Abstract
Microinjection of recombinant DNA into zygotic pronuclei has been widely used for producing transgenic mice. However, with this method, the insertion site, integrity, and copy number of the transgene cannot be controlled. Here, we present an integrase-based approach to produce transgenic mice via pronuclear injection, whereby an intact single-copy transgene can be inserted into predetermined chromosomal loci with high efficiency (up to 40%), and faithfully transmitted through generations. We show that neighboring transgenic elements and bacterial DNA within the transgene cause profound silencing and expression variability of the transgenic marker. Removal of these undesirable elements leads to global high-level marker expression from transgenes driven by a ubiquitous promoter. We also obtained faithful marker expression from a tissue-specific promoter. The technique presented here will greatly facilitate murine transgenesis and precise structure/function dissection of mammalian gene function and regulation in vivo.
Production of transgenic mice via microinjection of DNA into zygotic pronuclei (1–3) has served mammalian genetics for 30 years. Although still the predominant method used to produce transgenic mice, it has several limitations: the insertion site, integrity, and copy number of the transgene cannot be controlled. Insertion of DNA into different chromosomal loci at random could disrupt the function of endogenous genes. Transgenes generated in this manner can be influenced by the local chromatin environment (i.e., position effect) that can lead to transgene silencing or ectopic expression (4–7). Moreover, transgenic DNA concatemerized into a large array is subject to repeat-induced gene silencing (8).
Single-copy transgenesis in mice can be achieved with retroviruses (9) and transposons (10, 11), but these approaches integrate transgenes throughout the genome. As a result, the transgenes are subjected to the local chromatin environment and can cause endogenous gene disruption, although the mutagenic properties of transposons can be desirable for particular applications (10). These problems can be overcome by targeting the transgene to a specific chromosomal locus via homologous recombination in embryonic stem (ES) cells (12, 13). However, this method is significantly more laborious and time-consuming, as it involves creation of modified ES cells and mouse chimeras, as well as eventual germline transmission of the transgene.
Integrase enzymes from a variety of sources have been used to catalyze integration of transgenes in heterologous systems (14, 15). Integrases catalyze irreversible recombination between appropriate attB and attP sites (14, 16). ϕC31 integrase from a Streptomyces phage has previously been used for transgene integration in flies (17–19). In mice, ϕC31 integrase has been used to catalyze integration of circular DNA into pseudo-attP sites in the genome for gene therapy (20) or low-efficiency transgenesis (21), for transgenesis in mouse ES cells (22), and for removal of undesirable transgene portions or reporter activation (23, 24).
Here, we describe an integrase-mediated method for site-specific transgenesis in mice via pronuclear microinjection, with integration efficiencies as high as 40%. We use ϕC31 integrase to catalyze recombination between one or two attB sites in a recombinant DNA with one or more tandem attP sites that we previously inserted into specific loci in the mouse genome (Fig. 1). We show that the plasmid bacterial backbone within the transgene and nearby transgenic elements dramatically decrease expression of our transgenes, and that the absence of these elements results in global, high-level transgene expression from a ubiquitous promoter. Finally, we show that a promoter for the murine transcription factor Hb9 integrated into one of the predetermined loci drives proper tissue-specific marker expression.
Fig. 1.
Schematic summary for site-specific ϕC31 integrase-mediated transgenesis via pronuclear injection in mice. A single or three tandem attP sites were knocked into the Hipp11 (H11) or Rosa26 loci via homologous recombination in ES cells (Left; for details, see Fig. S1). Mice homozygous for one of the modified loci (H11 is shown as an example) served as embryo donors. A mix of DNA and in vitro transcribed ϕC31 mRNA was injected into a single pronucleus of each zygote. The integration of plasmid bacterial backbone (BB) that decreases the transgene expression was avoided either by injecting a minicircle DNA with a single attB site (top branch; in this case, ϕC31 catalyzes a typical integration reaction), or by injecting plasmid DNA where the gene of interest (e.g., GFP) was flanked by two attB sites (bottom branch; in this case, ϕC31 catalyzes a recombinase-mediated cassette exchange reaction). Right, Middle: Green fluorescence of a representative transgenic F0 embryo and the corresponding PCR results that indicate site-specific insertion. The red numbers for the PCR results correspond to the numbers on the H11P3-pCA-GFP transgene scheme below; the same PCR tests can be used for the transgene above. The particular embryo shown was obtained by cassette exchange. Inset: Bright-field image of the same embryo.
Results
Strategy and Proof of Principle for Integrase-Mediated Site-Specific Transgenesis.
To generate embryos containing attP sites for ϕC31 integrase-mediated transgenesis, we used standard homologous recombination-based methods in mouse ES cells (12, 25). We inserted three shortened tandem ϕC31 integrase attP sites (attPx3) or a single “full-length” attP site (14) into two loci: the Rosa26 locus on mouse chromosome 6 (26) and an intergenic Hipp11 (H11) locus on mouse chromosome 11 (27) (Fig. 1, Left, and SI Appendix, Fig. S1). The Rosa26 locus supports global expression of a single copy knock-in transgene driven by a combination of the CMV enhancer and the chicken β-actin promoter (pCA) (28, 29). Knock-in experiments confirmed that H11 supports high-level global gene expression from the pCA promoter and a higher rate of mitotic (interchromosomal) recombination compared with Rosa26 (27). The latter property suggested that H11 might allow better access to ϕC31 integrase than Rosa26. The modified ES cells were used to produce chimeric mice, and mice with germline-transmitted alleles were used to establish mouse colonies homozygous for the knock-in cassettes.
