Abstract
Wild organisms are under increasing pressure to adapt rapidly to environmental changes. Predicting the impact of these changes on natural populations requires an understanding of the speed with which adaptive phenotypes can arise and spread, as well as of the underlying mechanisms. However, our understanding of these parameters is poor in natural populations. Here we use experimental and molecular approaches to investigate the recent emergence of resistance in eastern populations of North American house finches (Carpodacus mexicanus) to Mycoplasma galliseptum (MG), a severe conjunctivitis-causing bacterium. Two weeks following an experimental infection that took place in 2007, finches from eastern US populations with a 12-y history of exposure to MG harbored 33% lower MG loads in their conjunctivae than finches from western US populations with no prior exposure to MG. Using a cDNA microarray, we show that this phenotypic difference in resistance was associated with differences in splenic gene expression, with finches from the exposed populations up-regulating immune genes postinfection and those from the unexposed populations generally down-regulating them. The expression response of western US birds to experimental infection in 2007 was more similar to that of the eastern US birds studied in 2000, 7 y earlier in the epizootic, than to that of eastern birds in 2007. These results support the hypothesis that resistance has evolved by natural selection in the exposed populations over the 12 y of the epizootic. We hypothesize that host resistance arose and spread from standing genetic variation in the eastern US and highlight that natural selection can lead to rapid phenotypic evolution in populations when acting on such variation.
Keywords: genetic basis of resistance, host–parasite co-evolution, immunosuppression, quantitative RT-PCR, emerging disease
A pressing question in modern biology is how quickly natural populations can respond to anthropogenic selection pressures (1). Integral to predicting evolvability is an understanding of the speed with which adaptive phenotypes can spread in a population and their underlying molecular bases. Adaptive phenotypic changes have been suggested to evolve rapidly in wild animals, sometimes within a few generations (2, 3), but few studies confirm that such changes arise through the selection of adaptive genotypes rather than from plastic consequences of gene-by-environment interactions (4). Although there is growing appreciation that phenotypic plasticity might play an important role in the evolution and spread of adaptive phenotypes, ultimately, evolution requires adaptive changes in gene frequencies (5, 6). Studies that have been able to tease evidence of evolution from other mechanisms of phenotypic change not only provide some of the most convincing evidence of evolution by natural selection, but also enhance our understanding of evolvability and its underlying processes (7, 8). However, identifying beneficial mutations and measuring their spread in response to a selective agent is not straightforward (8–10). An alternative may be to identify apparent evidence of evolutionary change and then measure phenotypic and accompanying molecular changes within an experimental framework designed to rule out other sources of influence.
Host–parasite systems represent dynamic interactions and so provide outstanding models for studying evolutionary change (11, 12). For example, when parasites represent novel and intense selection pressures, it is possible to document the spread of host resistance within populations over just a few generations (13), as has been recorded recently in eastern US house finches (Carpodacus mexicanus) (14–16). House finches are native to western North America, but in 1940 a founder population was introduced to the eastern United States near New York City. By 1990, house finches had spread throughout much of eastern North America and numbered over 100 million, although they remained geographically isolated from their western counterparts (17). In 1994, Mycoplasma gallisepticum (MG), a bacterium found in poultry (18) but not in songbirds (19), was detected in the eastern populations of house finches (20). MG causes respiratory tract and conjunctivitis infections, and as a result, many millions of eastern house finches died between 1994 and 1998 (14). Naturally infected captive finches confirmed mortality due to MG-induced conjunctivitis (21) (Fig. 1A). However, declines in eastern populations began stabilizing in 1998–1999 (14), experimental infections of 1999-born finches in Alabama showed precursory evidence of resistance (15, 16), and evidence from this 2007 research shows MG conjunctivitis reduced to endemic levels (Materials and Methods).
Fig. 1.
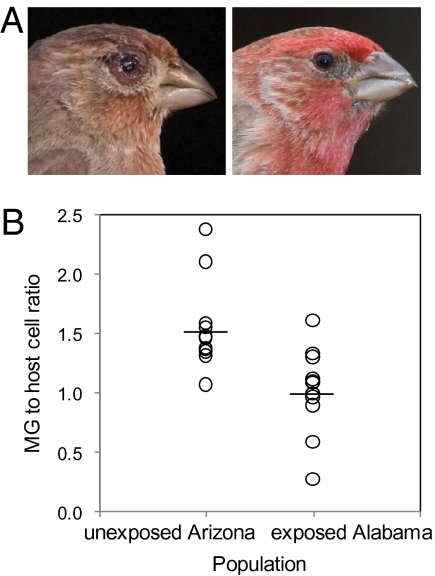
Symptoms of M. gallisepticum infection and MG load in the conjunctivae of house finches. (A) Naturally infected (Left) and healthy (Right) wild house finches. (B) Quantification of MG load in the conjunctiva of infected finches from Arizona and Alabama sampled in 2007, 2 wk postinfection. Raw values of MG load are expressed as a ratio of host cell number; horizontal lines indicate mean values of raw data.
