Abstract
Discriminating closely related molecules remains a major challenge in the engineering of binding proteins and inhibitors. Here we report the development of highly selective inhibitors of small ubiquitin-related modifier (SUMO) family proteins. SUMOylation is involved in the regulation of diverse cellular processes. Functional differences between two major SUMO isoforms in humans, SUMO1 and SUMO2/3, are thought to arise from distinct interactions mediated by each isoform with other proteins containing SUMO-interacting motifs (SIMs). However, the roles of such isoform-specific interactions are largely uncharacterized due in part to the difficulty in generating high-affinity, isoform-specific inhibitors of SUMO/SIM interactions. We first determined the crystal structure of a “monobody,” a designed binding protein based on the fibronectin type III scaffold, bound to the yeast homolog of SUMO. This structure illustrated a mechanism by which monobodies bind to the highly conserved SIM-binding site while discriminating individual SUMO isoforms. Based on this structure, we designed a SUMO-targeted library from which we obtained monobodies that bound to the SIM-binding site of human SUMO1 with Kd values of approximately 100 nM but bound to SUMO2 400 times more weakly. The monobodies inhibited SUMO1/SIM interactions and, unexpectedly, also inhibited SUMO1 conjugation. These high-affinity and isoform-specific inhibitors will enhance mechanistic and cellular investigations of SUMO biology.
Keywords: protein engineering, molecular recognition, posttranslational modification, antibody mimic, fibronectin type III domain
Proteomes contain many protein families whose members share high levels of sequence and structural similarity. Individual members of such protein families can have distinct functions, but such functional differences are often difficult to define. Affinity reagents and inhibitors capable of selectively targeting individual family members have great potential as tools for selectively manipulating and thus defining the unique functions of these proteins. However, the development of such highly selective reagents is a formidable challenge because of high homology among family members. In this work, we report the development of selective inhibitors of members of the small ubiquitin-like modifier (SUMO) family.
SUMOs are structurally similar to ubiquitin and are posttranslationally conjugated to other proteins resulting in a variety of functional modulations. In humans, there are four SUMO isoforms (SUMO1–4) (1). SUMO1 and SUMO2 share 41% sequence identity (72% similarity) but are functionally distinct (Fig. S1) (2, 3). SUMO2 and SUMO3, collectively referred to as SUMO2/3, share 97% sequence identity and are assumed to be functionally identical (1, 4). SUMO4’s relevance as a posttranslational modification is not clear (5, 6). Thus, most studies in SUMO biology have focused on SUMO1 and SUMO2/3. SUMOylation plays important roles in regulating diverse cellular processes including DNA repair, transcription, nuclear transport, and chromosome dynamics (1, 4). The dominant mechanism by which SUMOylation alters protein function appears to be through SUMO-mediated interactions with other proteins containing a short peptide motif known as a SUMO-interacting motif (SIM) (4, 7, 8).
There are few inhibitors of SUMO/SIM interactions, a deficiency that limits our ability to finely dissect SUMO biology. In the only reported example of such an inhibitor, a SIM-containing linear peptide was used to inhibit SUMO/SIM interactions, establishing their importance in coordinating DNA repair by nonhomologous end joining (9). This peptide sensitized cancer cells to radiation and chemotherapeutic-induced DNA damage, illustrating a therapeutic potential of SUMO/SIM inhibitors. These findings clearly establish the utility of SUMO/SIM inhibitors, but the peptide inhibitor suffers from two significant shortcomings. First, the peptide binds equally well to SUMO1 and SUMO2/3, making it impossible to differentiate the roles of each isoform. Second, the peptide has low affinity for SUMO (Kd ∼ 5 μM) (8). As a result, high concentrations of the peptide are required for inhibition. Most natural SIM peptides exhibit similarly low affinities and discriminate individual SUMO isoforms by approximately 10-fold or less (7, 10–13). Higher affinity reagents capable of selectively inhibiting the SIM interactions of individual SUMO isoforms could be powerful tools for better defining the functions of each isoform and potentially as more potent therapeutics. However, the development of such highly selective inhibitors presents a formidable challenge as the SIM-binding site is highly conserved among SUMO isoforms (Fig. 1A and Fig. S1) (14).
Fig. 1.