For most experiments, we integrated transgenes into the H11 locus because homozygous insertions into this locus are not predicted to disrupt any endogenous genes, and the resulting mice are completely healthy and fertile (27). We injected embryos homozygous for attP or attPx3 at the H11 locus (H11P or H11P3, respectively) with ϕC31 mRNA together with attB-pCA-GFP, a minicircle DNA that was generated by removal of the plasmid backbone to enable proper transgene expression (as detailed later). The attB-pCA-GFP minicircle contains a full-length attB site (14) and the sequence for a thermotolerant GFP (30, 31) driven by the ubiquitous pCA promoter (SI Appendix, SI Materials and Methods). We obtained integration of the transgene (Fig. 1, Top Right, and Table 1, rows 1–3; SI Appendix, Table S1, provides more details).
Table 1.
Efficiency of site-specific integration
| Row | DNA* | DNA type | DNA size, kb | Strain | Background | F0 (n) | SS F0 (n) | Significance¶ | SS, % (of F0) | R F0 (n) | R, % (of F0) |
| 1 | attB-pCA-GFP | Minicircle | ~3 | H11P | Mix | 21 | 1 | NS vs. row 2 | 4.8 | 1 | 4.8 |
| 2 | attB-pCA-GFP | Minicircle | ~3 | H11P3 | Mix | 39 | 4 | 10.3 | 1 | 2.6 | |
| 3 | attB-pCA-GFP | Minicircle | ~3 | H11P3 | FVB N4 | 15 | 6 | P < 0.05 vs. row 2 | 40.0 | 3 | 20.0 |
| 4 | attB-pCA-GFP-attB | Plasmid | ~6 | H11P3 | FVB N4 | 38‡ | 6‖ | — | 15.8 | 1 | 2.6 |
| 5 | attB-pCA-GFP, no RNA | Plasmid | ~6 | H11P3NVϕ | Mix | 32‡ | 0 | — | 0.0 | 5 | 15.6 |
| 6 | attB-pCA-GFP | Plasmid | ~6 | H11P3NVϕ | Mix | 64§ | 10 | NS vs. row 7 | 15.6 | 4 | 6.3 |
| 7 | attB-pCA-GFP | Plasmid | ~6 | H11PNVϕ | Mix | 30‡ | 2 | 6.7 | 0 | 0.0 | |
| 8 | attB-pCA-GFP-FRT5 | Plasmid | ~6 | H11P | Mix | 51 | 5 | NS vs. row 9 | 9.8 | 3 | 5.9 |
| 9 | attB-pCA-GFP-(FRT5)† | Plasmid | ~6 | H11P3 | Mix | 61 | 4 | 6.6 | 9 | 14.8 | |
| 10 | attB-pCA-GFP-FRT5 | Plasmid | ~6 | H11P3 | FVB N4 | 8 | 3 | P < 0.05 vs. row 9 | 37.5 | 1 | 10.3 |
| 11 | attB-pHB9-GFP-FRT5 | Plasmid | ~14 | H11P3 | FVB N4 | 66 | 2 | — | 3.0 | 2 | 3.0 |
| 12 | attB-pCA-GFP | Plasmid | ~6 | R26P3NVϕ | Mix | 22‡ | 2 | — | 9.1 | 2 | 9.1 |
Abbreviations: F0, embryos or animals obtained from injections; SS, site-specific integration; R, random integration; mix, mixed background of 129, C57BL/6 and DBA2; FVB N4, mice of the mixed background were outcrossed for 4 generations to the FVB strain and then intercrossed.
*All DNA was coinjected with ϕC31o mRNA, except for row 5.
†Both FRT and non-FRT versions of attB-pCA-GFP were used.
‡F0s were analyzed only as E10 or E11 embryos.
§F0s were analyzed either as E10 or E11 embryos or as live pups.
‖The six founders listed contained pCA-GFP without the bacterial backbone; five more founders with cassette exchange contained only the bacterial backbone. Therefore, the total number of founders with cassette exchange is 11 (29%).
¶Fisher's exact test. NS, not significant.
As an alternative to insertion of DNA at a single attP site, we also injected a plasmid in which pCA-GFP is flanked by two attB sites (pattB-pCA-GFP-attB) into H11P3 homozygous embryos. In principle, this approach should allow ϕC31 integrase to catalyze a recombinase-mediated cassette exchange reaction (Fig. 1, Bottom Right). We tested the integration of the circular pattB-pCA-GFP-attB plasmid into H11P3 and obtained successful cassette exchange as confirmed by PCR (Fig. 1, Bottom Right, and Table 1, row 4). In these experiments, we never detected insertion of a full plasmid.
The integrated transgenes were properly transmitted from founders to progeny (SI Appendix, Table S2). Both strategies resulted in broad and high-level GFP expression in pCA-GFP transgenic mice (Fig. 1, Right, Fig. 2D, and Fig. S1C, Bottom). These data provide the proof of principle for our site-specific integrase-mediated transgenesis in mice. In the subsequent sections, we describe in more detail the optimization process that led to these results.
