Abstract
Rearrangements of the MLL (ALL1) gene are very common in acute infant and therapy-associated leukemias. The rearrangements underlie the generation of MLL fusion proteins acting as potent oncogenes. Several most consistently up-regulated targets of MLL fusions, MEIS1, HOXA7, HOXA9, and HOXA10 are functionally related and have been implicated in other types of leukemias. Each of the four genes was knocked down separately in the human precursor B-cell leukemic line RS4;11 expressing MLL-AF4. The mutant and control cells were compared for engraftment in NOD/SCID mice. Engraftment of all mutants into the bone marrow (BM) was impaired. Although homing was similar, colonization by the knockdown cells was slowed. Initially, both types of cells were confined to the trabecular area; this was followed by a rapid spread of the WT cells to the compact bone area, contrasted with a significantly slower process for the mutants. In vitro and in vivo BrdU incorporation experiments indicated reduced proliferation of the mutant cells. In addition, the CXCR4/SDF-1 axis was hampered, as evidenced by reduced migration toward an SDF-1 gradient and loss of SDF-1–augmented proliferation in culture. The very similar phenotype shared by all mutant lines implies that all four genes are involved and required for expansion of MLL-AF4 associated leukemic cells in mice, and down-regulation of any of them is not compensated by the others.
Keywords: leukemic cells’ migration, bone marrow colonization of leukemic cells
Rearrangements of the MLL/ALL1 gene occur in 20% of acute lymphoblastic leukemias (ALL) and in 5–6% of acute myeloid leukemias (AML) (1, 2). A high percentage of ALLs with MLL rearrangement show biphenotypic traits. The epidemiology of MLL-associated leukemias is unique (3). They predominate infant acute leukemia, and account for the majority of therapy-related leukemias occurring in cancer patients treated with etoposide (VP-16) or doxorubicin (4). The very short latency of the disease (3, 4) and its aggressive nature suggest the involvement of a powerful oncogene. Most MLL rearrangements are due to reciprocal chromosome translocations that link MLL to any of >100 partner genes (5). This results in production of oncogenic MLL proteins, comprising the N-terminal MLL polypeptide fused in frame to the C-terminal fragment of a partner protein. The partner proteins most commonly found associated with MLL (AF4, AF9, ENL, AF10, ELL) are transcriptional elongation factors (6). Because these factors physically associate (6, 7), the function of the most common partner polypeptides within MLL fusion proteins appears to be the recruitment of the other elongation factors, and consequently the augmentation of target genes transcription (8–11).
Initial gene expression analysis of leukemic cells from ALL patients indicated up-regulation of HOXA9 and HOXA10 and their essential cofactor MEIS1 in cells with the t(4;11) chromosome translocation and MLL-AF4 (12). Subsequent gene expression profiling of leukemic cells with MLL rearrangements from patients showed up-regulation of MEIS1 and HOXA5–HOXA10, as well as other genes (13–15). Recently, application of ChIP-seq determined that MEIS1, HOXA7, HOXA9, and HOXA10 are among the 226 primary targets of MLL-AF4 in a human ALL cell line (8). Up-regulation of similar Hoxa genes and Meis1 were obtained in murine model systems (16–19). Many studies applying overexpression of HOXA9 and/or MEIS1 showed the requirement for both proteins in induction of acute leukemia in mice (20, 21). HOXA9 and MEIS proteins function within the same protein complex (with PBX) and occupy simultaneously the same gene targets (22, 23).
The consistent up-regulation of several HOXA genes in MLL-associated leukemias raised the issue whether HOXA5-10 specify a “HOX code” (18, 24), in which each of the members contributes to pathogenicity and is required for it. Experiments to resolve the issue involved induction of leukemia in mice knocked out for HOXA9. The results varied with the particular MLL fusion, and with the way by which the MLL fusion was introduced into the mutant mice (18, 25, 26). Because murine leukemic cells, in particular those involved in AML, might not reproduce all of the processes occurring in human ALL cells with MLL rearrangements, we decided to examine the biological effects of knockdown of HOX A7,9,10 or MEIS1 in the human leukemic cell line RS4;11, expressing MLL-AF4.
Results
Knockdown of HOXA Genes or MEIS1 Impairs Engraftment of RS4;11 Cells.