Sequences and properties of ySUMO-binding monobodies. (A) ySUMO structure colored by conservation score among ySUMO and hSUMO isoforms (34). (B) Schematic of the FN3 scaffold with β-strands A–G labeled and surface loops diversified in monobody libraries colored red. (C) Amino acid sequences of variable loops of ySUMO-binding monobodies with Kd values from SPR. Residues are colored as follows: Tyr (yellow), Ser (red), Gly (green), Arg and Lys (dark blue), His (blue), polar amino acids (C, N, T, Q,) (pale blue), hydrophobic amino acids (A, L, V, F, I, W, M, P) (white), and Asp and Glu (pink), residues originating from the vector template (not from mutagenesis) (gray). (D) SPR traces for ySMB-1 and ySMB-2 binding to ySUMO with kinetic parameters calculated from a best fit (solid line) of the raw data (dashed line) to a 1∶1 binding model. (E) Epitopes of ySMB-1 and ySMB-2 mapped from NMR chemical shift perturbation shown on the ySUMO structure. Nitrogen atoms of unambiguously assigned amides in the 1H-15N heteronuclear single quantum coherence spectrum (spheres) are colored according their perturbation level: shift of > 2 peak widths (red), 1–2 peak widths (orange), ∼1 peak width (yellow), < 1 peak width (unaffected) (white).
Our group has established the fibronectin type III domain (FN3) as an effective scaffold for generating binding proteins that we call “monobodies” (15). FN3 has three loops at one end of the molecule that are analogous to antibody complementarity determining regions (Fig. 1B). We design and construct combinatorial libraries that diversify the lengths and amino acid sequences of these loops and, from these libraries, we isolate monobodies capable of binding to a target of interest. A monobody that binds to the Src homology 2 (SH2) domain of Abelson tyrosine kinase (Abl) has recently been developed that targets the phosphopeptide binding site (16). Although this site is highly conserved among the 120 human SH2 domains, just as the SIM-binding site is highly conserved in SUMO proteins, this monobody was highly selective and proved a valuable tool for better defining the biological role of Abl SH2. This example validated the concept of using monobodies to selectively target even highly conserved functional sites in individual members of a protein family.
We have attempted to isolate monobodies that specifically target individual human SUMO isoforms as well as the yeast homolog of SUMO (ySUMO), which has approximately 45% sequence identity (approximately 67% similarity) with human SUMOs (hSUMOs) (Fig. 1A and Fig. S1). Although we successfully isolated numerous monobodies to ySUMO with mid-nanomolar Kd values (Fig. 1 C and D and Fig. S2), puzzlingly, we generated only weakly binding monobodies to hSUMO1 and hSUMO2/3 (Kd values in the micromolar range), suggesting shortcomings in our library design for these targets.
In this work, we determined the crystal structure of a ySUMO-binding monobody bound to ySUMO, which revealed the structural basis for our success in targeting ySUMO and for our difficulties in targeting hSUMOs. Guided by this knowledge, we developed a “SUMO-targeted” monobody library that successfully produced isoform-specific monobodies to hSUMO1. Functional studies demonstrated that these monobodies are highly selective inhibitors of hSUMO1/SIM interactions and also of hSUMO1 conjugation.
Results
Diverse Monobodies Recognize the SIM-Binding Site of ySUMO and Discriminate ySUMO from hSUMOs.
As a first step toward understanding how monobodies recognize ySUMO, we mapped the epitopes of two of the highest affinity ySUMO-binding monobodies, termed ySMB-1 and ySMB-2 (Fig. 1 C and D and Fig. S2), using NMR chemical shift perturbation. Despite distinct amino acid sequences in their variable loops (Fig. 1C), both monobodies bound to similar epitopes centered on the SIM-binding site (Fig. 1E). Binding of 33 other ySUMO monobodies was inhibited by ySMB-1, indicating that they too bound to the SIM-binding site (Fig. S3A). Like ySMB-1, most ySUMO-binding monobodies have polyserine sequences in the BC and DE loops that originate from incomplete mutagenesis of the template vector in library construction, suggesting that these loops do not contribute to binding (Fig. 1C and Fig. S2). Furthermore, many of these monobodies have an 11-residue FG loop with a centrally located acidic residue and flanking aromatic and hydrophobic residues (Fig. 1C and Fig. S2). Together, these results suggest that essentially all the ySUMO-binding monobodies recognize the SIM-binding site using a similar mode of interaction.