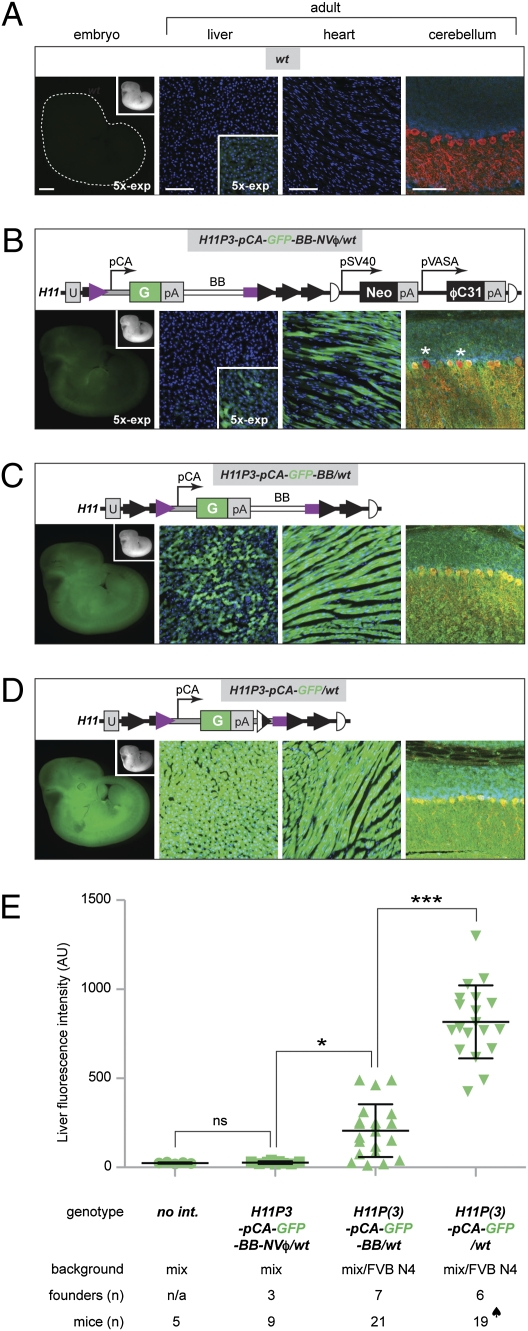
Fig. 2.
GFP expression in animals carrying site-specific pCA-GFP transgenes introduced by ϕC31 integrase-mediated transgenesis. (A–D) Representative fluorescence images from embryonic day 11 embryos and adult livers, hearts, and cerebella of N1 or N2 transgenic animals corresponding to the genotypes and schematics of transgenes shown (Upper). Embryos or same tissues were imaged under identical conditions, except that “5×-exp” designates fivefold longer exposure time than for the rest of the images in the same column. Whole-mount embryos were imaged for GFP fluorescence; corresponding bright-field images of each embryo are also shown (Insets). The livers and hearts are represented by epifluorescence images of 10-μm sections stained only by DAPI (blue). The green signal is GFP fluorescence. The cerebella are represented by confocal images of sections stained by anti-GFP antibody (green), anti-calbindin (red) for Purkinje cells, and DAPI (blue). Two Purkinje cells labeled by asterisks appear negative for GFP. ϕC31 attL and attR are the product of recombination of an attP (black arrows) and attB site (purple) and are therefore half black and half purple. Half circles represent FRT5 sites. Half white/half black triangle represent λ-integrase attB site created during minicircle production. pSV40, SV40 promoter. pVASA, VASA promoter. U, unique sequence. pCA, CMV enhancer and β-actin promoter. G, GFP. pA, polyA signal. BB, plasmid bacterial backbone. (Scale bars: 1 mm for embryos, 100 μm for tissue sections.) (E) Average fluorescence in arbitrary units (AU) in the GFP channel for liver sections from animals of genotypes shown below (no int., no integration; represents wt, H11P3NVϕ/wt, H11P3/wt, and H11P/wt genotypes). Each dataset is represented by mean ± SD. The numbers of individual animals and founders analyzed for each genotype are listed below the genotypes. When samples from multiple founders were combined to obtain an average, each founder was represented by the same number of animals except in the case labeled by a spade. The fluorescence intensities differ significantly among the groups by one-way ANOVA [F(3, 50) = 84.09, P < 0.0001]. Tukey's post-hoc test was used for pair-wise comparisons (ns, not significant; *P < 0.05 and ***P < 0.001).
Transgenic Integrase Did Not Enable Site-Specific Integration.
Our original knock-in cassettes contained a mammalian codon-optimized ϕC31 integrase (ϕC31o) (23) driven by a fragment of the mouse VASA promoter sufficient for germline expression (32), and a neomycin resistance gene, flanked by FRT5 sites (33) (SI Appendix, Fig. S1). This “NVϕ cassette” (i.e., Neo-VASA-ϕC31o) was designed to provide the integrase in embryos in situ. We injected mouse embryos homozygous for the H11P3NVϕ knock-in with a plasmid containing the attB-pCA-GFP transgene (pattB-pCA-GFP) but did not obtain any site-specific integration (0/32 F0s; Table 1, row 5) despite occasional random integrations. However, coinjecting pattB-pCA-GFP with ϕC31o mRNA into homozygous H11P3NVϕ embryos produced site-specific integrations (Table 1, row 6). Thus, the VASA promoter does not promote sufficient ϕC31o expression in situ to enable site-specific insertions.