The precursor B-cell line RS4:11, a classical biphenotypic cell line (27), was knocked down for the HOXA7, A9, A10 or MEIS1 genes. This was done by lentiviral-mediated transduction of constructs encoding short hairpin (sh) RNAs, designed to silence the genes (Table S1). The shRNAs sequences were selected to minimize homology to nontarget mRNAs and avoid perfect dsRNA stretches of >11 bp; also, no IFN response (28) was detected. We applied Western analysis to determine the extent of silencing affected by the different shRNA constructs. The identity of the HOXA/MEIS1 proteins was further confirmed by their comigration with the corresponding epitope-tagged (Flag or HA) proteins produced in 293T cells expressing the former (Fig. 1A). In most cases we found more than one construct encoding unique shRNA, potent in down-regulation of >80% of the protein; cell lines showing the most efficient knockdown were chosen for further work (Fig. 1A). As controls we used RS4;11 cells infected with the lentiviral vector, and cells expressing HOXA9 shRNA found to be nonpotent in elimination of the protein. Two recent studies indicated down-regulation of MEIS1 RNA (29, 30), as well as of RNAs of multiple HOXA genes (30), in MLL-rearranged cells knocked down for HOXA9. To examine whether similar cross-modulation is associated with MEIS1, HOXA7, and HOXA10 knockdowns, we analyzed the abundance of HOXA/MEIS1 RNAs by applying the technology of NanoString nCounter gene expression system, which captures and counts individual mRNA transcripts by their hybridization to a multicomplex probe library (31). Cultured lymphoid RS4;11 and SEM cells, both expressing MLL-AF4, were treated with MEIS1, HOXA7, HOXA10, or scrambled siRNAs and the extracted RNAs were reacted with a library containing probes for MEIS1 and HOXA and other mRNAs (SI Materials and Methods). Although cells treated with a particular siRNA were down-regulated for its expression, the abundance of the other HOXA (except HOXA5) or MEIS1 RNAs was not reduced (Fig. 1B).
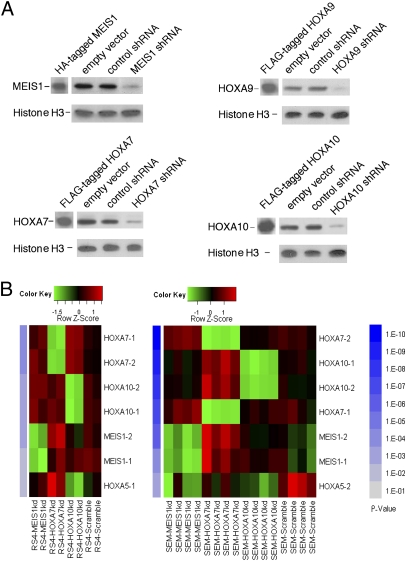
Fig. 1.
Knockdown of MEIS1 and HOXA by lentiviral-mediated delivery of shRNAs or by treatment with siRNAs. (A) Western analysis of MEIS1 and HOXA7, A9, and A10 proteins in nuclear extracts of RS4;11 cells expressing shRNAs directed against one of the four genes (constructs MEIS1_3a, HOXA9_1, HOXA7_1, HOXA10_4a; 3,1,4 specify the gene's sequence targeted, and a or b correspond to different plasmid clones of the same construct), or a vector construct, or a control construct. Identity of the proteins was confirmed by demonstrating their comigration with the epitope-tagged proteins expressed in 293T cells (A). Molecular mass (estimated by comparison of the electrophoretic mobility to that of BioRad protein standards) of MEIS1, HOXA7, HOXA9 and HOX A10 proteins are 49, 34, 36, and 50 kDa, respectively. Histone H3 was used as loading control. (B) Knockdown of MEIS1, HOXA7, or HOXA10 by treatment of RS4;11 and SEM cells with siRNAs does not show cross down modulation between the three mRNAs. NanoString nCounter gene expression system was used to quantitate abundance of mRNAs. Genes are ordered by their P values from top to bottom. Controls are cells treated with scramble siRNA. Two to four biological replicas were analyzed. Probes for two different regions of most genes were used (e.g., HOXA10-1, HOXA10-2). kd, knocked down. Additional details are provided in SI Materials and Methods.
We also considered the possibility that the shRNAs induced a general deleterious effect on the cells. However, we found this unlikely because shRNAs directed against different sequences of the same gene and downregulating its expression induced very similar biological and biochemical effects when contrasted with cells expressing shRNAs inefficient in silencing of the gene which behaved like vector-infected or noninfected cells (see Figs. 2 and 6A and Table S2).