Most ySUMO-binding monobodies exhibited negligible levels of binding to hSUMO1 or hSUMO2 in a phage enzyme-linked immunosorbent assay (ELISA) (Fig. 2A). Such high selectivity was unexpected, because the SIM-binding site is the most highly conserved surface between ySUMO and hSUMO proteins (Fig. 1A). Surface plasmon resonance (SPR) measurements showed that ySMB-1 (selective for ySUMO in ELISA) bound to ySUMO with a 82-nM Kd and to hSUMO1 with an approximate 54-µM Kd and exhibited no detectable binding to hSUMO2 (Fig. 2B), discriminating ySUMO from hSUMOs by more than 600-fold in affinity. ySMB-9 (nonselective in ELISA) bound to all three SUMO proteins. Although ySMB-9 surprisingly bound to hSUMO1 with higher affinity (68-nM Kd) than either ySUMO or hSUMO2 (Fig. 2B), it discriminated hSUMO2 by only approximately 70-fold, which is 10-fold less selective than ySMB-1. Notably, ySMB-9 does not have polyserine BC and DE loops like most other ySUMO-binding monobodies, and it also has a significantly shorter FG loop (Fig. 1B and Fig. S2). Although competition data suggested that ySMB-9 binds to the SIM-binding site (Fig. S2), its distinct sequence features suggest that it employs a different mode of interaction than most ySUMO-binding monobodies, leading to its lower specificity. Together, these findings demonstrate that the binding mode of most ySUMO-binding monobodies is particularly effective in discriminating ySUMO and hSUMOs despite binding to the highly conserved SIM-binding site. Thus, we reasoned that generating monobodies that bind to hSUMOs in a mode similar to the ySUMO-binding monobodies would yield clones with higher isoform selectivity toward hSUMOs.
Fig. 2.
Specificity of ySUMO-binding monobodies. (A) Binding of eight ySUMO-binding monobodies to ySUMO, hSUMO1, and hSUMO2 assayed using phage ELISA. Clone numbers are of the format ySMB-X in Fig. 1C and Fig. S2. (B) Equilibrium SPR measurements of ySMB-1 (Left) and ySMB-2 (Right) binding to ySUMO, hSUMO1, and hSUMO2. Equilibrium responses at multiple concentrations (Left) were fit with a simple 1∶1 binding model (Right).
Crystal Structure of the ySMB-1/ySUMO Complex.
To understand the structural basis for the isoform-selective recognition of the SIM-binding site, we determined the crystal structure of ySMB-1 in complex with ySUMO at 2.4-Å resolution (structural statistics in Table S1). Consistent with the NMR epitope mapping data, ySMB-1 bound to the SIM-binding site (Figs. 1E and 3A). The monobody formed the binding surface using a single variable loop (FG loop) and residues from the invariant FN3 scaffold (Fig. 3A). As inferred from their polyserine sequences, the BC and DE loops of ySMB-1 were not involved in direct contacts with ySUMO.
Fig. 3.
Crystal structure of the monobody ySMB-1/ySUMO complex. (A, Upper) ySUMO (gray surface/cartoon) and ySMB-1 (cyan cartoon) are shown with monobody paratope residues shown as sticks; FG loop residues (carbon atoms blue) and scaffold residues (carbon atoms yellow) are indicated. ySUMO is shown in the same orientation as in Fig. 1E. (Lower) An alternative view with the monobody paratope depicted as a surface. (B) Close-up of the ySMB-1/ySUMO interface. ySUMO (surface/sticks) is shown with residues comprising the hydrophobic center of the epitope colored pink and the charged/polar rim colored green. Monobody paratope residues are shown as in A. (C, Left and Center) Comparison of the binding modes of ySMB-1 (cyan) to ySUMO (gray) and the SIM of RanBP2 (yellow) to hSUMO1 (gray). Both form intermolecular β-sheets with their SUMO targets (expanded box). (Right) Overlay of the RanBP2 SIM (yellow) and SIM mimicking monobody residues (cyan) with the ySUMO surface (gray) shown.
Residues 78–85 of the ySMB-1 FG loop form a β-hairpin that provides 84% of the monobody binding surface with nonloop scaffold residues contributing the remainder (Fig. 3B and Fig. S4A). The edge of this hairpin docks along the hydrophobic center of the SIM-binding site forming an intermolecular β-sheet with ySUMO and closely mimicking the interaction mode of SIMs (Fig. 3 B and C) (7, 17, 18). SIMs generally contain a stretch of hydrophobic residues flanked by a stretch of acidic residues, e.g., DVLIVY in Ran-GTP binding protein 2 (RanBP2) and TLDIVD in protein inhibitor of activated STAT x (8, 9, 19). In ySMB-1, this motif is mimicked by the FG loop sequence DLYYSY (residues 80–85) (Figs. 1C, and 3 B and C). D80 of the monobody aligns with the “top” basic portion of the SIM-binding site in a similar orientation as a conserved acidic stretch in SIMs (7, 18) and Tyr residues line the hydrophobic tract where aliphatic residues are usually found in SIMs.
