Abstract
Cut-and-paste DNA transposable elements are major components of eukaryotic genomes and are grouped into superfamilies (e.g., hAT, P) based on sequence similarity of the element-encoded transposase. The transposases from several superfamilies possess a protein domain containing an acidic amino acid triad (DDE or DDD) that catalyzes the “cut and paste” transposition reaction. However, it was unclear whether this domain was shared by the transposases from all superfamilies. Through multiple-alignment of transposase sequences from a diverse collection of previously identified and recently annotated elements from a wide range of organisms, we identified the putative DDE/D triad for all superfamilies. Furthermore, we identified additional highly conserved amino acid residues or motifs within the DDE/D domain that together form a “signature string” that is specific to each superfamily. These conserved residues or motifs were exploited as phylogenetic characters to infer evolutionary relationships among all superfamilies. The phylogenetic analysis revealed three major groups that were not previously discerned and led us to revise the classification of several currently recognized superfamilies. Taking the data together, this study suggests that all eukaryotic cut-and-paste transposable element superfamilies have a common evolutionary origin and establishes a phylogenetic framework for all future cut-and-paste transposase comparisons.
Transposable elements (TEs) are fragments of DNA that can move to new genomic locations. The element originally discovered by Barbara McClintock over 60 y ago are referred to as “cut-and-paste” TEs that transpose via a double-strand DNA intermediate (1, 2). Cut-and-paste TEs are now recognized as a major component of most eukaryotic genomes. For example, they comprise ∼25% of the genomic DNA of Xenopus tropicalis (western clawed frog), ∼20% of Hydra magnipapillata (hydra), >19% of Aedes aegypti (yellow-fever mosquito), >13% of Oryza sativa ssp. japonica (rice), and >6% of Phytophthora infestans (potato late blight) (3–7). Even in the human genome, where they account for only 3% of genomic DNA, there are almost 400,000 individual elements (8).
Cut-and-paste TEs are characterized by a transposase gene that is flanked by a terminal inverted repeat (TIR) of variable length. The transposase catalyzes DNA cleavage during the “cut and paste” process, whereby the element is excised from the donor site (causing a double-strand break) and inserted elsewhere in the genome. The TE sequence could be restored to the empty donor site via host repair of the double-strand break, leading to an increase in copy number. Integration of the elements into a new genomic location usually generates a short target-site duplication (TSD) from host sequences (2–10 bp).
Eukaryotic cut-and-paste TEs are grouped into superfamilies (e.g., Tc1/mariner, hAT, P) primarily on the basis of sequence similarity of the transposase. As a rule of thumb, transposase sequences with an E-value less than 0.01 in BLASTP or PSI-BLAST searches are assigned to the same superfamily (9). In addition, the length of the TSD and often the terminal nucleotides of the TIR are also diagnostic of each superfamily (e.g., hAT TSD: 8-bp; Tc1/mariner TSD: “TA”). All superfamilies contain autonomous and nonautonomous members with autonomous elements encoding the protein products required for transposition; nonautonomous elements use transposase encoded by autonomous elements located elsewhere in the genome. Prior studies have classified eukaryotic cut-and-paste transposases into 19 superfamilies, including hAT, Tc1/mariner, CACTA (En/Spm), Mutator (MuDR), P, PiggyBac, PIF/Harbinger, Mirage, Merlin, Transib, Novosib, Rehavkus, ISL2EU, Kolobok, Chapaev, Sola, Zator, Ginger, and Academ [see Repbase (10); http://www.girinst.org/repbase/index.html]. Six of these 19 superfamilies (Mirage, Novosib, Rehavkus, ISL2EU, Kolobok, Academ) have been described only in Repbase and, as such, their validity and relationship to other superfamilies remains to be evaluated (11).