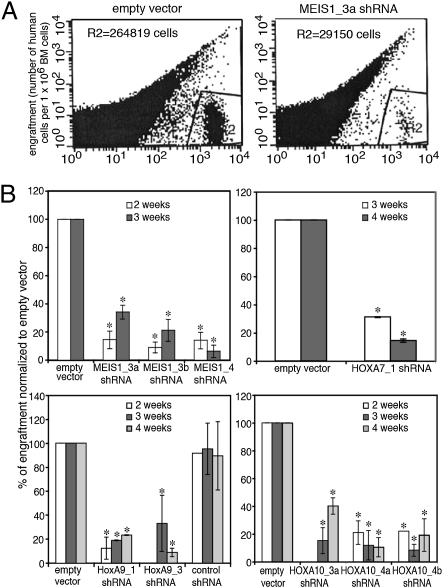
Fig. 2.
Impaired engraftment of RS4;11 cells knocked down for MEIS1 or HOXA into the BM of NOD/SCID mice. Cells were obtained from the BM at 2–4 wk after transplantation, and percentage of human cells was assayed using human-specific mAbs. (A) Examples of bone marrow obtained 3 wk post transplantation. Gate R2 shows human engrafting cells; number of the cells in R2 is indicated at the top of each image. (B) Percentage of engrafting mutant cells is compared with that of cells transfected with empty vector. *P < 0.05. Results correspond to average ± SD of five independent experiments performed in duplicate.
Fig. 6.
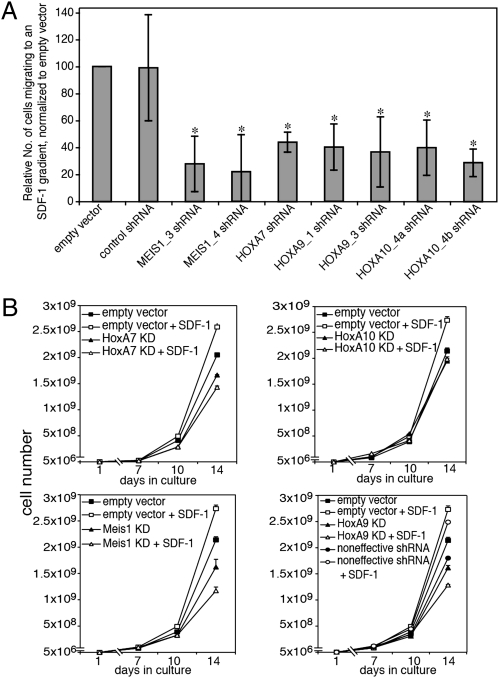
Reduced migration of RS4;11 mutants to an SDF-1 gradient, and loss of SDF-1–dependent growth augmentation in culture. (A) SDF-1 was added to lower chamber of transwells; manipulated RS4;11 cells were placed at the upper chamber and allowed to migrate toward the SDF-1 gradient. Percentage of cells migrated was evaluated by FACS. Results show the relative number of cells migrated, normalized to the number of vector-infected cells migrated, set as 100 (*P < 0.05). Results shown are average ± SD of five independent experiments. (B) Manipulated RS4;11 cells (shRNA constructs as in Fig. 4) were seeded into flasks and grown for 2 wk in RPMI containing 10% FCS, in the presence or absence of recombinant SDF1-α. Curves represent average ± SD of four different experiments. Constructs as in Fig. 4. KD, knocked down.
The transplanted NOD/SCID mice were killed at 1 d (16 h), 1–5 wk (mice injected with control cells), or 1–6 wk (mice injected with mutant cells). At 4 and 6 wk after injection of control and mutant cells, respectively, increasing numbers of animals appeared ill or succumbed to the malignancy. Initially, engraftment in the BM and spleen was determined. At 21 and 26 d, the frequency of intact RS4;11 cells was fivefold higher in the bone marrow (BM) compared with the spleen (Fig. S1). An even higher tendency to accumulate in the BM rather than in the spleen (10- to 20-fold) was observed for the manipulated cells. Therefore, in the next experiments, we focused on the fate of the cells in the BM. In this analysis, the RS4;11 cells were sorted by virtue of surface expression of CD45; all these cells were positive for GFP, indicating expression of the lentivirus. The results demonstrated that similar numbers of mutant and control cells populated the bone marrow during the first day after transplantation (Table S2). However, at 2–4 wk reduced engraftment of the mutant cells was indicated (Fig. 2 and Table S2). Only at 5 wk, when substantial numbers of mice injected with the control cells had already died, the frequency of the mutant cells in BM was not far off the frequency of the control cells in BM of the surviving animals (Table S2). These results implied that the knockdown cells did not vary from the control cells in arrival (homing) to the bone marrow, but were impaired during subsequent stages.