Because the NVϕ cassette did not perform as expected we removed it from the H11P3NVϕ allele by FLP-mediated recombination to generate the H11P3 allele (SI Appendix, Fig. S1, and SI Appendix, SI Materials and Methods). Similarly, the H11P allele was derived from H11PNVϕ. Coinjection of pattB-pCA-GFP and ϕC31o mRNA into homozygous H11P or H11P3 embryos produced site-specific integrants (Table 1, rows 8–10). Thus, we coinjected integrase mRNA with DNA for all subsequent experiments.
Bacterial Backbone and the NVϕ Cassette Affect Proper Transgene Expression.
Despite proper transmission of the site-specifically integrated transgenes produced from the pattB-pCA-GFP plasmid, the progeny of these transgenic founders exhibited a wide range of GFP expression levels as seen in whole-mount embryos (SI Appendix, Fig. S1C). Moreover, the GFP expression in the progeny was mosaic in several internal tissues, including the heart, brain, and particularly the liver (Fig. 2B and SI Appendix, Fig. S2). We first observed this variable and mosaic expression with H11P3NVϕ as the host. We suspected that the nearby germline-specific VASA promoter and/or other elements within the NVϕ cassette (e.g., the neomycin resistance gene) could affect the expression of pCA-GFP inserted at H11. Indeed, transgenic embryos containing pCA-GFP inserted at H11P3, which lack the NVϕ cassette, produced more uniform GFP expression (Fig. 2C and SI Appendix, Fig. S2). However, considerable variability of pCA-GFP expression still persisted especially in the livers of N1 or N2 animals derived from a number of transgenic founders (Fig. 2C and SI Appendix, Fig. S2).
It has been reported that plasmid bacterial backbone can decrease the expression of integrated and episomal transgenes (34–37). To test if this is the case in our system, we developed an in vitro method to produce minicircle DNA: circular DNA containing desired transgene elements, but devoid of the bacterial backbone (SI Appendix, Fig. S3 and SI Appendix, SI Materials and Methods). We injected the minicircle DNA into H11P3 embryos to produce transgenic animals (SI Appendix, Fig. S1, and Table 1). For simplicity, we designate hereafter the transgenic alleles derived from integration of the entire pCA-GFP plasmid (which contains the bacterial backbone) as pCA-GFP-BB (Fig. 2C), and the mice derived from integration of the pCA-GFP minicircle as pCA-GFP (Fig. 2D). Transgenic animals derived from the pCA-GFP minicircle exhibited higher and more uniform expression in embryos and all adult tissues examined (Fig. 2D and SI Appendix, Fig. S2). The removal of bacterial backbone from the pCA-GFP-BB transgene by crossing to our newly generated GFP-FLPo transgenic mice (SI Appendix, SI Materials and Methods) also resulted in elevated transgene expression (SI Appendix, Fig. S4). These data demonstrate that pCA-GFP at the H11 locus can express GFP ubiquitously in the absence of the NVϕ cassette and the bacterial backbone.
Because the greatest variability was observed in the liver (Fig. 2 and SI Appendix, Fig. S2), we used it as a model to determine the relative contributions of the NVϕ cassette and the bacterial backbone to GFP expression variability in these transgenic mice. In addition, to test for the possible differences between a single attP site and attPx3, we analyzed transgenic mice obtained by insertion into the H11P allele (Table 1, rows 1 and 8). Finally, to probe the effect of genetic background, we analyzed transgenes inserted into the H11P3 allele in mice that had been outcrossed to the FVB inbred strain for four generations (Table 1, rows 3 and 10). We compared the total GFP fluorescence of liver sections from different transgenic animals under identical conditions. We observed no statistically significant differences in GFP fluorescence between transgenes that differed only in the number of attP copies or in the genetic background of the mouse strain used for transgenesis (SI Appendix, Fig. S5). Therefore, we grouped all data according to the presence of the NVϕ cassette and/or the bacterial backbone (Fig. 2E). We found that, in the presence of both the NVϕ cassette and the bacterial backbone, GFP expression was detectable in the liver in a small number of cells and at a very low level (Fig. 2B), but total fluorescence was statistically indistinguishable from negative controls (Fig. 2E, second column vs. first column). In other organs analyzed (heart and brain), GFP expression was apparent but mosaic (Fig. 2B and SI Appendix, Fig. S2). When the NVϕ cassette was removed but the bacterial backbone was still present, average GFP fluorescence intensity became significantly higher (Fig. 2E, third column vs. second column). Finally, when the bacterial backbone was removed, average GFP fluorescence intensity became even higher (Fig. 2E, fourth column vs. third column). Thus, both the NVϕ cassette and the bacterial backbone significantly reduced transgene expression. The reduction of total fluorescence intensity could be caused by low-level of expression in every cell, absence of expression in a subset of cells, or a combination of the two. As is evident from Fig. 2 A–D and SI Appendix, Fig. S2, both mechanisms contributed to the reduced level of transgene expression in the presence of the NVϕ cassette and/or the bacterial backbone.