The crystal structure suggested a structural basis for isoform selectivity of ySUMO-binding monobodies and for our difficulties in generating monobodies to hSUMOs. Only 5 of the 16 residues in the ySMB-1 epitope are poorly conserved between ySUMO and hSUMOs (positions 25, 34, 36, 50, and 54) (Fig. S1). Three of these residues (N25, E34, and F36) form a cluster at one side of the interface that is highly buried, comprising 23% (147 Å2) of total ySUMO surface buried by the monobody (Figs. 3B and 4A). hSUMO1 contains N25K and F36H. hSUMO2 contains E34V and F36Q. Thus, any monobody that forms an interface similar to ySMB-1 is not likely to tightly bind to hSUMO1 or hSUMO2/3. Notably, this cluster is contacted in large part by scaffold residues in ySMB-1 (Y31, R33, and Y73) (Figs. 3C and 4A). Because these scaffold residues were not varied in our library and are anchored in a conformationally rigid β-sheet, nonconservative substitutions in the cluster in hSUMOs could not have been accommodated, making the generation of ySMB-1-like monobodies for hSUMOs impossible. These structural restraints would eliminate a potentially very large number of ySMB-1-like monobodies that have an FG loop otherwise capable of binding to hSUMOs. Thus, these observations strongly suggest that residues within the monobody scaffold serve as both positive design elements favoring ySUMO binding and negative design elements disfavoring binding to hSUMOs.
Fig. 4.
hSUMO1-binding monobodies from the SUMO-targeted library. (A) Design of the SUMO-targeted monobody library. (Left) ySMB-1 paratope residues (backbone sticks/spheres) are shown with FG loop (blue) and scaffold residues (yellow) indicated. ySUMO (white surface) is shown with ySMB-1 epitope residues as green sticks. ySUMO residue labels are colored according to conservation between ySUMO and hSUMOs: blue (completely conserved), green (conservative substitution), and red (nonconservative substitution). The residue types at each position in hSUMO1 and hSUMO2/3 are shown in parentheses. (Right) Amino acid diversity used in the SUMO-targeted library. The wild-type ySMB-1 residue is in brackets. (B) Amino acid sequences of hSUMO1-binding monobodies from the SUMO-targeted library. Kd values from SPR are also shown. At bottom, sequences of an additional hSUMO1 binding monobody (hS1MB-22) and a very similar ySUMO-binding monobody (ySMB-ST6) recovered from the SUMO-targeted library are shown. Residues are colored as in Fig. 1C. Representative SPR traces are shown at right. (C) Epitope of hS1MB-4 mapped from chemical shift perturbation shown on the hSUMO1 structure. Data are represented using the same scheme as in Fig. 1D. (D) Sequence conservation of ySUMO- and hSUMO1-binding monobodies shown as sequence logos (35, 36). The height of individual letters reflects how frequently that amino acid was recovered at that position, the letters stacked at a position are ordered from most to least frequently occurring and the overall height of a stack reflects the overall conservation level at that position. (E) Binding of hS1MB-22 and ySMB-ST6 to ySUMO and hSUMO1 measured by phage ELISA. (F) Contacts made by scaffold residues in the ySMB-1/ySUMO complex (Left) and in a modeled ySMB-1/hSUMO-1 complex (Right). Monobody (yellow sticks) and SUMO residues (gray surface/green sticks) are indicated. The ySMB-1/hSUMO1 complex was modeled by superposition of the ySUMO portion of the ySMB-1 complex with the hSUMO1 structure.
Structure-Guided Design of a SUMO-Targeted Monobody Library.
We hypothesized that the binding mode of ySMB-1 could be used as a template for designing isoform-specific monobody inhibitors of hSUMO/SIM interactions. Thus, we designed a library that was aimed at “reprogramming” ySMB-1 for binding to hSUMO proteins. We introduced amino acid diversity at each ySMB-1 paratope position that included the wild-type amino acid and other amino acid types that might allow effective complementation of any of the three SUMO proteins (Fig. 4A) (SI Materials and Methods). Notably, this library included diversity at previously invariant scaffold positions that participated in ySUMO binding. The number of independent clones in the constructed phage-display library was 2.0 × 109, giving reasonable coverage of the theoretical size of the design (1.6 × 1011).
Selection of Monobodies from the SUMO-Targeted Library.
After four rounds of library sorting against hSUMO1, hSUMO2, and ySUMO, 32 randomly chosen clones for each target were assayed for binding activity using phage ELISA. All clones tested positive for binding in the cases of ySUMO and hSUMO1 but none bound to hSUMO2. Five ySUMO-binding and 10 hSUMO1-binding monobodies were expressed as soluble proteins and assessed using SPR, all of which produced binding signals (Fig. 4B), consistent with phage ELISA results. For ySUMO, the monobodies exhibited Kd values similar to those of monobodies from the previous naïve library (39 nM to 3.3 μM) (Fig. 1C and Fig. S2 and S5). For hSUMO1, Kd estimates ranged from 118 nM to 3.6 μM (Fig. 4B). Thus, unlike our original library, the SUMO-targeted library readily produced monobodies with good affinity to both ySUMO and hSUMO1.