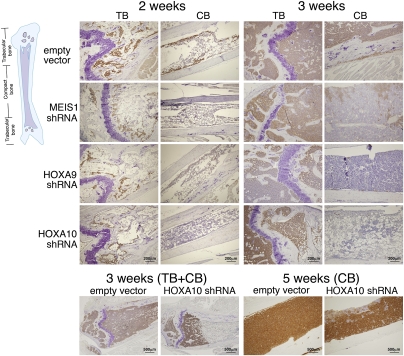
To compare the location of the control cells and of knockdown RS4;11 cells in the BM, immunohistochemical analysis was carried out on BM sections from the femurs and tibias of mice 1–5 wk after transplantation (Fig. 3). At 1 wk (not shown) and 2 wk (Fig. 3), both mutant and control cells were found in the trabecular bone (TB) area, composed of metaphysis and epiphysis, but not in the compact bone (CB, also termed diaphysis or bone shaft) area. At 3 wk after transplantation, a clear difference appeared between the control and mutant cells: whereas the former filled up both the TB and CB areas, the latter were localized nearly only to the TB region (Fig. 3). At 5 wk, the knockdown cells packed the TB and expanded into large zones within the CB (Fig. 3, bottom right). Thus, although the control human cells spread from the trabecular to the compact bone area between the second and third week post transplantation, the spread of the knockdown cells lingered by ∼2 wk. The borders between the TB areas of the bone inhabited by human cells, and the CB areas which were not, were quite sharp in some sections (Fig. 3, bottom left). This possibly reflected the presence of two discrete zones within the bone itself. Because previous studies have shown that SDF-1 (CXCL12), a key chemotactic factor, is expressed at higher levels in the epiphysis compared with the diaphysis (32, 33), we applied immunohistochemistry to examine the location of SDF-1 within the BM of mice transplanted with mutant or control RS4;11 cells (Fig. S2). Indeed, SDF-1 localized mostly to the trabecular area, but there was no difference in its abundance between BM containing control or mutant cells.
Fig. 3.
Immunohistochemical analysis of mutant and control RS4;11 cells in BM of transplanted NOD/SCID mice indicates a lag in spreading of the mutant into the CB area. BM sections were prepared at different times after transplantation. Human cells appear brown. Cells transplanted harbored empty vector, MEIS1_3b, HOXA9_1, or HOXA10_4a constructs.
Proliferation of Mutant RS4;11 Cells Is Decreased.
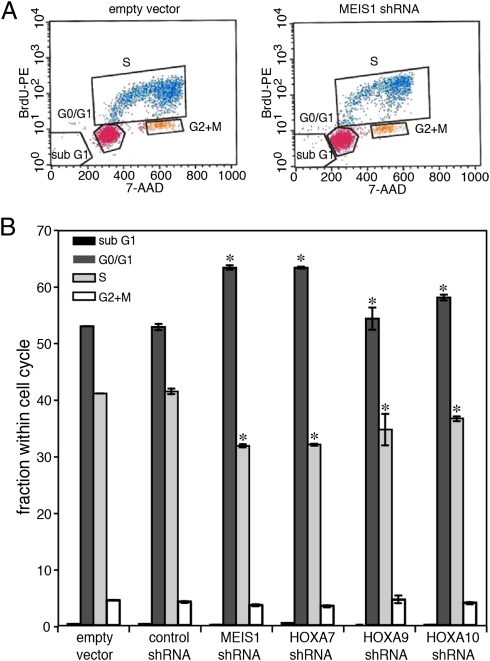
One potential reason for impairment in engraftment of the knocked-down cells is reduced proliferation. This was first examined in tissue culture. The cells were labeled with BrdU and subsequently analyzed by FACS for incorporation of BrdU and for distribution within the cell cycle (Fig. 4). The percentage of cells in the S phase was reduced in the mutant cells, and a higher percentage of them was identified in the G0/G1 phase. This indicated diminished proliferation of the knockdown cells. Importantly, no cells in sub G1 were observed in any of the lines, excluding the possibility that the alteration in growth was due to enhanced apoptosis. Also, apoptosis was not observed in experiments with Annexin (not shown). Next we performed in vivo BrdU labeling in mice 3 wk after their transplantation. FACS analysis (Fig. 5) indicated that the mutant cells were reduced in incorporation of BrdU and hence in proliferation. The lower proliferation in the BM is likely to explain, at least in part, the impaired engraftment of the knockdown cells, as well as the delayed spread to the compact bone area (Discussion).