MG is known for its ability to manipulate host immunity in poultry (22, 23). At the onset of infection, MG triggers the up-regulation of proinflammatory cytokines and chemokines (23, 24) and induces an inflammatory response (25), which can cause damage to host epithelia (26). In addition, MG can also have immunosuppressive effects (23), particularly on later stages of the immune response (24). For example, 1–2 wk postinfection, MG has been shown to be associated with a decline in the infiltration of T cells in the trachea (24) and a suppression of T-cell activity (22) in chicken (Gallus gallus domesticus). There is also evidence that infection with MG is associated with a reduced humoral antibody response to other pathogens (27, 28). These immunomodulatory properties, be they induced directly or indirectly by MG, allow MG to evade and suppress host defenses (29).
Changes in disease dynamics in eastern house finches appear to provide evidence of the rapid evolution of host resistance, but two other factors could drive the emergence of resistance: (i) phenotypic plasticity (including acquired immunity, maternal effects, and short/long-term condition-dependent effects) and (ii) attenuation of MG virulence. The aim of this study was to use experimental and molecular approaches in the above host–parasite system to test the basis for the apparent emergence of host resistance. We conducted an MG-infection experiment and compared MG loads and gene expression profiles 2 wk postinoculation in birds captured in 2007 from eastern (Alabama) and western (Arizona) US populations. We also compared these results to expression profiles previously published from a similar experiment conducted on birds captured in 2000 in Alabama (30). MG was first detected in Alabama in 1995 (31), but had never been reported in Arizona before 2009, despite long-term monitoring (32, 33). Alabama and Arizona are at a similar latitude, and sampling was conducted at three different suburban sites in both states. In Arizona, the sites were 1–2 km apart and the birds were captured over 3 d (these sites are hereafter referred to as the Arizona population), whereas, in Alabama, the sites were 10–103 km apart and the birds were captured over the course of a month (these sites are hereafter referred to as the Alabama population). Birds were kept in identical conditions on ad libitum food and water for 3 mo before the onset of the experiment. All 2007 experimental birds were inoculated with the same January 2007 Alabama strain of MG (Materials and Methods). The 2000 study (30) was conducted using birds from the same Alabama population as the current study but infected with a 1999 strain of MG and took place before the spread of resistance in Alabama (15, 16).
None of the birds that we used from either population had been exposed to MG during their lifetimes, as confirmed by both PCR and agglutination assays (Materials and Methods). This removes interpopulation differences in responses to infection caused by immune priming from prior exposure to MG. First, we tested whether birds from Alabama and Arizona in 2007 differed in their level of resistance to MG by quantifying MG load in the conjunctivae of birds 2 wk postinfection. Second, we assessed how birds from Alabama and Arizona differed in their response to infection by quantifying changes in gene expression, again after 2 wk. Third, we investigated how molecular responses to infection have changed over time by conducting a quantitative comparison of gene expression differences between birds captured from Arizona in 2007 and from Alabama in both 2000 and 2007 (30).
Results
Population Differences in MG Load Following Experimental Infection.
If MG loads are lower in birds from Alabama than in birds from Arizona following maintenance in identical ad libitum conditions for 3 mo and infection with the same strain of MG, this would support the hypothesis of rapid evolution of resistance. In addition, it would rule out the possibility that emergence of resistance in the Alabama population resulted solely from (i) short-term environmental effects, such as improvements to individual body condition, or (ii) reductions in the virulence of MG. After controlling for the confounding influence of the amount of host tissue sampled (general linear model: F1,21 = 9.41, P = 0.006), we found that MG load differed significantly between populations (F1,21 = 13.0, P = 0.002, R2 = 30%) (Fig. 1B). Birds from Alabama in 2007 already showed a 33% reduction in MG load in their conjunctivae 2 wk after experimental infection compared with birds from Arizona in 2007, a substantial difference given that mortality as a result of MG usually occurs 25–70 d after the onset of conjunctivitis (15). These results support the hypothesis that birds have evolved resistance to MG in Alabama, but we as yet cannot rule out a confounding influence of long-term (life-long) differences in individual body condition.
Population Differences in Gene Expression Patterns.
