Abstract
It is well known that the endoplasmic reticulum (ER) is capable of expanding its surface area in response both to cargo load and to increased expression of resident membrane proteins. Although the response to increased cargo load, known as the unfolded protein response (UPR), is well characterized, the mechanism of the response to membrane protein load has been unclear. As a model system to investigate this phenomenon, we have used a HeLa-TetOff cell line inducibly expressing a tail-anchored construct consisting of an N-terminal cytosolic GFP moiety anchored to the ER membrane by the tail of cytochrome b5 [GFP-b(5)tail]. After removal of doxycycline, GFP-b(5)tail is expressed at moderate levels (1–2% of total ER protein) that, nevertheless, induce ER proliferation, as assessed both by EM and by a three- to fourfold increase in phosphatidylcholine synthesis. We investigated possible participation of each of the three arms of the UPR and found that only the activating transcription factor 6 (ATF6) arm was selectively activated after induction of GFP-b(5)tail expression; peak ATF6α activation preceded the increase in phosphatidylcholine synthesis. Surprisingly, up-regulation of known ATF6 target genes was not observed under these conditions. Silencing of ATF6α abolished the ER proliferation response, whereas knockdown of Ire1 was without effect. Because GFP-b(5)tail lacks a luminal domain, the response we observe is unlikely to originate from the ER lumen. Instead, we propose that a sensing mechanism operates within the lipid bilayer to trigger the selective activation of ATF6.
Keywords: phospholipid biosynthesis, tail-anchored proteins, membrane biogenesis
Cells are capable of adjusting the dimensions, architecture, and molecular composition of their organelles to changing functional needs. The endoplasmic reticulum (ER) offers a well-studied example of such organelle plasticity. When exposed to altered conditions, it initiates signaling cascades that result in the adjustment of its size and composition to the new situation. Among these signaling pathways, the one triggered by increased demand on the lumenal folding machinery, known as the unfolded protein response (UPR), plays a prominent role and has been extensively investigated (1).
In yeast, the UPR is mediated by the ER transmembrane kinase/endonuclease Ire1p, whereas in mammalian cells, the pathway has evolved to a higher degree of complexity, with the participation of two additional transmembrane sensors: the basic leucine zipper (bZIP) transcriptional regulator activating transcription factor 6α/β (ATF6α/β) and the PKR-like ER kinase (PERK) (1). In the presence of unfolded proteins in the ER lumen, the three sensors induce changes in the activity of the transcriptional and translational apparatus that alleviate ER stress by (i) increasing the capacity of the lumenal folding machinery; (ii) enhancing ER-associated degradation; (iii) diminishing delivery of newly synthesized proteins to the ER lumen; and (iv) increasing phospholipid synthesis, with a resulting expansion of ER surface and volume. This fourth response (2) is elicited both by XBP1 (the transcription factor that is produced in response to Ire1 activation) and by the active cleaved form of ATF6α (3, 4).
As has been known for many decades (5), the ER adjusts its size not only to meet the requirements of the lumen but also to accommodate increased concentrations of ER resident membrane proteins. Well-known examples of membrane proteins that induce such ER expansion are cytochromes P450 (6, 7) and b(5) (8). These cytosolically exposed proteins are anchored to the ER membrane by a hydrophobic segment near the N terminus or C terminus, respectively, and lack a lumenal domain. Hence, the proliferation response they elicit cannot be triggered by a direct demand on the lumenal folding machinery and is more likely initiated by events occurring within the lipid bilayer. What these events are, however, and what signaling pathways they activate are poorly understood. In particular, it is unclear whether the UPR is involved. Indeed, there are contradictory reports on the requirement for Ire1p in the ER expansion induced by membrane proteins in yeast (2, 9–11). In mammals, membrane protein overexpression can induce the UPR (7, 12), but a causal relationship between UPR signaling and membrane protein-induced ER expansion has not been established. In addition, a high load of membrane proteins, such as that caused by many viral infections or transient transfections, can drive both the UPR and activation of the transcription factor NF-κB (12, 13). The latter response, known as the ER overload response, depends both on Ca2+ leakage from the ER lumen and on reactive oxygen species (ROS) and may represent a cellular defense mechanism rather than a signaling pathway causally linked to membrane proliferation. High loads of transfected ER resident membrane proteins also often cause striking architectural rearrangements of the organelle (14), but how these relate to the modulation of ER size in response to altered levels of endogenous membrane proteins is not clear.
To investigate mechanisms underlying the regulation of ER size in a physiological context, we have developed a model consisting of a HeLa-TetOff cell line inducibly expressing a GFP fusion protein, anchored to the ER membrane by the C-terminal tail of cytochrome b(5) (15). This tail-anchored protein, called GFP-b(5)tail, exposes the N-terminal GFP domain to the cytosol and only seven polar residues to the ER lumen. Gradual accumulation of the GFP reporter is accompanied by a near-twofold expansion of the ER, which, however, maintains its normal branched tubular/cisternal organization (15). Here, we have used this inducible system to investigate the signaling pathways that link membrane protein expression to phospholipid synthesis. We find that the ATF6 arm of the UPR is selectively activated by moderate levels of GFP-b(5)tail expression and is required for the ER proliferation response. These results raise questions about the mechanism of activation of ATF6 and its context-dependent role in transcriptional regulation.