Fig. 4.
Decreased proliferation of cultured manipulated RS4;11 cells. Cells in culture were labeled with BrdU and subjected to FACS analysis of the cell cycle. (A) Examples of the analysis. Cells harbored the empty lentiviral vector, or the MEIS1_3b, shRNA construct. (B) Percentage of cells at different stages of the cell cycle are compared with that of cells transfected with empty vector. *P < 0.05. Results shown are average ± SD of two independent experiments performed in quadruplicate.
Fig. 5.
Diminished proliferation of knockdown cells within BM of transplanted mice. Transplanted mice were labeled with BrdU. BM cells labeled with human-specific Ab were subjected to FACS analysis to determine fraction of human cells that have incorporated BrdU. (A) Example of analysis. Cells contained control constructs or knockdown constructs as in Fig. 4. M1 and M2 were determined according to the pattern of the IgG control, where M2 encompassed <1% of the total IgG counts. (B) Percentage of BrdU-positive cells within human cell population is compared with that obtained from BM of mice injected with vector-infected cells. *P < 0.05. Results are average ± SD of four independent experiments.
Flawed SDF-1–Dependent Chemotaxis and Proliferation of Mutant Cells.
Extensive experimental evidence has indicated the importance of the CXCR4/SDF-1 axis for BM engraftment of both normal stem/progenitor cells and leukemic cells (34–36). Therefore, we compared the control and knockdown cells for migration toward an SDF-1 gradient and for acceleration in proliferation in response to the presence of SDF-1 in the culture's medium. Migration of the mutant cells to an SDF-1 gradient was reduced by 56–78% compared with control cells (Fig. 6A). In parallel, growth kinetics assays indicated that inclusion of SDF-1 in the cultures' media enhanced proliferation of the control but not of the mutant cells (Fig. 6B), in which SDF-1 did not affect or reduce growth. Thus, knockdown of of the genes altered response to SDF-1. In an attempt to pinpoint the stage at which the CXCR4/SDF-1 axis was impaired, the cells were compared for abundance of the CXCR4 receptor on their surface; the abundance was found to be similar in the mutants and controls (Fig. S3). Furthermore, comparison of the amounts of GTP-bound RAC, an activated effector of CXCR4/SDF-1 (37), did not show a difference between HOXA10 and MEIS1 knockdowns and control cells (Fig. S4). Finally, the extent of total or activated (phosphorylated) AKT, ERK1/2, and PKCzeta, downstream mediators of RAC-GTP (38), did not vary (not shown) between HOXA7, HOXA9, HOXA10, and MEIS1 mutants and control cells, prior or subsequent to treatment with SDF-1 (the amount of pAKT, but not pERK1/2, increased after treatment of all cells with 200 ng/mL of SDF-1 for 2 min). In addition to the CXCR4/SDF-1 axis, other pathways are involved in engraftment and retention of stem/precursor cells and of leukemic cells in the BM (39, 40). These pathways mediate interactions of the cells with BM niches, and include adhesive interactions between the α4β1 and α5β1 integrin proteins on the cells' surface and fibronectin and /or VCAM-1 (both produced in the niche), the interaction between the cells receptor c-Kit and the niche's stem cell factor (SCF) ligand, and interaction between CD44 on the cells' surface and hyaluronan (HA) in the niche. To this end, we compared the mutants and control cells for the presence on their surface of VLA4, VLA5 (components of α4β1 and α5β1, respectively), the integrin LFA-1, c-Kit, and CD44. c-Kit was not present on the cells' surface, and the abundance of the other proteins was similar on mutant and control cells (Fig. S5). In addition, adhesion of the mutant and control cells to fibronectin, or VCAM1 was roughly equivalent, and adhesion of the mutants to HA, was somewhat increased (Fig. S6).
Discussion
By knocking down HOXA7 or HOXA9 or HOX10 or MEIS1 in RS 4;11 cells, we found impaired leukemogenicity in NOD/SCID mice of each of the cell lines. Although knockdown of HOXA9 was previously shown to down-regulate the transcription of other HOXA genes and MEIS1 (30), such cross–down-regulation was not found here after knockdown of HOXA7, HOXA10, or MEIS1. The four mutant cell lines showed similar biological and biochemical features culminating in an impaired capacity to proliferate in the NOD/SCID mice. This suggests that the four genes are all involved in expansion of leukemic MLL-AF4–associated cells in vivo, and loss of each cannot be compensated by the intact other three. Further insights to the precise functions of the genes should emerge from a comparative analysis of the primary and secondary targets of the four proteins in leukemic cells with the t(4;11) abnormality, together with analysis of infant ALL cases positive for t(4;11) and expressing MEIS1, but lacking expression of HOXA (41, 42).