H11 Can Support Tissue-Specific Expression.
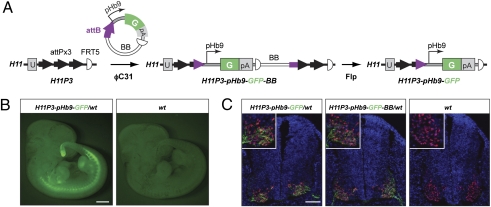
To test if a tissue-specific promoter can provide appropriate expression using our transgenesis method, we integrated pattB-pHb9-GFP-FRT5 into H11P3 (Table 1, row 11). This plasmid contains an approximately 9-kb promoter fragment from the murine transcription factor Hb9 gene that has been shown to be sufficient to direct appropriate tissue- and cell-specific expression in transgenic animals (38). We examined tissue-specific marker expression before and after removal of the bacterial backbone by using the GFP-FLPo transgene (SI Appendix, SI Materials and Methods and Fig. 3A). In agreement with the reported expression pattern (38), we observed GFP expression in motor neurons in the ventral spinal cord and the tail tip (Fig. 3B). In this case, the removal of bacterial backbone did not appear to affect the expression level of the transgene (Fig. 3C). Double-labeling with endogenous Hb9 protein confirmed the motor neuron-specific expression of the transgene (Fig. 3C). These experiments indicate that our integrase-based strategy can be used for faithful tissue-specific expression of transgenes.
Fig. 3.
GFP expression in embryos carrying a single copy of pHb9-GFP transgene site-specifically integrated in H11. (A) Schematic representation of the generation of H11P3-pHb9-GFP allele. After site-specific integration of the plasmid pBT366 (SI Appendix, SI Materials and Methods), the bacterial backbone (BB) was removed by crossing to the GFP-FLPo transgenic line (SI Appendix, SI Materials and Methods). The embryos that inherited only the Hb9 allele but not the Flpo transgene were tested for GFP expression. (B) GFP expression in a whole-mount representative embryonic day 11 embryo containing the H11P3-pHb9-GFP allele. A WT littermate is also shown (Right). (Scale bar, 1 mm.) (C) Immunofluorescence of sections from embryonic day 11 spinal cords at limb level with anti-GFP signal in green, anti-Hb9 signal in red, and DAPI in blue. Insets: Magnified bottom left portions of each image containing Hb9-positive nuclei. (Scale bar, 100 μm.)
Integration Efficiency.
We compared the integration efficiency for attP-modified loci, expressed as the percentage of F0 animals with site-specific integrations obtained from the total number of F0s (Table 1; SI Appendix, Table S1, provides more details). Although in pooled data (SI Appendix, Table S3), H11P3 (three copies of shortened attP) appeared somewhat more efficient than H11P (one copy of the full-length attP), the efficiencies of site-specific insertions into these two loci were statistically indistinguishable (SI Appendix, Table S3, compare rows 1 vs. 2; and Table 1, compare rows 1 vs. 2, 6 vs. 7, and 8 vs. 9). In contrast, outcrossing the H11P3 mice to the FVB strain for four generations (FVB N4) significantly increased the integration efficiency to approximately 40% (Table 1, compare rows 2 vs. 3 and 9 vs. 10; and SI Appendix, Table S3, compare rows 2 vs. 3). This efficiency is comparable to or better than the efficiency of traditional transgenesis with random integration. Circular DNAs with sizes from 3 to 6 kb appeared to have similar efficiencies of integration (Table 1), but larger DNA (14 kb) showed decreased integration efficiency (∼3%; Table 1, row 11). The efficiency of cassette exchange by using H11P3 is approximately 30%, but because identical attB sites in the plasmid and identical attP sites in the genome were used, cassette exchange could result in either integration of the transgene of interest or the bacterial backbone. Therefore, only half of the cassette-exchange insertions (∼16%) contained GFP and the other half contained the plasmid backbone (Table 1, row 4).
Although we used circular DNA for injections, we also observed insertions at locations other than our intended attP sites (Table 1). In 20 of 23 founders that transmitted their site-specific transgenes to the progeny, the site-specific integrants contained a single-copy transgene and did not contain a second random insertion as judged by PCR and quantitative PCR (SI Appendix, SI Materials and Methods). In rare cases, when site-specific and random integration occurred in the same transgenic founder, the two distinct transgene integrations could be readily segregated in the N1 progeny.
Site-Specific Integration into Rosa26.
To test whether ϕC31-mediated integration is applicable to other genomic loci, we injected pattB-pCA-GFP into embryos homozygous for R26P3NVϕ (attPx3+NVϕ integrated into the Rosa26 locus). We obtained site-specific integrants (Table 1, row 12). We have removed the NVϕ cassette from R26P3NVϕ by using Flpo, and have recently created homozygous R26P3 mice to provide a second locus for integrase-mediated transgenesis.