NMR chemical shift perturbation assays validated that a newly generated hSUMO1-binding monobody, termed hS1MB-4, targeted the SIM-binding site (Fig. 4C). Binding of 15 other hSUMO1-binding monobodies was inhibited by hS1MB-4 as tested in ELISA, strongly suggesting that all these hSUMO1-binding monobodies targeted the SIM-binding site as intended (Fig. S3B).
The amino acid sequences of 44 hSUMO1-binding clones and 40 ySUMO-binding clones revealed that monobodies to both targets contained FG loop sequences highly similar to ySMB-1 (Fig. 4D), suggesting that a ySMB-1-like binding mode was maintained in these monobodies and that ySMB-1-like FG loop sequences are effective for binding to both ySUMO and hSUMO1. In contrast, scaffold residues were sharply different in monobodies to the two targets (Fig. 4D). The wild-type ySMB-1 scaffold residues were highly conserved among ySUMO-binding monobodies, but in hSUMO1-binding monobodies, the wild-type amino acid was never recovered at position 33 and only infrequently recovered at position 31. These results strongly support our hypothesis that isoform selectivity in ySUMO-binding monobodies arises from contacts made by the nonloop regions of the monobody scaffold. Consistent with this mechanism, in a pair of monobodies with nearly identical FG loop sequences, hS1MB-22 and ySMB-ST6, we observed that ySMB-ST6 containing the wild-type scaffold residues bound only to ySUMO, whereas hS1MB-22 containing altered scaffold residues bound to both ySUMO and hSUMO1 (Fig. 4 B and E). Taken together, these results illustrate the importance of altering nonloop residues in the FN3 scaffold in order to facilitate binding to hSUMO1.
Modeling a ySMB-1 interface with hSUMO1 provides a clear rationale for the observed mutations at scaffold positions in the hSUMO1-binding monobodies. N25K and F36H substitutions in hSUMO1 with respect to ySUMO result in a likely electrostatic and steric clash between R33 of the monobody scaffold and K25 of hSUMO1 as well as a loss of a close, edge-plane aromatic interaction between Y73 of the monobody and F36 in ySUMO (Fig. 4F). Notably, the most favored amino acid types at position 33 in hSUMO1-binding monobodies were Ala and Glu, either of which should resolve a clash with K25, supporting this molecular mechanism for binding specificity.
hSUMO1-Binding Monobodies Are Isoform Specific.
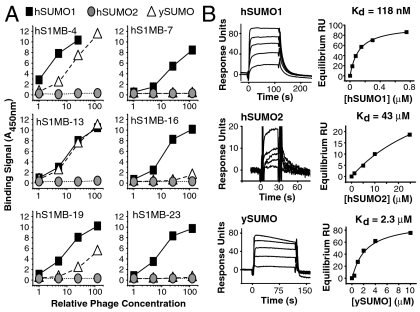
hSUMO1-binding monobodies had varied ability to discriminate hSUMO1 and ySUMO as assessed by phage ELISA (Fig. 5A and Fig. S6A). There were several clones (e.g., hS1MB-7, 16, and 23) that showed no detectable binding to ySUMO, representing at least 100-fold weaker binding to ySUMO than to hSUMO1 (Fig. 5A and Fig. S6A). The difference in the affinity of hS1MB-4 to ySUMO and hSUMO1, as measured by SPR, was approximately 20-fold, validating the phage ELISA experiment that gave an approximate 10-fold difference (Fig. 5B and Fig. S6B). No distinct features were evident between the sequences of clones that did and did not discriminate ySUMO (Fig. S6B), suggesting that the mechanism of ySUMO/hSUMO1 discrimination is complex, likely involving several positions, and varied across different clones. As expected from the failure of our library to generate monobodies to hSUMO2, the hSUMO1-binding monobodies showed no measurable binding to hSUMO2 in phage ELISA (Fig. 5A and Fig. S6A), and the affinity of hS1MB-4 to hSUMO2 determined by SPR was very weak (Kd = 43 μM; Fig. 5B), corresponding to 360-fold discrimination between hSUMO1 and hSUMO2. Taken together, these data demonstrate that the SUMO-targeted library enabled us to generate diverse monobodies that have high affinity and high specificity to hSUMO1.
Fig. 5.
Specificity of hSUMO-1-binding monobodies. (A) Binding curves derived from phage ELISA of six hSUMO1-binding monobodies binding to ySUMO, hSUMO1, and hSUMO2. Data for additional monobodies are shown in Fig. S6A. Serial dilutions of phage containing culture supernatant (titer approximately 108/mL) were used. Absorbance values were scaled to 1-cm path length. (B) Equilibrium SPR measurements of hS1MB-4 binding to ySUMO, hSUMO1, and hSUMO2. Equilibrium responses at multiple concentrations (Left) were fit to a simple 1∶1 binding model (Right).
New Monobodies Inhibit the SUMO1/SIM Interaction and SUMO1 Conjugation.