A catalytic domain signified by an acidic amino acid triad, known as the “DDE/D” motif, has been unambiguously identified in the transposases from 11 of the 19 currently recognized superfamilies [hAT (12, 13), Tc1/mariner (14), Mutator (15), PIF/Harbinger (16), Merlin (17), Transib (18), Chapaev (19), PiggyBac (20), Sola (21), Zator (21), Ginger (22)]. The DDE/D motif consists of two aspartic acid (D) residues and a glutamic acid (E) residue or a third D, located in a conserved core that forms a characteristic RNase H-like fold of mixed α-helices and β-strands (β1-β2-β3-α1-β4-α2/3-β5-α4-α5/6) (reviewed in ref. 11). The first D is located on β1, the second D is on or just after β4, and the third D/E appears on or just before α4 (11). For members of two superfamilies (Hermes: hAT; Mos1: Tc1/mariner) the 3D structure of the DDE/D triad forms a catalytic pocket containing two divalent metal ions that assist in the various nucleophilic reactions during DNA cleavage (13, 23). For the other eight superfamilies, including the extensively studied P and CACTA, the catalytic domain remains either undetermined or ambiguous. Characterization of the catalytic domain in these superfamilies is a necessary prerequisite to understanding both their transposition mechanism and evolutionary origin. For example, absence of a DDE/D domain could indicate that a superfamily uses a distinct protein domain to catalyze DNA cleavage that may have evolved independently from those with the DDE/D domain.
The initial objective of this study was to identify the putative catalytic domains of all currently recognized superfamilies. This objective was accomplished by collecting diverse TEs of each superfamily from a wide range of organisms and performing multiple alignments of the transposase amino acid sequences. In the course of the analysis, we found that all superfamilies not only have a DDE/D domain, but also a superfamily-specific “signature string” consisting of multiple highly conserved amino acid residues and motifs within the DDE/D domain. When these residues were used as phylogenetic characters, we could infer the evolutionary relationships among superfamilies and revise the classification of several currently recognized superfamilies. Finally, we surveyed the taxonomic distribution of each superfamily in the revised classification along 50 major branches of the eukaryotic tree of life.
Results
Identification of the DDE/D Domain in All Superfamilies.
A major impediment to identifying the DDE/D domain in 8 of the 19 transposase superfamilies has been the lack of TE sequence from a wide range of organisms. To remedy this situation, we collected sequences for each superfamily from selected genomes that represent the diversity of eukaryotes and performed multiple alignments of the deduced protein sequences. To this end, we first analyzed autonomous TEs previously deposited in Repbase (see Methods) and assessed which superfamilies were inadequately represented and which were not. For example, no additional collections of Tc1/mariner and hAT superfamily members were needed, as each was represented by over 450 Repbase entries from a wide range of organisms. For all other superfamilies, we first identified the most conserved region from Repbase entries and used these regions as queries to annotate homologous TEs from genomes not currently represented in Repbase (see details in Methods). For these new collections, it was not necessary nor feasible to annotate all related TEs in the ∼160 genomes that were selected to represent the major lineages of the eukaryotic tree of life. Instead, we took a taxon sampling approach. For example, if there are >10 species in the fungal clade “Sordariomycetes” that have Mutator elements, we selected only one species to annotate, with the assumption that the Mutator elements in one species are likely to represent the diversity of the whole clade.
The newly annotated TE sequences were appended to the original Repbase entries and multiple-alignment was performed. We then trimmed the regions that were not conserved at the ends of the alignment and eliminated redundant elements by retaining only one member from a group of elements with >40% identity over the conserved transposase region (Methods). In this way we produced an alignment profile (Dataset S1) representing the diversity of each superfamily with a minimum number of sequences.
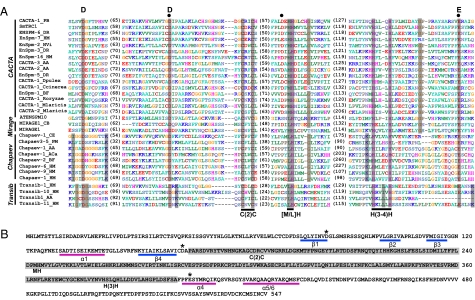
Examination of the conserved blocks in the alignment profiles revealed conserved DDE/D triads in the transposases of all eight superfamilies where the DDE/D domain was undetermined (i.e., Mirage, ISL2EU, Rehavkus, Novosib, Kolobok, and Academ) or where identification was inconclusive (i.e., CACTA, P) (Fig. 1 and Figs. S1–S6). As an example, Fig. 1A shows the alignment of the DDE domains of transposases from superfamilies CACTA, Mirage, Chapaev, and Transib, which are considered together because the conserved blocks that encompass the DDE triad could be aligned across all four groups. The DDE triad identified here (Fig. 1A, noted above the alignment) corresponds to D84, D164, and E406 of the SmTRC1 transposase (24) (GenBank: AM268206). A previous study based on SmTRC1 also identified D84 and D164 as part of the putative DDE triad, but the E residue proposed in that study (E136) is located between the two Ds and is not conserved (not shown in Fig. 1A, as it is located in a variable region). A more recent report in Repbase suggests D164, D241, and E406 as the DDE triad (25) (marked by black dots in Fig. 1A). Again, this is unlikely, as the second D is not as highly conserved in our alignment (Fig. 1A). The DDE triad identified by our alignment is conserved across all four superfamilies. Furthermore, the predicted secondary structure of the SmTRC1 DDE domain using PSIPRED (26) assumed a classic RNase H-like fold, with the first D on β1, the second D right after β4, and the E on α4 (Fig. 1B).