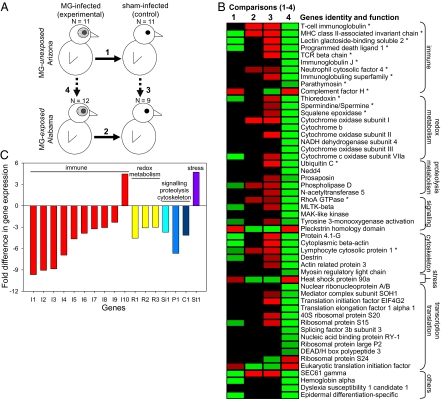
Investigating patterns of gene expressions following experimental infection can elucidate pathogen-induced changes and host responses. Transcript levels were quantified using a microarray printed with cDNA clones selected from two substraction suppression hybridization libraries enriched in clones differentially expressed between MG-infected and control house finches 2 wk postinfection (Materials and Methods and SI Materials and Methods). Differences in transcript levels were tested between infected vs. control birds in Arizona in 2007 (comparison 1); infected vs. controls in Alabama in 2007 (comparison 2); control birds from Arizona vs. Alabama in 2007 (comparison 3); and infected birds from Arizona vs. Alabama in 2007 (comparison 4) (Fig. 2A). Overall, after correcting for false discovery rates, 162 clones were found to be differentially expressed significantly; sequencing and blast searches in GenBank for vertebrate homologs revealed a subset of 52 genes of known function that were differentially expressed across at least one of these four comparisons (Fig. 2B; see SI Materials and Methods and Table S1 for details on gene functions).
Fig. 2.
Comparisons and patterns of splenic gene expression. (A) Schematic of the four analytical comparisons made with gene expression data. (B) Heat map of gene expression patterns in comparisons 1–4 (Fig. 2A). Red and green indicate significantly higher and lower expression levels, respectively, with bright colors reflecting at least a threefold difference in magnitude and values in black indicating no difference. Comparisons in each of the four columns shown for first treatment/population vs. second one are outlined in Fig. 2A. The 52 genes included showed differential expression in at least one comparison (1–4) and were of known identity and function (see Table S1 for full details). Asterisks indicate genes with direct and auxiliary immune functions. (C) Fold difference in expression levels of immune (n = 10), immune-related (n = 6), and stress (n = 1) genes in comparison 4. Genes shown were differentially expressed and known to have direct immune (I1–I10), indirect immune (R1–R3; Si1, P1, C1), or stress (St1) functions (Table S1). Negative values represent lower expression in infected birds from Arizona relative to those from Alabama. Red (I1–I10): immune genes (T-cell Ig and mucin domain containing-4; MHC class II-associated invariant chain I1; lectin galactoside-binding soluble-2-protein; programmed death ligand 1; TCR β-chain; Ig J; neutrophil cytosolic factor-4; Ig superfamily member 4A isoform a; parathymosin; and complement factor-H). Yellow (R1–R3): redox metabolism genes (thioredoxin; spermidine/spermine N1-acetyltransferase variant 1; and squalene epoxidase). Light (Si1), medium (P1), and dark (C1) blue: signal transduction (RhoA GTPase), proteolysis (ubiquitin C), and cytoskeleton (lymphocyte cytosolic protein) genes, respectively. Purple (St1): stress gene (heat-shock protein 90a). The stress gene was included because it was one of the few up-regulated in comparison 4, suggesting that birds from Arizona were more stressed by the infection. Gene I10, complement factor-H, is expected to have increased expression in infected Arizona finches and is not anomalous with the other immune genes (see Discussion).
Birds from the two populations in 2007 showed significant differences in both the number and the direction of expression changes following infection. First, a greater percentage in Alabama (38%) than in Arizona (21%) of the 52 genes of known function showed postinfection expression changes (comparison 1 vs. comparison 2; two-sample binomial test = 1.93, P = 0.05). This difference was generated by a greater percentage of genes in Arizona than in Alabama being down-regulated (80% of 20 vs. 27% of 11 genes; Fisher exact test, P = 0.007). Second, although 67% of the 52 genes were differentially expressed between control birds of the two populations (comparison 3), this increased to all 52 genes being differentially expressed between experimental birds of the two populations (comparison 4), representing a significant increase in between-population expression differences following infection (two-sample binomial test = −4.51, P < 0.001). Again, this difference was generated by a greater percentage of genes being expressed at lower levels in Arizona vs. Alabama (90% vs. 10%; two-sample binomial test = 8.24, P < 0.001).
The results above are largely driven by the differential expression of functionally relevant immune genes. Of the 52 genes showing differential expression in at least one of the four comparisons above (Fig. 2 A and B), we identified 16 that are known to be linked to immunity: 10 with direct immune function and 6 with auxiliary immune function (Fig. 2C and Table S1). Given that (i) MG has immunosuppressive effects on later stages of host immunity (i.e., after 1–2 wk; see Introduction), (ii) the microarray consisted of clones differentially expressed between infected and control birds 2 wk postinfection, and (iii) we examined transcriptional changes occurring 2 wk postinfection, the hypothesis of recently evolved resistance would predict population differences in susceptibility to immunosuppression and in the ability of birds to mount an immune response against MG. More precisely, it would predict that the evolution of resistance to MG would be associated with a postinfection up-regulation of genes involved in immunity or immune activation among finches from Alabama in 2007 and down-regulation of those genes in finches from Arizona in 2007.