Discussion
Here we describe an efficient method for producing transgenic mice containing an intact, single-copy transgene integrated into a predetermined locus via pronuclear injection. Our method is considerably simpler than transgenesis using homologous recombination in ES cells and offers many technical advantages compared with the current method of random integration of transgenes via pronuclear injection (1–3). Transgenes produced from our site-specific integration method are intact, have a defined copy number and chromosomal environment, and do not disrupt endogenous genes (at least at the H11 locus). These properties will facilitate many transgenesis-based experiments and will increase their reliability and efficiency. For example, the relationships between amino acid sequences or domain structures of a protein and its in vivo biological functions can be more reliably compared if a series of transgenes encoding different variants of a protein are expressed at the same level. The regulatory elements that control gene expression can also be systematically dissected when reporter transgenes from the same integration site are compared. Subtle differences in levels or patterns of transgene expression that would be overwhelmed by positional effects and differences in copy numbers in randomly integrated transgenes are more appropriately compared by using site-specific integration of transgenes.
Recently, two other approaches for site-specific transgenesis in mice using pronuclear injection were reported (39–41). One approach relied on zinc-finger nucleases to create site-specific double stranded breaks that were repaired by injected recombinant DNA via homologous recombination (39, 41). The two reports using this approach achieved transgene integration with a frequency of approximately 2.5% (two events among 80 embryos) into the Rosa26 locus (39), and approximately 5% (two of 40 for GFP insertion) into the Mdr1a locus (41). Both reports have yet to demonstrate germline transmission and proper adult expression of the transgenes, although proper germline transmission for the same technique in rats was reported (41). The other approach used the Cre recombinase to catalyze cassette exchange in the Rosa26 locus and a tissue-specific locus, H2-Tw3, at an average frequency of approximately 4.3%, with proper expression and germline transmission of the transgenes (40). One drawback of the Cre-based method is that it cannot be used to generate Cre-activated transgenes (containing loxP-STOP-loxP), which are frequently used for reporting Cre activity and perturbing gene function in cells in which Cre is expressed. The integration frequencies of these studies and the present study cannot be easily compared, as the studies used different strains, loci, and constructs. However, our approach with the FVB strain and the H11 locus consistently produces higher integration efficiencies with 3- to 6-kb plasmids than either of the other two approaches.
The present study also revealed that plasmid bacterial backbone and a nearby transgenic cassette (NVϕ) have profound effects on the expression reliability of the GFP transgenes driven by a ubiquitous promoter. We did not observe any obvious change in HB9-GFP transgene expression upon removal of the bacterial backbone; this observation could be the consequence of small numbers of animals compared, or a result of the possibility that the bacterial backbone could have different effects on different promoters. The effect of bacterial backbone has been reported for randomly integrated and episomal transgenes (34–37), and other native bacterial sequences like the lacZ gene or the neomycin resistance gene have been linked to variegation in transgenic animals (40, 42–44). Our experiments based on site-specific integration enabled us to systematically and quantitatively characterize these effects for single-copy chromosomally integrated transgenes.
We have generated mice that allow integration at two defined loci, the widely used Rosa26 locus (26) and the new Hipp11 locus (27), which support high-level ubiquitous expression of integrated transgenes. We describe three different approaches to create transgenes devoid of the bacterial backbone: (i) use of minicircle DNA for transgenesis, (ii) flanking the gene of interest with two attB sites in a plasmid to enable cassette exchange, (iii) removing the bacterial backbone from a transgene generated from a plasmid by crossing the transgenic mice to GFP-Flpo transgenic mice. Although currently only half of the cassette exchange events are desirable, the cassette exchange strategy removes the bacterial backbone without the need to produce minicircle DNA or to remove the plasmid backbone by subsequent crossing to GFP-Flpo mice. Therefore, the cassette exchange may be the approach of choice because of its combination of convenience and good integration efficiency. A future improvement could use two mutually noncompatible pairs of attB and attP sites to control for the direction of insertion. In addition, to expand the application of our method for producing transgenic mouse models, we are in the process of introducing the attP sites into the frequently used C57BL/6 genetic background. In summary, the present study facilitates murine transgenesis, highlights the requirements for gene expression reliability in mammals, and provides an efficient system for studies of gene expression and function in vivo.
Materials and Methods
Recombinant DNA.
We used standard methods of recombinant DNA to construct all plasmids used in this study. Construction details are described in SI Appendix, SI Materials and Methods.
Gene Targeting in Mouse ES Cells.
We used standard techniques to modify mouse ES cells (45). SI Appendix, SI Materials and Methods, provides more details.
Mouse Breeding and Maintenance.
All experimental procedures were carried out in accordance with the Administrative Panel on Laboratory Animal Care protocol and the institutional guidelines by the Veterinary Service Center at Stanford University.
The F1 attP knock-in animals obtained from the cross of chimeras to B6D2F1/J females (stock no. 100006; Jackson Laboratories) were crossed to each other to establish homozygous knock-in mouse lines. These lines were maintained by intercrosses between homozygous animals. To outcross the mice to FVB (Charles River), we started from a homozygous transgenic male and bred him and his transgenic male progeny to FVB females for a total of four generations. During the outcrossing, we preferentially selected transgenic mice of white coat color. The fourth generation outcrossed mice (FVB N4) were crossed to each other to make homozygous males and females that were subsequently used to produce zygotes for microinjection. The FVB N4 homozygous line was subsequently maintained by homozygous crosses. For testing transgenic founders we crossed the founder (F0) animals to WT CD1 mice (Charles River) to generate the N1 generation. For the N2 and N3 generations, we continued crossing to CD1. We have generated homozygous mice from the founder E1 (SI Appendix, Table S2), and they were healthy and fertile.