The main organ targeted by the injected RS4;11 cells was the bone marrow and not the spleen, and the mutant cells arrived to the target at similar time and efficiency as the control cells. Both mutant and control cells initially inhabited the trabecular area of the bone, wherein the control cells proliferated considerably more rapidly. In addition to their slower proliferation, the knockdown cells were delayed in their spread into the compact bone. The preferred initial arrival site and growth in the trabecular area of both types of cells might be due to the higher concentration of SDF-1 in that region of the bone. The delayed translocation of the mutant cells into the compact bone could reflect the longer time required for them to reach a threshold density for overflowing into the compact bone, or a shortage within the mutant cells of a substance necessary for growth in that compartment.
We identified, in our mutants, impairment in two parameters known to play roles in the engraftment of hematopoietic cells into the bone marrow: proliferation, and response to the SDF-1 chemokine. Both HOXA and MEIS1 mutants exhibited reduced proliferation within the bone marrow of transplanted mice and a shift in the cell cycle of cultured cells toward smaller numbers in S phase and diminished multiplication. The defects we observed in our mutant cells in proliferation in vitro and in engraftment/proliferation in mice that have undergone transplantation and irradiation are reminiscent of well-known results of Lawrence et al. (43), who showed that Hoxa9 knockout murine progenitor marrow cells homed normally to the bone marrow but exhibited reduction in proliferation in culture, as well as in in vivo repopulation ability. Also, fetal liver cells from Meis1 knockout mice were found to be impaired in engraftment and repopulation of irradiated mice (44). In this context, it should be noted that numerous studies applying overexpression or down-regulation approaches to normal hematopoietic stem cells (45) or leukemia stem cells (17) showed a role of HOX and MEIS1 genes in self-renewal (which involves expansion of cells and suppression of their differentiation potential). Direct linkage between MEIS1, cell proliferation, and the cell cycle has emerged very recently from microarray analysis (46, 47), suggesting MEIS1 regulation of genes acting in DNA replication and in cell-cycle entry (Cdk2, Cdk6, CdkN3, Cdc7, cyclin D3, and others). Furthermore, several HOX proteins were shown to bind DNA replication origins in vivo and interact with the replication regulator geminin (ref. 48 and references therein). Thus, the silencing of MEIS1 and HOXA might slow cell cycle entry and inhibit initiation of replication, respectively.
The second biological function found to be impaired in the mutant cells was the response to the SDF-1 chemokine. The flaw in the CXCR4-SDF-1 axis was shown here by in vitro assays of SDF-1-dependent migration and growth. Surprisingly, homing of the mutant cells to the bone marrow 16 h post transplantation (dependent on the CXCR4-SDF-1 axis) was similar to that of the control cells. We note, however, that hematopoietic progenitors knocked out for Hoxa9, although defective in engraftment, exhibited normal homing (43); moreover, in addition to the CXCR4/SDF-1 axis, the VLA-4/VCAM-1 pathway was also found operative in BM homing of precursor cells and could compensate for the loss of the former (49). Our intensive attempts to pinpoint the defective component within the CXCR4/SDF-1 pathway were not successful. However, because this pathway interacts with other routes such as Wnt (50) and Notch (51), the critical depleted component might reside within those routes or others. Because proliferation of the mutants in culture is slowed down also in the absence of SDF-1, we consider it likely that the defect in proliferation and the defect in response to SDF-1 are due to alterations in expression of different MEIS1/HOXA targets.
Two recent reports used the shRNA knockdown approach to show roles for MEIS1 (46) and HOXA9 (30) in survival and leukemia induction of mouse MLL-AF9 knockin cells transplanted into C57Bj/6 mice, and human SEMK2 cells (expressing MLL-AF4) transplanted into SCID-beige mice, respectively. Most mice injected with MEIS1 knockdown cells survived even after 170 d. In the second system, the transplanted parental SEMK2 cells populated the spleens, but the knockdown cells failed to grow in the mice. In both systems, cultured human cells expressing the shRNAs were slowed in their growth, and exhibited cell cycle arrest and apoptosis. We suggest that the milder effects shown by our knockdown cells in vitro and their ability to grow, although more slowly, in the transplanted mice is due to a less robust silencing of the target genes or to the differences in the cells studied. The preferred homing and engraftment into the bone marrow and not to the spleen in our experiments is likely due to the variance in the cell lines or in the recipient mouse strains.