In accordance with these predictions, of 11 immune-related genes differentially expressed between infected and control birds across both populations, 5 of 6 were down-regulated in Arizona and 5 of 5 were up-regulated in Alabama (comparisons 1 and 2; Fisher exact test, P = 0.015). In addition, of the 10 genes with direct immune function and the 6 genes with auxiliary immune function, 90% and 100%, respectively, displayed lower expression levels in infected birds from Arizona vs. Alabama (comparison 4) (one-sample binomial test = 14.25, P < 0.001). Taken together, these results independently suggest that MG infection is associated with the suppression of immunity in house finch hosts and that birds from Alabama are able to mount a more robust immune response to MG at the molecular level than birds from Arizona.
Population Changes in Gene Expression Patterns.
We used quantitative comparisons of our 2007 expression patterns with those of a 2000 Alabama study to further test hypotheses regarding the emergence of resistance in eastern US house finches (Fig. S1). Evidence against the MG-attenuation hypothesis as the only driver for the emergence of resistance would again be supported if expression patterns between infected and control birds in 2007 in Arizona (comparison 1) and in Alabama (comparison 2) were more similar to each other than to those of infected vs. control birds in Alabama in 2000, because the latter study used an earlier, potentially more virulent strain of MG. If long-term changes to individual body condition accounted for the emergence of resistance in eastern finches, then we would expect expression differences between infected and control birds to be a function of the site of origin; differences should be more similar between years within Alabama than between Alabama in 2000 and Arizona in 2007. By contrast, the hypothesis that resistance to MG involved genetic evolution in the host would be supported if expression changes between infected and controls in Alabama in 2000 (i) differed from those in the same population in 2007 (comparison 2) and resembled those in Arizona in 2007 (comparison 1) and (ii) resembled the expression differences between infected birds from Alabama and Arizona in 2007 (comparison 4). The first prediction arises because birds from Alabama in 2007 were expected to be resistant, whereas birds in Alabama in 2000 and in Arizona in 2007 were not. The second prediction arises because if birds from Alabama in 2007 had evolved resistance, infected finches from both Alabama in 2000 and Arizona in 2007 should display lower expression levels than control birds from Alabama in 2000 and infected birds from Alabama in 2007, respectively.
Overall, 14 genes were identified as being differentially expressed postinfection in both the 2000 and 2007 studies. Of these, 11 were down-regulated and 3 were up-regulated in 2000 (30). Whereas 7 of the 14 genes showed expression changes in the same direction when comparing infected vs. control birds from Alabama in 2000 and from Arizona in 2007 (comparison 1), none of the gene expression changes were in the same direction when considering the Alabama population in 2007 (comparison 2) (Fig. S1B). Thus, responses to infection were more similar between eastern and western birds with little or no evolved resistance to MG than among birds captured from the same sites but at different stages of the epizootic (Fisher exact test, P = 0.003). In addition, 12 of 14 genes up- or down-regulated following infection in Alabama in 2000 showed a reversed direction of expression difference when comparing infected birds between Alabama and Arizona in 2007 (comparison 4) (one-sample binomial test = 2.40, P = 0.016; Fig. S1B). In other words, infected birds from a population before the spread of resistance to MG (Alabama 2000) expressed genes at lower levels than did control birds from the same population. Similarly, infected birds from a population that had never experienced MG (Arizona 2007) expressed genes at lower levels than infected birds from a population that had apparently evolved resistance to MG (Alabama 2007). Taken together, these results rule out MG-attenuation or long-term differences in body condition as likely explanations for the emergence of resistance in eastern house finches, but fully support all predictions of the evolution of resistance hypothesis.
Discussion
Two weeks following an experimental infection conducted on wild-caught house finches in 2007, we found that finches from populations in the eastern United States (Alabama), with 12 y of exposure to the conjunctivitis-causing bacterium MG in the wild, harbored 33% less MG in their conjunctivae than finches from populations in the western United States (Arizona), which had never experienced the disease. Furthermore, we detected distinct transcriptional responses between populations, both in terms of the number and the direction of expression changes, in response to experimental MG infection. In particular, infected birds from Arizona in 2007 showed significant down-regulation and reduced expression of immune-related genes compared with infected birds from Alabama in 2007. A comparison with a previous macroarray analysis of gene expression following similar experimental conditions (30) suggested that these transcriptional changes have evolved over the past 12 y in eastern finches and hence have accompanied the spread of resistance to MG.
Suggestions of rapid evolution based on phenotypic changes at the population level can often be attributed to phenotypic plasticity rather than to adaptive changes in gene frequencies (4, 34). Phenotypic plasticity could account for the emergence of resistance in populations of eastern house finches if individuals were able to acquire immunity during their lifetimes and pass it on to following generations (35), or if environmental conditions in the recent past (i.e., after 2000) were more conducive to resistance in the short or long term. Our experimental setup in conjunction with measurements of phenotypes at the organismal and molecular level allowed us to distinguish between competing hypotheses that could potentially explain the emergence of resistance in eastern populations of house finches.