Preparation of mRNA and DNA for Microinjection.
Capped mRNAs for ϕC31o and Flpo were generated by using a mMESSAGEmMACHINE in vitro transcription kit (Ambion) according to the manufacturer's instructions from BamHI-digested pBT317 (SI Appendix, SI Materials and Methods) and BssHII-digested pFlpo (23), respectively. The integrity of the RNA was assessed by electrophoresis on a 1% agarose gel. Before loading on the gel, the RNA was denatured by using the loading buffer provided in the Ambion kit according to the manufacturer's instructions.
Plasmid DNA was prepared using a modified Qiagen miniprep procedure and was subsequently extracted with phenol/chloroform (SI Appendix, SI Materials and Methods). The DNA was diluted to 6 ng/μL by sterile microinjection TE buffer (0.1 mM EDTA, 10 mM Tris, pH 7.5) and was kept at −80 °C until the injection. The DNA was tested to be RNase-free by incubation with an in vitro transcribed RNA at 37 °C for 1 h and then by analyzing the mix on a 1% agarose gel. Before loading on the gel, the RNA was denatured as described earlier. SI Appendix, SI Materials and Methods provides details on preparation of minicircle DNA.
Microinjection for Generation of Site-Specific Integrants.
Microinjection was performed with an established setup at the Stanford Transgenic Facility. Superovulated homozygous attP-containing females were bred to corresponding males to generate homozygous attP-containing zygotes. A DNA/mRNA mix of interest was microinjected into a single pronucleus and cytoplasm of each zygote by using a continuous flow injection mode. The surviving zygotes were implanted into oviducts of pseudopregnant CD1 (Charles River) recipient mothers. All injection mixes contained 3 ng/μL DNA and 48 ng/μL of in vitro transcribed ϕC31o mRNA in microinjection TE buffer (0.1 mM EDTA, 10 mM Tris, pH 7.5). The injection mixes were prepared fresh before each injection by mixing equal volumes of 6 ng/μL DNA solution and 96 ng/μL mRNA solution.
PCR.
To test F0 animals for site-specific and random insertions, we performed three PCRs: one for the 5′-end junction, one for the 3′-end junction, and one internal to the transgene. These PCRs cannot detect random insertions that occurred in mice with site-specific insertions. For that purpose, see PCR analysis of N1 generation below. For testing integration into H11P or H11P3 alleles, we used PCR1, PCR2, and PCR6 (SI Appendix, SI Materials and Methods). To test if a particular site-specific insertion into H11P or H11P3 originated from appropriate cassette exchange or minicircle insertion, we used PCR3 and PCR4 or PCR4′ (SI Appendix, SI Materials and Methods). These PCRs demonstrated that all injections of minicircle DNA produced only minicircle insertions, suggesting that contamination of minicircle preps with full-length plasmid was negligible.
To test germline transmission of both site-specific and random insertions to N1 animals, we performed three PCRs on the N1 progeny: one for the 5′-end junction, one for the 3′-end junction, and one with internal primers. Correlation of 100% between the GFP-specific and site-specific integration PCRs on DNA from N1 animals suggested that the corresponding F0 founder most likely contained only a single site-specific insertion. This conclusion was reinforced by quantitative PCR (SI Appendix, SI Materials and Methods) for GFP to show that a selected number of N1 animals indeed had a single-copy transgene.
SI Appendix, SI Materials and Methods (46, 47), provides additional information on PCR procedures used in this study, and SI Appendix, Table S4 provides primer sequences.
Tissue Preparation and Immunohistochemistry.
The procedures were performed essentially as described (29). SI Appendix, SI Materials and Methods, includes further details.
Quantification of GFP Fluorescence in Liver Sections.
At least three individual images were taken from randomly chosen 10-μm sections for each liver by a camera connected to a fluorescence microscope (Nikon) with a 20× objective. The regions of interest were consistently chosen to contain minimal number of large blood vessels, so that the majority of every image would be covered by hepatocytes. All images were taken with the same exposure time (5 ms), same gain, and during two consecutive days of imaging. At this condition, even the samples with brightest fluorescence had no saturated pixels. Total fluorescence for each image was calculated by using ImageJ. Averaged total fluorescence from all images of the same liver was plotted on a graph (SI Appendix, Fig. S2). The fluorescence images shown in figures represent the same fields that were used for the measurements, but exposed four times longer for easier visualization.
Animal and Reagent Availability.
Plasmids (containing attB sites or integrase cDNA) and H11P3 and R26P3 homozygous frozen embryos and mice will be distributed through Applied StemCell, Inc. (www.appliedstemcell.com), for prices comparable to those of other distributors (e.g., Addgene for plasmids, Jackson Labs for mice). Applied StemCell will also provide services for making customized, integrase-mediated site-specific transgenic mice.