Fig. 1.
DDE domains of CACTA, Mirage, Chapaev, and Transib elements. (A) Alignment shown is after redundancy elimination. Distances between the conserved blocks are indicated in the number of amino acid residues. Conserved residues within each superfamily (and between superfamilies) are highlighted in gray. The DDE triad identified here is marked with letters above the alignment; the DDE triad for CACTA elements identified in ref. 25 is marked with black dots. Three additional conserved motifs discussed in the text, C(2)C, [M/L]H, and H(3-4)H, are also noted. (B) Predicted secondary structure of the DDE domain of the SmTRC1 transposase (GenBank: AM268206). Asterisks indicate the DDE triad. α-Helices and β-strands are highlighted with pink and blue bars, respectively. Note that the position of “α2/3-β5” of the typical “β1-β2-β3-α1-β4-α2/3-β5-α4-α5/6” fold remains unclear. The inserted domain (highlighted in gray) between the second D and the E residue is rich in α-helices.
The results for the other 11 superfamilies are consistent with previous studies (e.g., Transib and Chapaev in Fig. 1A; PIF/Harbinger in Fig. S2A; Mutator in Fig. S3; PiggyBac in Fig. S7). The DDE/D alignment profiles generated for Tc1/mariner, Merlin, hAT, Sola, Zator, and Ginger are available from Dataset S1.
“Signature String” for Each Superfamily.
The multiple alignments also revealed other highly conserved residues or motifs (i.e., occurring in >90% of the sequences in the alignment profile) within the DDE/D domain. Using the CACTA superfamily again as an example, there are three highly conserved motifs [C(2)C, [M/L]H, and H(3-4)H] that are located at approximately the same position between the second D and the E residues (Fig. 1A, labeled by black bars below the alignment). A glycine (G) 4 aa downstream of the [M/L]H motif and another G 8 aa downstream of the H(3-4)H motif are also conserved. These conserved residues and motifs, including the approximate spacing between them, form a “signature string” that is specific to each superfamily (illustrated in Fig. 2; also see Table S1).
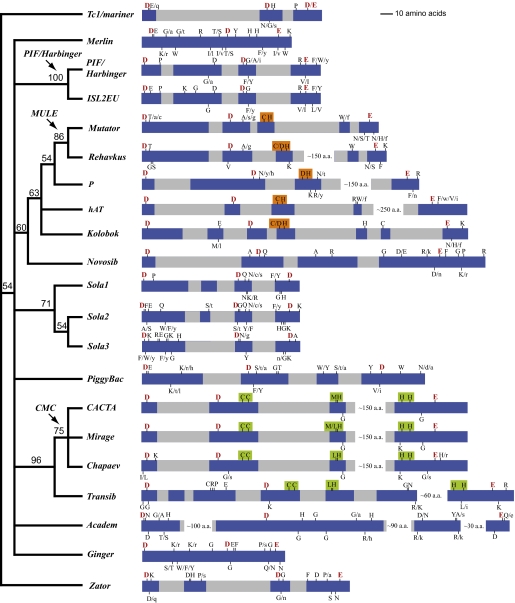
Fig. 2.
An unrooted consensus tree of the transposase superfamilies inferred from the presence or absence of the highly conserved residues in the signature strings. Bootstrap values are at the nodes. The arrows with labels indicate superfamily clusters merged in our revised classification. Shown on the right is a schematic representation of the DDE/D domain and the signature string for each superfamily. Conserved blocks are highlighted in blue, variable regions are in gray. White gaps are regions not drawn to scale. The DDE triads are highlighted in red. Alternative residues are marked by slashes; lowercase indicates that a residue occurs in <10% of the sequences in the alignment profile. The [C/D](2)H motif is highlighted in orange; the C(2)C, [M/L]H, and H(3-4)H motifs are highlighted in green.