Materials and Methods
For engraftment experiments, RS4;11 cells infected with viruses expressing shRNAs were injected i.v. 24 h after sublethal irradiation. At various time points single cell suspensions were prepared from the BM of transplanted mice and engraftment was assayed by flow cytometry. BrdU incorporation in cultured cells and in vivo was assayed by flow cytometry analysis. Cell migration assays were done in transwells, and immunochemical staining was performed on mouse BM sections. Details of all methods, constructs, and antibodies used are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are indebted to Professor Ronen Alon for his support and numerous discussions. We also thank Drs. Jeff Palatini, Cecillia Fernandez, and Yaron Vagima, as well as Amir Schajnovitz, Kfir Lapid, and Ziv Schulman for their help, insightful advice, and valuable suggestions. This work was supported by grants from the US-Israel binational Science Foundation, the Israel Cancer Research Fund, the Hal and Marlene Spitz Family Philanthropic Fund, the D. Levinson Estate and Bogen Charitable Trust, the Weizmann-Chaim Sheba Medical Center Collaboration, the Leona M. and Harry B. Helmsley Charitable Trust, and National Institutes of Health Grants CA 128609 and RO1GM075141.
Footnotes
The authors declare no conflict of interest.
2Deceased October 8, 2009.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103154108/-/DCSupplemental.
References
- 1.Canaani E, et al. ALL-1/MLL1, a homologue of Drosophila TRITHORAX, modifies chromatin and is directly involved in infant acute leukaemia. Br J Cancer. 2004;90:756–760. doi: 10.1038/sj.bjc.6601639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Somervaille TC, Cleary ML. Grist for the MLL: How do MLL oncogenic fusion proteins generate leukemia stem cells? Int J Hematol. 2010;91:735–741. doi: 10.1007/s12185-010-0579-8. [DOI] [PubMed] [Google Scholar]
- 3.Greaves M. Molecular genetics, natural history and the demise of childhood leukaemia. Eur J Cancer. 1999;35:1941–1953. doi: 10.1016/s0959-8049(99)00296-8. [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Relling MV. Topoisomerase II inhibitor-related acute myeloid leukaemia. Br J Haematol. 2000;109:13–23. doi: 10.1046/j.1365-2141.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 5.Meyer C, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 6.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 7.Erfurth F, Hemenway CS, de Erkenez AC, Domer PH. MLL fusion partners AF4 and AF9 interact at subnuclear foci. Leukemia. 2004;18:92–102. doi: 10.1038/sj.leu.2403200. [DOI] [PubMed] [Google Scholar]
- 8.Guenther MG, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller D, et al. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin C, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozovskaia T, et al. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4 : 11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong SA, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 14.Yeoh EJ, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 15.Rozovskaia T, et al. Expression profiles of acute lymphoblastic and myeloblastic leukemias with ALL-1 rearrangements. Proc Natl Acad Sci USA. 2003;100:7853–7858. doi: 10.1073/pnas.1132115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 17.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar AR, et al. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- 19.Cano F, Drynan LF, Pannell R, Rabbitts TH. Leukaemia lineage specification caused by cell-specific Mll-Enl translocations. Oncogene. 2008;27:1945–1950. doi: 10.1038/sj.onc.1210818. [DOI] [PubMed] [Google Scholar]
- 20.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 21.Sitwala KV, Dandekar MN, Hess JL. HOX proteins and leukemia. Int J Clin Exp Pathol. 2008;1:461–474. [PMC free article] [PubMed] [Google Scholar]
- 22.Shen WF, et al. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang GG, Pasillas MP, Kamps MP. Persistent transactivation by meis1 replaces hox function in myeloid leukemogenesis models: Evidence for co-occupancy of meis1-pbx and hox-pbx complexes on promoters of leukemia-associated genes. Mol Cell Biol. 2006;26:3902–3916. doi: 10.1128/MCB.26.10.3902-3916.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton SJ, et al. Continuous MLL-ENL expression is necessary to establish a “Hox Code” and maintain immortalization of hematopoietic progenitor cells. Cancer Res. 2005;65:9245–9252. doi: 10.1158/0008-5472.CAN-05-1691. [DOI] [PubMed] [Google Scholar]