Supplementary Material
Acknowledgments
We thank Yanfeng Li, Jennifer Lin, Hong Zeng, Ying Jiang, and Carlota Manalac for technical support; Michele Calos for plasmids; Russell Fernald for real-time PCR machine; Silvia Arber for anti-Hb9 antibody; Dritan Agalliu for help with embryo dissections; and Kazunari Miyamichi and Tim Mosca for comments on the manuscript. This work is supported by National Institutes of Health Grant R01-NS050835. B.T. was a Damon Runyon Fellow and was supported by Damon Runyon Cancer Research Foundation Grant DRG-1819-04. S.H. was supported by postdoctoral fellowships from the European Molecular Biology Organization (ALTF 851-2005), Human Frontier Science Program Organization (LT00805/2006-L), and Swiss National Science Foundation (PA00P3_124160). L.L. is an investigator of The Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: Y.C.-T. is a consultant and co-founder of Applied StemCell, Inc.
*This Direct Submission article had a prearranged editor.
See Commentary on page 7659.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019507108/-/DCSupplemental.
References
- 1.Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci USA. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon JW, Ruddle FH. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214:1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- 3.Brinster RL, et al. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27:223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milot E, et al. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 5.Pedram M, et al. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol Cell Biol. 2006;26:1865–1878. doi: 10.1128/MCB.26.5.1865-1878.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Q, et al. Telomeric transgenes are silenced in adult mouse tissues and embryo fibroblasts but are expressed in embryonic stem cells. Stem Cells. 2007;25:3085–3092. doi: 10.1634/stemcells.2007-0478. [DOI] [PubMed] [Google Scholar]
- 7.Williams A, et al. Position effect variegation and imprinting of transgenes in lymphocytes. Nucleic Acids Res. 2008;36:2320–2329. doi: 10.1093/nar/gkn085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 9.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 10.Ding S, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Mátés L, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 12.Doetschman T, et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 13.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 14.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keravala A, et al. A diversity of serine phage integrases mediate site-specific recombination in mammalian cells. Mol Genet Genomics. 2006;276:135–146. doi: 10.1007/s00438-006-0129-5. [DOI] [PubMed] [Google Scholar]
- 16.Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 19.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olivares EC, et al. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat Biotechnol. 2002;20:1124–1128. doi: 10.1038/nbt753. [DOI] [PubMed] [Google Scholar]
- 21.Hollis RP, et al. Phage integrases for the construction and manipulation of transgenic mammals. Reprod Biol Endocrinol. 2003;1:79. doi: 10.1186/1477-7827-1-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat Biotechnol. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- 23.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS ONE. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sangiorgi E, Shuhua Z, Capecchi MR. In vivo evaluation of PhiC31 recombinase activity using a self-excision cassette. Nucleic Acids Res. 2008;36:e134. doi: 10.1093/nar/gkn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 26.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 27.Hippenmeyer S, et al. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron. 2010;68:695–709. doi: 10.1016/j.neuron.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 30.Siemering KR, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 31.Okada A, Lansford R, Weimann JM, Fraser SE, McConnell SK. Imaging cells in the developing nervous system with retrovirus expressing modified green fluorescent protein. Exp Neurol. 1999;156:394–406. doi: 10.1006/exnr.1999.7033. [DOI] [PubMed] [Google Scholar]
- 32.Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seibler J, Bode J. Double-reciprocal crossover mediated by FLP-recombinase: A concept and an assay. Biochemistry. 1997;36:1740–1747. doi: 10.1021/bi962443e. [DOI] [PubMed] [Google Scholar]
- 34.Townes TM, Lingrel JB, Chen HY, Brinster RL, Palmiter RD. Erythroid-specific expression of human beta-globin genes in transgenic mice. EMBO J. 1985;4:1715–1723. doi: 10.1002/j.1460-2075.1985.tb03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 36.Chen ZY, He CY, Meuse L, Kay MA. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 2004;11:856–864. doi: 10.1038/sj.gt.3302231. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki M, Kasai K, Saeki Y. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J Virol. 2006;80:3293–3300. doi: 10.1128/JVI.80.7.3293-3300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arber S, et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 39.Meyer M, de Angelis MH, Wurst W, Kühn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtsuka M, et al. Pronuclear injection-based mouse targeted transgenesis for reproducible and highly efficient transgene expression. Nucleic Acids Res. 2010;38:e198. doi: 10.1093/nar/gkq860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui X, et al. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 42.Montoliu L, Chávez S, Vidal M. Variegation associated with lacZ in transgenic animals: a warning note. Transgenic Res. 2000;9:237–239. doi: 10.1023/a:1008995730285. [DOI] [PubMed] [Google Scholar]
- 43.Cohen-Tannoudji M, Babinet C, Morello D. lacZ and ubiquitously expressed genes: Should divorce be pronounced? Transgenic Res. 2000;9:233–235. doi: 10.1023/a:1008916910392. [DOI] [PubMed] [Google Scholar]
- 44.Fiering S, et al. Targeted deletion of 5’HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 45.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, et al. Visualizing the distribution of synapses from individual neurons in the mouse brain. PLoS ONE. 2010;5:e11503. doi: 10.1371/journal.pone.0011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.