Some components of the signature strings are shared by multiple superfamilies. For example, the G residue 3 to 4 aa downstream of the [M/L]H motif is shared by CACTA, Mirage, Chapaev, but not by Transib (Fig. 1A), but the C(2)C, [M/L]H, and H(3-4)H motifs are shared by all four superfamilies (black bars in Fig. 1A; also see Fig. 2, highlighted in green). A C(2)H motif was found in both hAT (12) and Mutator (27), and a slightly different version of this motif, [C/D](2)H, was found in P, Rehavkus, and Kolobok (Fig. 2, highlighted in orange).
Evolutionary Relationships Among Superfamilies and Their Revised Classification.
The notion that certain conserved residues or motifs are shared by some superfamilies but not by others motivated us to systematically exploit these signature string components as phylogenetic characters to infer relationships among superfamilies. In brief, we coded the presence or absence state of all highly conserved residues and motifs in the signature strings as “1” or “0”, respectively, and then performed phylogenetic analysis on this binary character matrix (available on request) using the Parsimony criterion (see Methods). Our phylogenetic analysis revealed several close relationships among different superfamilies, which led us to revise the classification of some of the currently recognized superfamilies.
Previously, cut-and-paste TE superfamilies were usually classified based on transposase sequence similarity, where E-values less than 0.01 in BLASTP or PSI-BLAST searches identified members of the same superfamily (9). Here we employ a phylogenetic approach to complement the BLAST-based method by using shared derived characters to define monophyletic superfamilies. Specifically, we use the signature strings of the DDE/D domains as the primary feature and use the TSD and TIR as additional characters.
Our analysis revealed three major groups that were not previously discerned. The first contains PIF/Harbinger and ISL2EU (Fig. 2), two superfamilies that share multiple conserved residues besides the DDE triad (highlighted in gray in Fig. S2A). Their similar TSDs are also consistent with this relationship. Although the previously characterized PIF/Harbinger elements usually generate “TWA” (W = A or T) duplications upon insertion (16), we found PIF/Harbinger elements from fungi and Chromalveolates protists that frequently generate “AWT” duplications (two examples are shown in Fig. S2B). On the other hand, ISL2EU elements usually generate “AT” TSDs (Fig. S2B). On the basis of this close relationship, we lumped ISL2EU into the well-established PIF/Harbinger superfamily.
The second major group consists of Mutator, Rehavkus, P, hAT, Kolobok, and Novosib. All but Novosib share a [C/D](2)H motif 15 to 45 aa downstream of the second D of the DDE triad. This motif is always right after β5 in the predicted secondary structure (12, 14) (Figs. S1B and S4B). Novosib is included in this group because it shares similarities in the conserved block containing the E residue with the P and hAT superfamilies, including an “F” 2 aa downstream of the E and a “D” 2 aa upstream of the E residue (Figs. S1A and S6A). The grouping of these six superfamilies is also consistent with their unusually long TSDs. For example, the predominant TSDs for hAT, P, and Novosib are 8 bp, and 9 to 10 bp for Mutator and Rehavkus. The only exception is Kolobok, which usually produces a “TTAA” TSD (28), similar to PiggyBac and Sola3 (21). The support for this major group is relatively weak (Bootstrap value is 60%) (Fig. 2) because of the small number of shared conserved residues. As such, we consider this grouping tentative because the possibility remains that convergent evolution may explain their shared features.
On the other hand, within the second major group a smaller monophyletic group composed of Mutator and Rehavkus is strongly supported (Fig. 2). In addition to the multiple shared conserved residues in the signature string (Fig. S3, highlighted in gray), both TSD and TIR features support this relationship. Rehavkus produce 9-bp TSDs, similar to Mutator (9–10 bp), and most importantly, both Rehavkus and Mutator tend to have longer TIRs (hundreds of nucleotides) than other superfamilies. On the basis of this evidence, we lumped Rehavkus into the well-established Mutator superfamily and applied the previously used name MULE (Mutator-like element) (29) for the united superfamily.