First, the lack of previous exposure to MG of the actual birds used in this study meant that differences in MG-load or gene expression changes following experimental infection could not be explained by acquired immunity. An alternative explanation, however, is that infected mothers transmit antibodies against MG to developing offspring (35), somehow conferring on them an early or more long-lasting advantage against MG. Although such maternal effects could facilitate the spread of MG resistance following an evolution of resistance, if maternal effects preceded the evolution of resistance, we would expect gene expression profiles at the two time points in Alabama to be more similar to each other than to Arizona in 2007. To the contrary, expression profiles in Alabama in 2000 were more similar to those in Arizona in 2007 than to those in Alabama in 2007. Furthermore, evidence of immunosuppression in Arizona in 2007 and in Alabama in 2000 suggested that the transmission of maternal antibodies against MG is unlikely to have driven changes in disease dynamics in the wild (see below). Second, by maintaining all birds in identical conditions for 3 mo before the onset of the experiment, we removed the possibility that the interpopulation differences in MG load and gene expression in 2007 could be caused by short-term condition dependence. Nevertheless, the 3-mo acclimatization period would not necessarily entirely eliminate all differences in long-term condition indices arising from differing developmental conditions between sites (36). If ecological differences between Alabama and Arizona influence house finch immunity and gene expression, we would again expect expression profiles within Alabama to be more similar. As indicated above, this was not the case, suggesting that interpopulation differences in responses to MG infection were independent of any differences in ecological conditions. Finally, the greater similarity in expression patterns between Arizona in 2007 and Alabama in 2000 indicated that attenuation of MG between 2000 and 2007 could not exclusively explain our results either. Taken together, the best-supported explanation for our results is the evolution of host resistance by natural selection in eastern house finches over the 12-y period from the fall of 1995 to early 2007.
Examination of our gene expression profiles further supported this conclusion. MG is well known for its complex immunomodulatory effects in poultry, which include the suppression of important immune processes a week or two after MG inoculation (22, 23). Consistent with the evolution of reduced susceptibility to immunomodulation, Alabama finches showed greater up-regulation (or increased expression compared with infected Arizona finches) of immune-related genes, 2 wk postinfection. Overall, all of the immune genes that were differentially expressed between infected vs. controls in Alabama in 2007 were up-regulated, whereas 83% of the differentially expressed immune genes between infected vs. controls in Arizona in 2007 were down-regulated. In addition, all but one of the immune-related genes was expressed at higher levels in the infected birds from Alabama vs. Arizona. One gene (hCG40889 or complement factor H) revealed an illuminating exception to this pattern. Complement factor H restricts the activation of the complement cascade to protect host cells and tissues (Table S1), and unlike the other 15 immune-related genes that are all involved with counteracting infections (Table S1), this gene exhibited an expression direction opposite to that expected (37). Under MG-induced immunosuppression, an opposite expression pattern of complement factor H relative to other immune-related genes identified would be expected. Thus, the apparent exception is actually consistent rather than anomalous, and our results strongly suggest that birds from Alabama in 2007 have evolved resistance to infections with MG and are able to counter MG-induced immunosuppression, an observation with important implications for the evolution of immunity in vertebrates (38).
Evolution can arise through the emergence and subsequent selection of a novel mutation or through selection on existing (standing) variation in the population (39). Experiments with Escherichia coli reveal that adaptive mutations typically arise over hundreds or thousands of generations (8). Our evidence that eastern house finches evolved resistance within 12 y suggests that genetic variability in resistance to MG existed at the time of outbreak. Selection by MG would then have produced a shift in allelic frequency reflected in the change in gene expression in the eastern US finches over time, resulting in population-level changes in resistance to MG. In addition to helping us understand the evolvability of wild populations, our results may also help us predict the impact of an outbreak of MG that would reach Arizona. Given that the eastern US finch population originated from western US birds, it is reasonable to assume that standing variation for resistance is present in Arizona. Furthermore, in our infection experiments, the MG load detected in the conjunctivae of 2 of the 11 experimentally infected Arizona birds ranked among those of the 10 Alabama birds (after excluding an individual from Alabama that showed no signs of resistance to MG; Fig. 1B). A simple extrapolation suggests that at least 2 in every 11 birds (∼20%) would be likely to resist an MG outbreak in Arizona, which is close to the estimated 30% that survived the outbreak in Alabama (http://birds.audubon.org/historical-results), but this will also depend on the virulence of MG (40).
In conclusion, there are important implications from the observation that house finches exposed to MG have evolved resistance through changes in the expression of functionally relevant genes within only 12 y. Few studies have shown that adaptive phenotypes can spread rapidly in wild vertebrate populations (3, 10). In addition, we show that such a spread is associated with changes in functionally relevant gene expression, an observation predicted by current evolutionary theory (34), but previously confined to selection experiments in the laboratory (7). Furthermore, although differences in gene expression have been hypothesized to indicate evolution in wild populations (41–43), the validity of this scenario requires evidence that observed differences have changed over time in response to an identified selective agent and also have functional significance (44). Our study lends weight to the suggestion that differences in gene expression in the wild can reflect adaptive evolution (41–43) and indicates that population evolvability can be extremely rapid where sufficient standing variation exists.