- 25.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- 27.Stong RC, Korsmeyer SJ, Parkin JL, Arthur DC, Kersey JH. Human acute leukemia cell line with the t(4;11) chromosomal rearrangement exhibits B lineage and monocytic characteristics. Blood. 1985;65:21–31. [PubMed] [Google Scholar]
- 28.Cullen BR. Enhancing and confirming the specificity of RNAi experiments. Nat Methods. 2006;3:677–681. doi: 10.1038/nmeth913. [DOI] [PubMed] [Google Scholar]
- 29.Hu YL, Fong S, Ferrell C, Largman C, Shen WF. HOXA9 modulates its oncogenic partner Meis1 to influence normal hematopoiesis. Mol Cell Biol. 2009;29:5181–5192. doi: 10.1128/MCB.00545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faber J, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 32.Sun YX, et al. Skeletal localization and neutralization of the SDF-1(CXCL12)/CXCR4 axis blocks prostate cancer metastasis and growth in osseous sites in vivo. J Bone Miner Res. 2005;20:318–329. doi: 10.1359/JBMR.041109. [DOI] [PubMed] [Google Scholar]
- 33.Xie Y, et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature. 2009;457:97–101. doi: 10.1038/nature07639. [DOI] [PubMed] [Google Scholar]
- 34.Kim CH, Broxmeyer HE. In vitro behavior of hematopoietic progenitor cells under the influence of chemoattractants: Stromal cell-derived factor-1, steel factor, and the bone marrow environment. Blood. 1998;91:100–110. [PubMed] [Google Scholar]
- 35.Peled A, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 36.Burger JA, Bürkle A. The CXCR4 chemokine receptor in acute and chronic leukaemia: A marrow homing receptor and potential therapeutic target. Br J Haematol. 2007;137:288–296. doi: 10.1111/j.1365-2141.2007.06590.x. [DOI] [PubMed] [Google Scholar]
- 37.Cancelas JA, Jansen M, Williams DA. The role of chemokine activation of Rac GTPases in hematopoietic stem cell marrow homing, retention, and peripheral mobilization. Exp Hematol. 2006;34:976–985. doi: 10.1016/j.exphem.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Thomas EK, Cancelas JA, Zheng Y, Williams DA. Rac GTPases as key regulators of p210-BCR-ABL-dependent leukemogenesis. Leukemia. 2008;22:898–904. doi: 10.1038/leu.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwasaki H, Suda T. Cancer stem cells and their niche. Cancer Sci. 2009;100:1166–1172. doi: 10.1111/j.1349-7006.2009.01177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papayannopoulou T, Scadden DT. Stem-cell ecology and stem cells in motion. Blood. 2008;111:3923–3930. doi: 10.1182/blood-2007-08-078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trentin L, et al. Two independent gene signatures in pediatric t(4;11) acute lymphoblastic leukemia patients. Eur J Haematol. 2009;83:406–419. doi: 10.1111/j.1600-0609.2009.01305.x. [DOI] [PubMed] [Google Scholar]
- 42.Stam RW, et al. Gene expression profiling-based dissection of MLL translocated and MLL germline acute lymphoblastic leukemia in infants. Blood. 2010;115:2835–2844. doi: 10.1182/blood-2009-07-233049. [DOI] [PubMed] [Google Scholar]
- 43.Lawrence HJ, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hisa T, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauvageau G, Iscove NN, Humphries RK. In vitro and in vivo expansion of hematopoietic stem cells. Oncogene. 2004;23:7223–7232. doi: 10.1038/sj.onc.1207942. [DOI] [PubMed] [Google Scholar]
- 46.Kumar AR, et al. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Argiropoulos B, et al. Linkage of the potent leukemogenic activity of Meis1 to cell-cycle entry and transcriptional regulation of cyclin D3. Blood. 2010;115:4071–4082. doi: 10.1182/blood-2009-06-225573. [DOI] [PubMed] [Google Scholar]
- 48.Salsi V, et al. HOXD13 binds DNA replication origins to promote origin licensing and is inhibited by geminin. Mol Cell Biol. 2009;29:5775–5788. doi: 10.1128/MCB.00509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonig H, Priestley GV, Papayannopoulou T. Hierarchy of molecular-pathway usage in bone marrow homing and its shift by cytokines. Blood. 2006;107:79–86. doi: 10.1182/blood-2005-05-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh MC, et al. Activation of Wnt5A signaling is required for CXC chemokine ligand 12-mediated T-cell migration. Blood. 2009;114:1366–1373. doi: 10.1182/blood-2008-08-175869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams CK, et al. Regulation of CXCR4 by the Notch ligand delta-like 4 in endothelial cells. Cancer Res. 2008;68:1889–1895. doi: 10.1158/0008-5472.CAN-07-2181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.