The third major group comprises CACTA, Mirage, Chapaev, and Transib, which all share three highly conserved motifs, C(2)C, [M/L]H, and H(3-4)H (Fig. 2, highlighted in green). Evidence from the terminal nucleotides of TIRs also supports this grouping: all four superfamilies have the conserved terminal nucleotides “CMC” (M = A or C). Within this group, CACTA, Mirage, and Chapaev are more closely related and form a subgroup that can be readily distinguished from Transib (Fig. 2). CACTA, Mirage, and Chapaev share an additional “G” 3 to 4 aa downstream of the [M/L]H motif (Fig. 1A) that is not shared by Transib. Likewise, there are multiple conserved residues unique to Transib (e.g., a “K” residue 1 aa downstream of the second D) (Fig. 1A). Moreover, the TSD lengths of the CACTA-Mirage-Chapaev subgroup overlap in a continuum (CACTA is 2–3 bp, Mirage is 2 bp, and Chapaev is 3–4 bp), whereas Transib has distinct 5-bp TSDs (18). Because of these differences, we combined CACTA, Mirage, and Chapaev into a CMC (initials of the three old names) superfamily but retained Transib as a distinct superfamily.
Finally, our analysis led to modification in the classification of some of the remaining superfamilies. When first described, the superfamily complex Sola1-Sola2-Sola3, was grouped into a single superfamily, Sola (21). However, we found it necessary to maintain the three lineages as separate superfamilies because, aside from the DDD triad, there is not a single amino acid residue that is conserved across all three groups. In contrast, after splitting there are multiple conserved residues supporting the monophyly of each group (Fig. 2). The potential affinity between Tc1/mariner and Zator (21) is not strongly supported by our analysis but is consistent with the tree topology (Fig. 2; note the “hidden” relationship between Tc1/mariner and Zator because of the unrooted nature of the tree). Because these two superfamilies can be readily distinguished from their signature strings and TSD features (Table S1), we have maintained them as distinct superfamilies. The remaining superfamilies, PiggyBac, Merlin, Ginger, and Academ, are not closely related to each other or to any other superfamilies in our analysis.
Taking these data together, our revised classification system contains 17 superfamilies. The signature string, TSD feature, and TIR terminal motif for each of the 17 superfamilies are summarized in Table S1.
Superfamily Distribution Across the Eukaryotic Tree of Life.
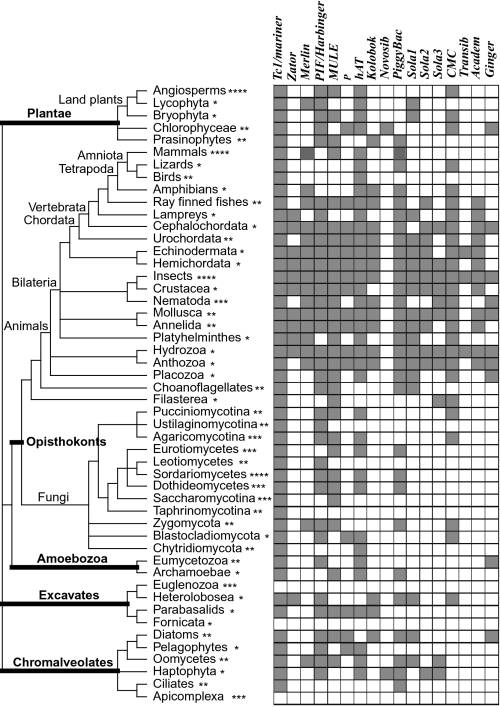
The distribution of the 17 superfamilies along 50 major eukaryotic lineages (represented by ∼160 selected genomes) is shown in Fig. 3. The presence of a superfamily in a genome was determined by TBLASTN searches using the DDE/D domain as query (see Methods).
Fig. 3.
Taxonomic distribution of the 17 superfamilies across the eukaryotic tree of life. Gray and white boxes indicate presence and absence, respectively. The illustrated tree was drawn according to refs. 32 and 33 and the Tree of Life webpage (http://tolweb.org/tree/). The five represented eukaryotic supergroups are highlighted in thickened lines. The asterisks after each terminal branch indicates the number of genomes representing that branch: *, 1 genome; **, 2 to 5 genomes; **, 6 to 10 genomes; ****, over 10 genomes.
Our survey has significantly expanded the taxonomic distribution of 7 of the 17 superfamilies (Table S2). For example, until this study the well-established PIF/Harbinger superfamily was not known to occur in Amoebozoa or Excavates. Similarly, P elements were recovered from fungi and Chromalveolates. The distribution of Merlin was extended into both fungi and plants. Kolobok, which had only been reported in animals (28), was found in three additional eukaryotic supergroups (Plantae, Excavates, and Chromalveolates).