Materials and Methods
In early 2007, male birds were captured in Alabama and Arizona and immediately transported to aviaries at Auburn University by plane (Arizona) or car (Alabama). Throughout, males were caged in pairs in two identical temperature-controlled rooms and fed and watered ad libitum. Birds from Alabama and Arizona were kept in separate rooms for the first months to monitor for signs of MG. Following quarantine, prior exposure to MG was investigated using serum plate agglutination assay and amplification of MG DNA from choanal and conjunctival swabs (SI Materials and Methods). Overall, 12 birds were removed from the study; no birds used in this study were found to be currently or previously infected with MG. Birds from each population were randomly assigned to sham-inoculated control (n = 11 Arizona vs. n = 9 Alabama) or MG-inoculated experimental (n = 11 Arizona vs. n = 12 Alabama) treatments. Infected birds were inoculated with 20 μL of culture containing 1 × 104 to 1 × 106 color-changing units−1 of a January 2007 Alabama MG isolate, whereas controls were given the same volume of sterile SP4 medium. Infected and control birds were kept in separate rooms in identical conditions. Two weeks posttreatment, birds were euthanized, and spleens and conjunctivae were immediately removed and stored in RNAlater (Ambion) at −80 °C (SI Materials and Methods). Protocols were approved by Auburn University Institutional Animal Care and Use Committee permit (#2007-1179) and by an Auburn University Institutional Biological Use Authorization (#243).
We determined the levels of MG in one of the two conjunctivae selected at random from each infected bird using TaqMan qRT-PCR amplification of the mgc2 gene (SI Materials and Methods). We then determined splenic transcript levels using a cDNA microarray (SI Materials and Methods). The microarrays were printed with a selection of cDNA clones from two substraction suppression hybridization libraries enriched in clones differentially expressed between MG-infected and control house finches 2 wk postinfection (n = 16,512 clones) (30). Of these, 220 were previously identified as significantly differentially expressed between infected and controls using a macroarray approach (30). The microarray constructed here consisted of unique amplicons of these 220 clones, as well as 694 randomly selected clones from the enriched libraries (SI Materials and Methods). We determined the gene ontology category and function of differentially expressed genes using Harvester (http://harvester.fzk.de/harvester/). Minimum Information About a Microarray Experiment is available at http://www.ecoex-moulis.cnrs.fr or by request to C.B.
Microarray Analysis.
We normalized the log base-2 measurements of mean fluorescence intensities for each dye channel in each spot on the array using R software (http://www.r-project.org) and a Matlab interface (MArray), which allows results to be graphically presented and normalized (45) (SI Materials and Methods). Normalized signal ratios were then fitted to the linear model for microarray data (LIMMA) in an R Bioconductor package; LIMMA is similar to a general linear model but provides false-discovery-rate–adjusted probability values of differential expression. This approach controls for multiple comparisons in microarray data, substantially reducing the probability of discovsering false positives (type I errors) (46). The model followed the following format: Yijc = μ + Ai + Bj + ABij + εijc, where Yijc is the log2 measurement for a particular clone (c), from a particular treatment (i) and a particular population of origin (j), μ is the parametric mean, A and B correspond to the single-factor effects (treatment and population of origin, respectively), AB is the two-way interaction between the two main effects, and ε is the residual between the data and the model. Microarray results were validated using independent estimates measured with qRT-PCR (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Kevin McGraw for logistics in Arizona, Frank F. Bartol, Christian Daly, and Mato Lagator for technical advice/help and Janis Antonovics, Tim Coulson, Arkhat Abzhanov, Niclas Backström, and two anonymous referees for constructive comments. Work was funded by a Biogrant from the Office for Vice President for Research at Auburn University; by funds from the Auburn University Center for Environmental and Cellular Signal Transduction; by Harvard University funds (to S.V.E. and C.B.); by a Harvard University Milton Grant (to S.V.E.); by Marie Curie Reintegration Grant FP7-PEOPLE-IRG-2008 #239257 (to C.B.); by the Centre National de la Recherche Scientifique (to C.B.); by an Alabama EPSCoR grant (to S.L.B.); and by a Royal Society University Fellowship Scheme (to A.F.R.).
Footnotes
The authors declare no conflict of interest.
Database deposition: Sequences reported in this paper have been deposited in the GenBank database (accession nos. bankit1324533, 1324536, 1324538, 1324542, 1324554, and GW346076–GW346170).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018580108/-/DCSupplemental.
