 10.1002/jcp.21451. [DOI] [PubMed] [Google Scholar]
- Ragsdale DS, Scheuer T, Catterall WA. Frequency and voltage-dependent inhibition of type IIA Na+ channels, expressed in a mammalian cell line, by local anesthetic, antiarrhythmic, and anticonvulsant drugs. Mol Pharmacol. 1991;40:756–765. [PubMed] [Google Scholar]
- Rambeck B, May T, Juergens U. Serum concentrations of carbamazepine and its epoxide and diol metabolites in epileptic patients: the influence of dose and comedication. Ther Drug Monit. 1987;9:298–303. doi: 10.1097/00007691-198709000-00008. [DOI] [PubMed] [Google Scholar]
- Rhodes TH, Lossin C, Vanoye CG, Wang DW, George AL., Jr Noninactivating voltage-gated sodium channels in severe myoclonic epilepsy of infancy. Proc Natl Acad Sci U S A. 2004;101:11147–11152. doi: 10.1073/pnas.0402482101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarao R, Gupta SK, Auld VJ, Dunn RJ. Developmentally regulated alternative RNA splicing of rat brain sodium channel mRNAs. Nucleic Acids Res. 1991;19:5673–5679. doi: 10.1093/nar/19.20.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Liu Z, Nomura Y, Goldin AL, Dong K. Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels. J Neurosci. 2002;22:5300–5309. doi: 10.1523/JNEUROSCI.22-13-05300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate SK, Depondt C, Sisodiya SM, Cavalleri GL, Schorge S, Soranzo N, Thom M, Sen A, Shorvon SD, Sander JW, Wood NW, Goldstein DB. Genetic predictors of the maximum doses patients receive during clinical use of the anti-epileptic drugs carbamazepine and phenytoin. Proc Natl Acad Sci U S A. 2005;102:5507–5512. doi: 10.1073/pnas.0407346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate SK, Singh R, Hung CC, Tai JJ, Depondt C, Cavalleri GL, Sisodiya SM, Goldstein DB, Liou HH. A common polymorphism in the SCN1A gene associates with phenytoin serum levels at maintenance dose. Pharmacogenet Genomics. 2006;16:721–726. doi: 10.1097/01.fpc.0000230114.41828.73. [DOI] [PubMed] [Google Scholar]
- Yang YC, Kuo CC. Inhibition of Na(+) current by imipramine and related compounds: different binding kinetics as an inactivation stabilizer and as an open channel blocker. Mol Pharmacol. 2002;62:1228–1237. doi: 10.1124/mol.62.5.1228. [DOI] [PubMed] [Google Scholar]
- Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich F, Stogmann E, Bonelli S, Baumgartner C, Mueller JC, Meitinger T, Zimprich A, Strom TM. A functional polymorphism in the SCN1A gene is not associated with carbamazepine dosages in Austrian patients with epilepsy. Epilepsia. 2008;49:1108–1109. doi: 10.1111/j.1528-1167.2008.01549_4.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.