Discussion
Our results indicate that all eukaryotic cut-and-paste transposase superfamilies detected to date have the DDE/D domain, suggesting a common evolutionary origin of the “cut-and-paste” transposition mechanism. Two major factors enabled us to identify the DDE/D domain in superfamilies where previously it remained undetermined or inconclusive. The first factor is broad sampling. To capture the diversity of transposases, new elements from each superfamily (except Tc1/mariner and hAT, as mentioned in Results) were annotated along major lineages of the eukaryotic tree of life, especially formerly underrepresented protist lineages. Increasing transposase diversity has greatly assisted in manifesting the most conserved amino acid blocks and highly conserved residues within these blocks. For example, our identification of the PiggyBac DDD triad differs slightly from a recent report (21) (marked by asterisks in Fig. S7 and corresponding to D268, D346, and D450 of the Trichoplusia ni PiggyBac transposase), where the third D (D450) is not universally conserved after adding newly annotated TEs from Chromalveolates protists. Instead, D447 is universally conserved in our alignment (Fig. S7). Our results are consistent with a previous experimental study (20), where mutations of D268, D346, and D447 completely abolished the catalytic activity of the transposase and mutation of D450 had no detectable effect on transposition in vitro.
The second factor contributing to the success of finding previously unidentified DDE/D domains is the comparison of the signature strings between related superfamilies. It is only after finding that signature strings of some superfamilies share similarities that we could use this strategy. For example, a recent attempt to identify the DDE domain of P elements revealed four highly conserved acidic residues (30), which is consistent with our results (Fig. S1A, three labeled by letters and one with an arrow). However, multiple-alignment of diverse sequences alone is not sufficient to distinguish which three of the four residues comprise the catalytic triad. In comparison with the related hAT and Mutator elements, we deduced that one of the “D” residues is part of the [C/D](2)H motif (Fig. 2, highlighted in orange) rather than the DDE motif, and the other three conserved residues, corresponding to D230, D303, and E531 of the P transposase from D. melanogaster (UniProt: Q7M3K2), form the putative DDE triad (Fig. 2 and Fig. S1A). This inference is strongly supported by the predicted secondary structure: the DDE domain has the classical “β1-β2-β3-α1-β4-α2/3-β5-α4-α5/6” fold, with an inserted domain between β5 and α4 (Fig. S1B). The critical GTP-binding motif of the Drosophila P element, NKSD (Fig. S1B, residue 376–379) (31), is located within this inserted domain. The fact that this motif is not conserved in P elements from non-Drosophila species suggests that the GTP-binding activity might be a cryptic feature that has evolved only in Drosophila P elements.
It should be noted that our assignment of the DDE triad for the Novosib superfamily is tentative, because thus far only seven Novosib elements from two genomes have been detected (Fig. S6). In contrast, all other superfamilies were represented in our analyses by diverse sequences from numerous genomes representing multiple domains of eukaryotic life, and therefore, the identified DDE/D domains are robust.
Evolutionary Relationships Among Superfamilies and Revised Superfamily Classification.
This study is unique in analyzing evolutionary relationships among all eukaryotic cut-and-paste transposase superfamilies. This analysis is important for two major reasons. First, we have evaluated the six superfamilies (Mirage, Novosib, Rehavkus, ISL2EU, Kolobok, Academ) that have been previously described only in Repbase and, as such, they have not been adequately vetted nor has their relationships to well-established superfamilies been determined. Our analysis revealed close affinities between Mirage and CACTA, between ISL2EU and PIF/Harbinger, and between Rehavkus and Mutator, which suggest that Mirage, ISL2EU, or Rehavkus may not be distinct superfamilies. On the other hand, Novosib, Kolobok, and Academ were sufficiently distinct from all other superfamilies. Second, our analysis established a phylogenetic framework of all known eukaryotic cut-and-paste transposase superfamilies. All future candidate novel transposase superfamilies can be compared with this framework to evaluate their distinctness. Although our analyses focused on eukaryotic elements, the same strategy of identifying signature strings and constructing phylogenetic relationships based on conserved residues could be readily applied to prokaryotic elements, of which the majority are cut-and-paste TEs and possess a DDE domain in their transposases (11).
We took a two-step procedure to revise the superfamily classification. The first step was to recognize monophyletic groups based on phylogenetic relationships inferred from the transposase signature strings. The second step was to compare the TSD and TIR features to determine which superfamilies should be combined and which should not. For example, we have included CACTA, Mirage, and Chapaev in the CMC superfamily but have retained Transib as its own superfamily because the CMC group has TSDs that overlap in a continuum (2–4 bp), whereas Transib has distinct 5-bp TSDs (Table S1). There is a practical reason why this second step is necessary to revise superfamily classifications. Most cut-and-paste TEs in eukaryotic genomes are nonautonomous elements without coding capacity. For these elements, distinctive TSD and TIR features are the only characters available to make superfamily assignments.