References
- 1.Walther GR, et al. Ecological responses to recent climate change. Nature. 2002;416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- 2.Reznick DN, Shaw FH, Rodd FH, Shaw RG. Evaluation of the rate of evolution in natural populations of guppies (Poecilia reticulata) Science. 1997;275:1934–1937. doi: 10.1126/science.275.5308.1934. [DOI] [PubMed] [Google Scholar]
- 3.Grant PR, Grant BR. Evolution of character displacement in Darwin's finches. Science. 2006;313:224–226. doi: 10.1126/science.1128374. [DOI] [PubMed] [Google Scholar]
- 4.Badyaev AV. Maternal effects as generators of evolutionary change: A reassessment. Ann N Y Acad Sci. 2008;1133:151–161. doi: 10.1196/annals.1438.009. [DOI] [PubMed] [Google Scholar]
- 5.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th Ed. Harlow, Essex, UK: Longmans Green; 1996. [Google Scholar]
- 6.Fisher RA. The Genetical Theory of Natural Selection. 2nd Ed. Oxford: Oxford University Press; 1958. [Google Scholar]
- 7.Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett RDH, Rogers SM, Schluter D. Natural selection on a major armor gene in threespine stickleback. Science. 2008;322:255–257. doi: 10.1126/science.1159978. [DOI] [PubMed] [Google Scholar]
- 10.Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. On the origin and spread of an adaptive allele in deer mice. Science. 2009;325:1095–1098. doi: 10.1126/science.1175826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.May RM, Anderson RM. Epidemiology and genetics in the coevolution of parasites and hosts. Proc R Soc Lond B Biol Sci. 1983;219:281–313. doi: 10.1098/rspb.1983.0075. [DOI] [PubMed] [Google Scholar]
- 12.Decaestecker E, et al. Host-parasite ‘Red Queen’ dynamics archived in pond sediment. Nature. 2007;450:870–873. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- 13.Best SM, Kerr PJ. Coevolution of host and virus: The pathogenesis of virulent and attenuated strains of myxoma virus in resistant and susceptible European rabbits. Virology. 2000;267:36–48. doi: 10.1006/viro.1999.0104. [DOI] [PubMed] [Google Scholar]
- 14.Hochachka WM, Dhondt AA. Density-dependent decline of host abundance resulting from a new infectious disease. Proc Natl Acad Sci USA. 2000;97:5303–5306. doi: 10.1073/pnas.080551197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farmer KL, Hill GE, Roberts SR. Susceptibility of a naïve population of house finches to Mycoplasma gallisepticum. J Wildl Dis. 2002;38:282–286. doi: 10.7589/0090-3558-38.2.282. [DOI] [PubMed] [Google Scholar]
- 16.Roberts SR, Nolan PM, Hill GE. Characterization of Mycoplasma gallisepticum infection in captive house finches (Carpodacus mexicanus) in 1998. Avian Dis. 2001;45:70–75. [PubMed] [Google Scholar]
- 17.Hill GE. House finch (Carpodacus mexicanus) In: Poole A, editor. The Birds of North America Online. Ithaca, NY: Cornell Lab of Ornithology; 1993. Retrieved from the Birds of North America Online at http://bna.birds.cornell.edu/bna.html/species/046. [Google Scholar]
- 18.Stipkovits L, Kempf I. Mycoplasmoses in poultry. Rev Sci Tech. 1996;15:1495–1525. doi: 10.20506/rst.15.4.986. [DOI] [PubMed] [Google Scholar]
- 19.Ley DH, Berkhoff JE, Levisohn S. Molecular epidemiologic investigations of Mycoplasma gallisepticum conjunctivitis in songbirds by random amplified polymorphic DNA analyses. Emerg Infect Dis. 1997;3:375–380. doi: 10.3201/eid0303.970318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ley DH, Berkhoff JE, McLaren JM. Mycoplasma gallisepticum isolated from house finches (Carpodacus mexicanus) with conjunctivitis. Avian Dis. 1996;40:480–483. [PubMed] [Google Scholar]
- 21.Luttrell MP, Stallknecht DE, Fischer JR, Sewell CT, Kleven SH. Natural Mycoplasma gallisepticum infection in a captive flock of house finches. J Wildl Dis. 1998;34:289–296. doi: 10.7589/0090-3558-34.2.289. [DOI] [PubMed] [Google Scholar]
- 22.Ganapathy K, Bradbury JM. Effects of cyclosporin A on the immune responses and pathogenesis of a virulent strain of Mycoplasma gallisepticum in chickens. Avian Pathol. 2003;32:495–502. doi: 10.1080/0307945031000154099. [DOI] [PubMed] [Google Scholar]
- 23.Mohammed J, et al. Chemokine and cytokine gene expression profiles in chickens inoculated with Mycoplasma gallisepticum strains Rlow or GT5. Vaccine. 2007;25:8611–8621. doi: 10.1016/j.vaccine.2007.09.057. [DOI] [PubMed] [Google Scholar]