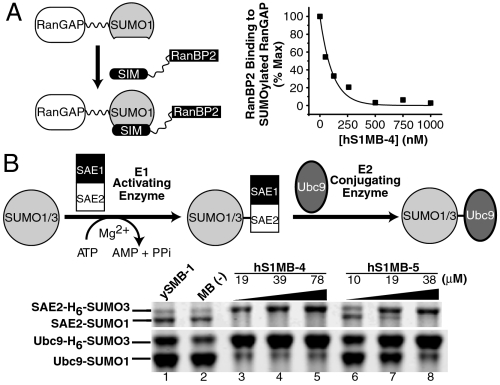
To investigate the potential utility of hSUMO1-specific monobodies as tools for studying SUMO biology, we examined their effects on three major processes: SUMO/SIM interactions, SUMOylation, and deSUMOylation. hS1MB-4 completely inhibited the SIM-mediated interaction between SUMO1-RanGAP (Ran GTPase activating protein) and RanBP2 (4, 20, 21) in a dose-dependent manner (Fig. 6A), further validating that these monobodies bind to the SIM-binding site as intended and demonstrating their efficacy as inhibitors of SUMO/SIM interactions.
Fig. 6.
(A, Left) Schematic of SIM-containing RanBP2’s interaction with SUMOylated RanGAP (modified with hSUMO1). (Right) Binding of RanBP2 to SUMO1-RanGAP in the presence of monobody hS1MB-4 in ELISA. (B, Upper) Schematic of the E1- and E2-dependent steps in the SUMOylation cascade. Covalently linked intermediates are formed sequentially between SUMO and E1 (SAE1/2) and E2 (Ubc9). (Lower) SDS-PAGE of SUMOylation reactions carried out in the presence of hS1MB-4 (lanes 3–5) and hS1MB-5 (lanes 6–8). Lanes 1 and 2 are negative controls with ySMB-1 and without a monobody, respectively. All reactions contained SAE1/2, Ubc9, and both hSUMO1 and hSUMO3 as substrates. Bands corresponding to the SAE2-SUMO and Ubc9-SUMO covalent intermediates for each isoform are indicated. His6-tagged SUMO3 (H6-SUMO3) was used to distinguish hSUMO1 from hSUMO3 on the gel.
We next examined effects of monobodies on SUMOylation by monitoring the in vitro formation of covalent complexes between SUMOs and the SUMO E1-activating (SAE1/SAE2) and E2-conjugating (Ubc9) enzymes of the SUMO conjugation cascade (Fig. 6B). In this assay, both hSUMO1 and hSUMO3 were present as substrates, enabling us to directly assess the isoform specificity of the monobodies. In the absence of a monobody or in the presence of the ySUMO-specific ySMB-1 monobody, E1 and E2 were conjugated with both hSUMO1 and hSUMO3 (Fig. 6B, lanes 1 and 2). In contrast, in the presence of either hS1MB-4 or hS1MB-5, conjugation of hSUMO1 was inhibited at the E1-dependent step, whereas hSUMO3 conjugation was enhanced (Fig. 6B, lanes 3–8). Because hSUMO1 and hSUMO3 compete for the same E1-activating enzyme, the enhancement of hSUMO3 conjugation is most likely because hSUMO1 was effectively eliminated as a competitor and thus the E1 enzyme was more available to hSUMO3. The potent inhibition of hSUMO1 conjugation by the monobodies was remarkable, because a SIM-based peptide inhibitor did not inhibit this process (9).
Superposition of the ySMB-1/ySUMO complex structure with the crystal structure of the E1/hSUMO1 complex (22) suggests that a monobody binding to hSUMO1 in a manner similar to ySMB-1 would not cause steric clashes with the structurally well-defined regions of E1. Rather, the monobody would be positioned in the trajectory of a long disordered loop in the SAE1 subunit (residues 175–205) (Fig. S7A). As a result, we speculate that steric clashes between the monobody and the SAE1 loop prevent binding of a monobody/hSUMO1 complex to E1, thus inhibiting SUMOylation at the E1-dependent step. The previously reported inhibitor based on a SIM peptide is much smaller and would not likely cause such a steric hindrance, explaining why it did not inhibit SUMOylation (9). Interestingly, hS1MB-4 was significantly more effective than hS1MB-5 in inhibiting SUMOylation (Fig. 6B), although their Kd values for hSUMO1 only differ by about twofold and their sizes are essentially identical (Fig. 4B). This difference in inhibition efficacy could be explained by subtle variations in the spatial arrangement of the two monobodies when bound to hSUMO1, consistent with the proposed mechanism.