Taxonomic Distribution of Superfamilies.
The mapping of the superfamily presence or absence along the eukaryotic tree of life (32, 33) revealed that 15 of the 17 superfamilies exist in at least two of the five eukaryotic supergroups surveyed here (Fig. 3 and Table S2). Because there is little evidence for the horizontal transfer of TEs between eukaryotic supergroups, this distribution strongly supports the view that the origin of most, if not all, superfamilies predates the divergence of eukaryotic supergroups (34).
The superfamily distribution map also has an important use in practice, as it provides an approximation of the cut-and-paste TE landscape of newly sequenced genomes. For example, to identify cut-and-paste TEs in a “Sordariomycetes” fungal genome, a prior expectation would be to find Tc1/mariner, PIF/Harbinger, MULE, hAT, and PiggyBac elements (Fig. 3). As such, this distribution map, together with the DDE/D domain alignment profiles (Dataset S1, which represent the diversity of each superfamily and can be directly used as query for blast searches), could serve as a guide for the annotation of cut-and-paste TEs in all eukaryotic genomes that remain to be sequenced.
Methods
Sequence Analyses.
All autonomous cut-and-paste TE sequences deposited in Repbase (10) (http://www.girinst.org/repbase/index.html) were downloaded in June 2010. Transposase coding sequences were predicted with GeneMark.hmm (http://exon.biology.gatech.edu/eukhmm.cgi) or GENSCAN (http://genes.mit.edu/GENSCAN.html). Multiple-alignments were performed using MUSCLE (http://www.ebi.ac.uk/Tools/muscle/index.html) with default parameters. Aligned sequences were manually inspected in Se-al v.2.0a11 (http://tree.bio.ed.ac.uk/software/seal/) and variable regions at the ends of the alignments were trimmed. A representative “alignment profile” was generated from the remaining conserved part by eliminating redundant elements; that is, only one element was selected to represent a group of elements that are >40% identical. If the group contains an element with demonstrated transposase activity (e.g., MuDR from maize), this element was selected. If an active element was not available, elements with multiple highly similar copies (e.g., >99% nucleotide identity) with intact predicted transposases were chosen. Secondary structure of representative transposases were predicted using PSIPRED (26).
Transposon Annotation.
The alignment profile of each superfamily (except Tc1/mariner and hAT), generated by sequence analysis as described above, was used as query to search the selected genomes by TBLASTN (35), as implemented in the TARGeT pipeline (36), with an E-value cutoff of 0.01. Genomes were selected using the National Center for Biotechnology Information (NCBI) genome project database (http://www.ncbi.nlm.nih.gov/genomes/leuks.cgi) and the Tree of Life webpage (http://tolweb.org/tree/) as guides to represent as many major eukaryotic lineages as possible. Flanking DNA sequences with 10 kb upstream and downstream of the matched region from TBLASTN searches were retrieved. The ends of a putative element were determined by aligning two closely related elements with their 20-kb flanking sequences, using NCBI-BLAST 2 SEQUENCES (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi) on the NCBI server. Usually, the breakpoint of a pair-wise alignment is the boundary of a full-length element, which can be subsequently refined by identifying the TIRs and TSDs around the breakpoint. One full-length element was then used to retrieve other similar copies through BLASTN search using TARGeT and a majority-rule consensus sequence was constructed to represent this family. The newly annotated TE consensus sequences were then subjected to the same sequence analyses as described above and the final DDE/D alignment profile was generated.
Character Coding and Phylogenetic Analysis.
The highly conserved residues and motifs in the signature strings were coded as binary characters. The presence or absence states were represented as “1” or “0”, respectively. Conserved motifs (e.g., [M/L]H) are weighted twice as much as a single amino acid residue. Phylogenetic analysis was performed using the Parsimony criterion as implemented in PAUP* v4.0b10 (37). Heuristic searches were performed with 1,000 random stepwise addition replicates and TBR branch swapping with the MULTREES on. Nodal support was determined by bootstrap analyses of 500 replicates.
Superfamily Distribution.