- 24.Gaunson JE, Philip CJ, Whithear KG, Browning GF. Lymphocytic infiltration in the chicken trachea in response to Mycoplasma gallisepticum infection. Microbiology. 2000;146:1223–1229. doi: 10.1099/00221287-146-5-1223. [DOI] [PubMed] [Google Scholar]
- 25.Gaunson JE, Philip CJ, Whithear KG, Browning GF. The cellular immune response in the tracheal mucosa to Mycoplasma gallisepticum in vaccinated and unvaccinated chickens in the acute and chronic stages of disease. Vaccine. 2006;24:2627–2633. doi: 10.1016/j.vaccine.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Ley DH. Mycoplasma gallisepticum infection. In: Calnell BW, Barnes HJ, Beard CW, McDougald LR, Saif YM, editors. Diseases of Poultry. 11th Ed. Ames, IA: Iowa State Press; 2003. pp. 722–743. [Google Scholar]
- 27.Matsuo K, Kuniyasu C, Yamada S, Susumi S, Yamamoto S. Suppression of immunoresponses to Haemophilus gallinarum with nonviable Mycoplasma gallisepticum in chickens. Avian Dis. 1978;22:552–561. [PubMed] [Google Scholar]
- 28.Naylor CJ, Al-Ankari AR, Al-Afaleq AI, Bradbury JM, Jones RC. Exacerbation of Mycoplasma gallisepticum infection in turkeys by rhinotracheitis virus. Avian Pathol. 1992;21:295–305. doi: 10.1080/03079459208418844. [DOI] [PubMed] [Google Scholar]
- 29.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Farmer K, Hill GE, Edwards SV. A cDNA macroarray approach to parasite-induced gene expression changes in a songbird host: Genetic response of house finches to experimental infection by Mycoplasma gallisepticum. Mol Ecol. 2006;15:1263–1273. doi: 10.1111/j.1365-294X.2005.02753.x. [DOI] [PubMed] [Google Scholar]
- 31.Nolan PM, Hill GE, Stoehr AM. Sex, size, and plumage redness predict house finch survival in an epidemic. Proc R Soc Lond B Biol Sci. 1998;265:961–965. [Google Scholar]
- 32.Dhondt AA, et al. Dynamics of mycoplasmal conjunctivitis in the native and introduced range of the host. EcoHealth. 2006;3:95–102. [Google Scholar]
- 33.Toomey MB, Butler MW, McGraw KJ. Immune-system activation depletes retinal carotenoids in house finches (Carpodacus mexicanus) J Exp Biol. 2010;213:1709–1716. doi: 10.1242/jeb.041004. [DOI] [PubMed] [Google Scholar]
- 34.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford: Oxford University Press; 2003. [Google Scholar]
- 35.Boulinier T, Staszewski V. Maternal transfer of antibodies: Raising immuno-ecology issues. Trends Ecol Evol. 2008;23:282–288. doi: 10.1016/j.tree.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Metcalfe NB, Monaghan P. Compensation for a bad start: Grow now, pay later? Trends Ecol Evol. 2001;16:254–260. doi: 10.1016/s0169-5347(01)02124-3. [DOI] [PubMed] [Google Scholar]
- 37.de Córdoba SR, de Jorge EG. Translational mini-review series on complement factor H: Genetics and disease associations of human complement factor H. Clin Exp Immunol. 2008;151:1–13. doi: 10.1111/j.1365-2249.2007.03552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulenburg H, Kurtz J, Moret Y, Siva-Jothy MT. Introduction: Ecological immunology. Philos Trans R Soc B Biol. Sci. 2009;364:3–14. doi: 10.1098/rstb.2008.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Hawley DM, et al. Common garden experiment reveals pathogen isolate but no host genetic diversity effect on the dynamics of an emerging wildlife disease. J Evol Biol. 2010;23:1680–1688. doi: 10.1111/j.1420-9101.2010.02035.x. [DOI] [PubMed] [Google Scholar]
- 41.Cheviron ZA, Whitehead A, Brumfield RT. Transcriptomic variation and plasticity in rufous-collared sparrows (Zonotrichia capensis) along an altitudinal gradient. Mol Ecol. 2008;17:4556–4569. doi: 10.1111/j.1365-294X.2008.03942.x. [DOI] [PubMed] [Google Scholar]
- 42.Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- 43.Abzhanov A, et al. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 44.Fay JC, Wittkopp PJ. Evaluating the role of natural selection in the evolution of gene regulation. Heredity. 2008;100:191–199. doi: 10.1038/sj.hdy.6801000. [DOI] [PubMed] [Google Scholar]
- 45.Wang JB, Nygaard V, Smith-Sørensen B, Hovig E, Myklebost O. MArray: Analysing single, replicated or reversed microarray experiments. Bioinformatics. 2002;18:1139–1140. doi: 10.1093/bioinformatics/18.8.1139. [DOI] [PubMed] [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.