Neither hS1MB-4 nor hS1MB-5 affected deSUMOylation as assayed in vitro by monitoring sentrin(SUMO)-specific protease 1 (SENP1) cleavage (23) at the hSUMO1 C-terminal diglycine sequence (Fig. S7B). Superposition of the ySMB-1/ySUMO structure with the structure of hSUMO1 bound to SENP1 (24) suggests no apparent clashes between the monobody and protease and that a monobody binding similarly to ySMB-1 would not inhibit the SENP1/hSUMO1 interaction, thus rationalizing our results.
Discussion
The structure-guided design of the SUMO-targeted library enabled us to generate diverse monobodies with good affinity to hSUMO1, without additional affinity maturation steps while also specifying their mode of interaction. These features make structure-guided library design a highly useful strategy. Although random mutagenesis methods (e.g., error-prone PCR) could have identified the important mutations at scaffold positions, such methods would not have provided the valuable mechanistic insights afforded by structural characterization. Clearly, the requirement of a starting crystal structure can be a major bottleneck in this approach. However, we have found that monobodies can act as effective “crystallization chaperones” for producing diffraction quality crystals of monobody-target complexes (25), suggesting that this requirement would not be prohibitively challenging for many systems. We anticipate that structure-guided library design would be effective in generating monobodies to diverse members of other protein families.
Despite success with hSUMO1, the SUMO-targeted library failed to produce monobodies with good affinity to hSUMO2. The SUMO-targeted library should, by design, contain a large number of SIM-mimicking FG loops, which in isolation should not effectively discriminate hSUMO1 and hSUMO2/3. Thus, this failure strongly suggests that there are negative design elements against SUMO2 binding still present in the SUMO-targeted library. More peripheral scaffold positions that were not diversified in the SUMO-targeted library could be important for binding hSUMO2. Iterative improvements could address these shortcomings in our current design.
The monobodies generated here have the highest affinity among SUMO/SIM inhibitors reported, although the binding mechanism of these monobodies is very similar to that of lower affinity SIM peptides (9, 18). The ySMB-1/ySUMO structure showed that the monobody and SIM interfaces are similar in buried surface area, shape complementarity values (26) and hydrophobicity (Table S2). Thus, none of these features alone can account for the increased monobody affinity. Instead, we propose that the conformational constraint of a SIM-like motif within the monobody scaffold reduces entropy loss for monobody binding resulting in increased affinity over SIM peptides. The approximate 10–50-fold higher affinity for monobodies (Kd as low as 118 nM) compared to SIM peptides (Kd ∼ 4–6 μM; ref. 18) is similar in magnitude to affinity increases for cyclic and disulfide constrained peptides over their linear equivalents (27–29), supporting our view.
The monobodies generated in this work discriminate SUMO isoforms by > 350-fold. Because these monobodies are also genetically encodable and function inside cells (16), they are likely to be powerful tools for dissecting SUMO biology. Although some natural SIMs have been reported to preferentially bind to one SUMO isoform over another (10–13), these SIMs discriminate individual isoforms by only about 10-fold, significantly less than the monobodies. We observed that contacts made by the FN3 scaffold with nonconserved residues at the periphery of the SIM-binding site were largely responsible for conferring isoform selectivity (Fig. 4 D–F). These contacts cannot be formed by SIM peptides, illustrating the challenge in achieving high isoform specificity with linear peptides or small molecules. Analogous to these observations, a monobody for an SH2 domain made similar scaffold-mediated contacts with peripheral, nonconserved residues, whereas the FG loop mimicked the binding mode of a phosphopeptide to the highly conserved phosphopeptide binding site (16). This modular combination of a conformationally constrained ligand mimic formed by the FG loop and a specificity filter formed by scaffold residues could make the monobody system a uniquely effective platform for generating selective inhibitors to many families of peptide binding domains where small molecule- and peptide-based inhibitors have largely failed to achieve high selectivity.
Although monobodies can be considered antibody mimics, intermolecular β-sheet formation and strong involvement of scaffold residues in the binding surfaces of SUMO-binding monobodies exemplify structural features that are not commonly observed in conventional antibodies. Interestingly, this prominent role of the FN3 scaffold represents an emerging trend in how monobodies recognize their targets. Of four target proteins for which the structure of a monobody/target complex has been determined, only one target, maltose binding protein, is recognized by the three variable loops of the monobody in the anticipated antibody-mimicking mode envisioned by conventional FN3 library designs (15, 30, 31). The other three targets are recognized using the FG loop/scaffold binding mode [refs. 16 and 32; Protein Data Bank (PDB) ID 2OCF]. This work showed that the use of the FG loop/scaffold binding surface was critical for binding to SUMOs, and we anticipate that the same is true for many other targets. Library design strategies based on the FG loop/scaffold surface are likely to be useful complements to existing, loop-directed strategies and are being investigated against diverse targets.