The final DDE/D alignment profile of each superfamily was used as query for TBLASTN searches using TARGeT. A match that covers >50% of the query (i.e., the DDE/D domain) with an E-value <0.01 was considered as a candidate element. More than 10 copies of such elements in a genome were scored as presence without further annotation. Elements with fewer than 10 copies were inspected to verify TE features, including TSD, TIR, and the signature residues. Presence required verification of two of the three features.
Supplementary Material
Acknowledgments
We thank C. Feschotte for critical reading of the manuscript. This research was supported by the University of Georgia Research Foundation and a grant from the National Science Foundation Plant Genome Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104208108/-/DCSupplemental.
References
- 1.Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- 2.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 3.Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumoto T, et al. International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 7.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 8.Pace JK, 2nd, Feschotte C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: Structure and evolution. Annu Rev Genomics Hum Genet. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- 10.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 11.Hickman AB, Chandler M, Dyda F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit Rev Biochem Mol Biol. 2010;45:50–69. doi: 10.3109/10409230903505596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou LQ, et al. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 13.Hickman AB, et al. Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol. 2005;12:715–721. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- 14.Robertson HM. The Tc1-mariner superfamily of transposons in animals. J Insect Physiol. 1995;41:99–105. [Google Scholar]
- 15.Hua-Van A, Capy P. Analysis of the DDE motif in the Mutator superfamily. J Mol Evol. 2008;67:670–681. doi: 10.1007/s00239-008-9178-1. [DOI] [PubMed] [Google Scholar]
- 16.Zhang XY, Jiang N, Feschotte C, Wessler SR. PIF- and Pong-like transposable elements: Distribution, evolution and relationship with Tourist-like miniature inverted-repeat transposable elements. Genetics. 2004;166:971–986. doi: 10.1534/genetics.166.2.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feschotte C. Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences. Mol Biol Evol. 2004;21:1769–1780. doi: 10.1093/molbev/msh188. [DOI] [PubMed] [Google Scholar]
- 18.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapitonov VV, Jurka J. Chapaev—A novel superfamily of DNA transposons. Repbase Reports. 2007;7:774–774. [Google Scholar]
- 20.Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27:1097–1109. doi: 10.1038/emboj.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bao WD, Jurka MG, Kapitonov VV, Jurka J. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol Biol Evol. 2009;26:983–993. doi: 10.1093/molbev/msp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao WD, Kapitonov VV, Jurka J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob DNA. 2010;1:3. doi: 10.1186/1759-8753-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson JM, et al. Mechanism of Mos1 transposition: Insights from structural analysis. EMBO J. 2006;25:1324–1334. doi: 10.1038/sj.emboj.7601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeMarco R, Venancio TM, Verjovski-Almeida S. SmTRC1, a novel Schistosoma mansoni DNA transposon, discloses new families of animal and fungi transposons belonging to the CACTA superfamily. BMC Evol Biol. 2006;6:89. doi: 10.1186/1471-2148-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapitonov VV, Jurka J. Zebrafish En/Spm DNA transposons. Repbase Reports. 2008;8:750–750. [Google Scholar]
- 26.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33(Web Server issue):W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalvet F, Grimaldi C, Kaper F, Langin T, Daboussi MJ. Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol Biol Evol. 2003;20:1362–1375. doi: 10.1093/molbev/msg155. [DOI] [PubMed] [Google Scholar]
- 28.Kapitonov VV, Jurka J. Kolobok, a novel superfamily of eukaryotic DNA transposons. Repbase Reports. 2007;7:113–113. [Google Scholar]
- 29.Le QH, Wright S, Yu ZH, Bureau T. Transposon diversity in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:7376–7381. doi: 10.1073/pnas.97.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapitonov VV, Jurka J. First examples of protozoan P DNA transposons. Repbase Reports. 2009;9:2162–2162. [Google Scholar]
- 31.Mul YM, Rio DC. Reprogramming the purine nucleotide cofactor requirement of Drosophila P element transposase in vivo. EMBO J. 1997;16:4441–4447. doi: 10.1093/emboj/16.14.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keeling PJ, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Hampl V, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han YJ, Burnette JM, 3rd, Wessler SR. TARGeT: A web-based pipeline for retrieving and characterizing gene and transposable element families from genomic sequences. Nucleic Acids Res. 2009;37:e78. doi: 10.1093/nar/gkp295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), Version 4b10. Sunderland, Massachusetts: Sinauer Associates, Inc.; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.