Materials and Methods
NMR epitope mapping, phage-display selection, phage ELISA, SPR experiments, and protein production were performed as previously described (15, 16, 30, 33). The ySMB-1/ySUMO crystal structure was determined by molecular replacement using previously determined structures (PDB ID codes 1FNA and 2EKE) as search models. Effects on SUMO1-RanGAP/RanBP2 interaction were assessed by ELISA in the presence of hS1MB-4. Effects on SUMOylation and deSUMOylation were assessed in vitro by monitoring SUMO conjugation to E1 and E2 SENP1 cleavage in the presence of monobodies. Further information can be found in the SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Erica Duguid for assistance with X-ray structure determination, Dr. Joseph Sachleben for assistance in NMR spectroscopy, and Dr. Anthony Kossiakoff for use of the BIAcore2000 instrument. This work was supported in part by the National Institutes of Health Grants R01-GM72688, R01-GM090324, and R21-CA132700 and by the University of Chicago Cancer Research Center. R.N.G. was supported in part by T32 GM007183-32A1. We acknowledge the use of the DNA sequencing core facility at the University of Chicago. We thank the staff of the Life Sciences Collaborative Access Team (LS-CAT) beamline at the Advanced Photon Source. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors for the ySMB-1/ySUMO complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3QHT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102294108/-/DCSupplemental.
References
- 1.Gareau JR, Lima CD. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 3.Vertegaal AC, et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 5.Bohren KM, Gabbay KH, Owerbach D. Affinity chromatography of native SUMO proteins using His-tagged recombinant UBC9 bound to CO2+-charged talon resin. Protein Expr Purif. 2007;54:289–294. doi: 10.1016/j.pep.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 7.Kerscher O. SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li YJ, Stark JM, Chen DJ, Ann DK, Chen Y. Role of SUMO:SIM-mediated protein-protein interaction in non-homologous end joining. Oncogene. 2010;29:3509–3518. doi: 10.1038/onc.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang PC, et al. Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J Biol Chem. 2010;285:5266–5273. doi: 10.1074/jbc.M109.088088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 12.Sekiyama N, et al. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J Biol Chem. 2008;283:35966–35975. doi: 10.1074/jbc.M802528200. [DOI] [PubMed] [Google Scholar]
- 13.Zhu J, et al. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J Biol Chem. 2008;283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chupreta S, Holmstrom S, Subramanian L, Iniguez-Lluhi JA. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol. 2005;25:4272–4282. doi: 10.1128/MCB.25.10.4272-4282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 16.Wojcik J, et al. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat Struct Mol Biol. 2010;17:519–527. doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 19.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem. 2000;275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 20.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 21.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatham MH, Hay RT. FRET-based in vitro assays for the analysis of SUMO protease activities. Methods Mol Biol. 2009;497:253–268. doi: 10.1007/978-1-59745-566-4_17. [DOI] [PubMed] [Google Scholar]
- 24.Shen L, et al. SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat Struct Mol Biol. 2006;13:1069–1077. doi: 10.1038/nsmb1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koide S. Engineering of recombinant crystallization chaperones. Curr Opin Struct Biol. 2009;19:449–457. doi: 10.1016/j.sbi.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 27.Millward SW, Fiacco S, Austin RJ, Roberts RW. Design of cyclic peptides that bind protein surfaces with antibody-like affinity. ACS Chem Biol. 2007;2:625–634. doi: 10.1021/cb7001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Neil KT, et al. Identification of novel peptide antagonists for GPIIb/IIIa from a conformationally constrained phage peptide library. Proteins. 1992;14:509–515. doi: 10.1002/prot.340140411. [DOI] [PubMed] [Google Scholar]
- 29.Nam NH, Ye G, Sun G, Parang K. Conformationally constrained peptide analogues of pTyr-Glu-Glu-Ile as inhibitors of the Src SH2 domain binding. J Med Chem. 2004;47:3131–3141. doi: 10.1021/jm040008+. [DOI] [PubMed] [Google Scholar]
- 30.Gilbreth RN, Esaki K, Koide A, Sidhu SS, Koide S. A dominant conformational role for amino acid diversity in minimalist protein-protein interfaces. J Mol Biol. 2008;381:407–418. doi: 10.1016/j.jmb.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci USA. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koide A, Abbatiello S, Rothgery L, Koide S. Probing protein conformational changes in living cells by using designer binding proteins: Application to the estrogen receptor. Proc Natl Acad Sci USA. 2002;99:1253–1258. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koide A, Koide S. Monobodies: Antibody mimics based on the scaffold of the fibronectin type III domain. Methods Mol Biol. 2007;352:95–109. doi: 10.1385/1-59745-187-8:95. [DOI] [PubMed] [Google Scholar]
- 34.Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 35.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.