Results
Moderate Levels of GFP-b(5)tail Expression Induce Proliferation of the ER, Which Retains Normal Architecture and Ca2+ Storage Function.
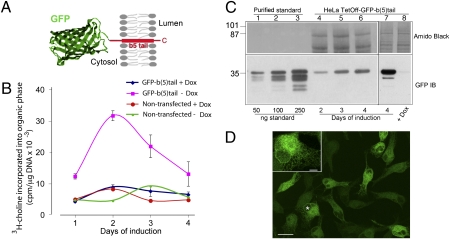
Our previous EM analysis showed that the expression of GFP-b(5)tail (construct illustrated in Fig. 1A) for 3 d induces a near-twofold expansion of the ER surface area (15). To analyze this phenomenon biochemically, we assayed phosphatidylcholine (PtdCho) synthesis at 24-h intervals by metabolic labeling with 3H-choline in induced and noninduced cells and, in parallel, determined the levels of GFP-b(5)tail expression by immunoblotting.
Fig. 1.
Moderate levels of GFP-b(5)tail induce PtdCho synthesis. (A) Schematic representation of GFP-b(5)tail [a detailed description of the construct is presented elsewhere (15)]. (B) PtdCho synthesis in induced or noninduced cells. GFP-b(5)tail–transfected and nontransfected HeLa-TetOff cells, grown in the presence or absence of Dox, were incubated with 3H-choline on the indicated days. (C) Evaluation of GFP-b(5)tail expression induced by Dox removal. (Lower) Immunoblot of increasing amounts of standard protein (lanes 1–3) and of 40 μg of total protein from cells grown in the absence of Dox for the indicated times (lanes 4–6). A heavily overexposed blot of the 4-d sample in comparison with cells grown in the presence of Dox is shown in lanes 7 and 8. (Upper) Portion of the two blots stained with Amido Black to check for protein load. (D) Confocal analysis of HeLa-TetOff cells expressing GFP-b(5)tail after 3 d of exposure to Dox-free medium. (Inset) Cell indicated with the asterisk at higher magnification. (Scale bars: 20 μm; Inset, 5 μm.)
As shown in Fig. 1B, doxycycline (Dox) removal strongly stimulated PtdCho synthesis, with a near-fourfold increase in induced vs. noninduced cells at 2 d. Removal of Dox had no effect on nontransfected HeLa-TetOff cells. Notably, the stimulation of PtdCho synthesis was highest at the early time points, when GFP-b(5)tail levels were still low (Fig. 1C).
The concentration of GFP-b(5)tail in the induced cells was estimated by comparison with known amounts of a purified standard recombinant protein. As shown in the immunoblot of Fig. 1C, after Dox removal, GFP-b(5)tail increased over time, but comparison with the standard showed that its levels were modest at all times. At 2 d of induction, the GFP-b(5)tail band generated from 40 μg of total lysate protein was less intense than the signal produced by 50 ng of standard, indicating that GFP-b(5)tail constitutes ∼0.1% of the total cellular proteins. Considering that the ER represents about 10–15% of the total cell mass (16), GFP-b(5)tail can be estimated to constitute ∼1% of the total ER protein. This level of expression is similar to that of abundant endogenous ER membrane proteins in tissues under physiological conditions (17, 18).
When GFP-b(5)tail is acutely overexpressed, it causes architectural rearrangements of the ER that are easily discernible by fluorescence microscopy (14). In agreement with its modest concentration, however, the construct did not alter ER architecture in the HeLa-TetOff cell line (15) (Fig. 1D and Fig. S1).
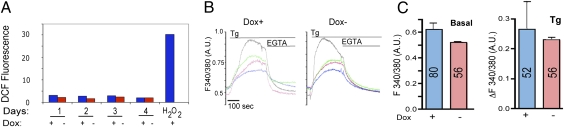
Overexpression of membrane proteins may cause ROS generation and Ca2+ leakage from the ER lumen, so we checked whether either of these parameters was altered in the induced cells. ROS generation, measured by dichlorofluorescein (DCF) fluorescence, was unchanged in cells grown with or without Dox but was observed if the cells were treated with H2O2 (Fig. 2A). Similarly, recordings on single cells, loaded with the Ca2+ indicator Fura-2 pentacetoxy methylester (FURA-2), revealed similar levels of basal [Ca2+] (Fig. 2C). We tested the capacity of the ER Ca2+ store by treating cells with the inhibitor of the ER Ca2+ pump, thapsigargin (Tg). The extent of Tg-induced release varied considerably among individual cells, as illustrated in the traces of Fig. 2B; however, the average amplitude of the peak release was similar in the two cultures (Fig. 2C, Right). Thus, the ER membrane of cells expressing GFP-b(5)tail appears to function normally as a permeability barrier.
Fig. 2.
Absence of ROS generation or Ca2+ leakage from the ER in GFP-b(5)tail–expressing cells. (A) Determination of ROS production in cells grown with or without Dox for the indicated days. H2O2 was added to one sample immediately before carrying out the fluorimeter readings. Shown are the 30-min readings corrected for the values recorded at the beginning of the registration, according to the equation [(F30 − F0)/F0] × 100. (B and C) Intracellular [Ca2+] measurements on induced and noninduced cells. (B) Representative 340:380 traces of single cells (each shown in a different color) from the two cultures, under basal conditions (first few seconds of each trace), during exposure to Tg and after addition of EGTA. (C) Average 340:380 values ± SD under basal conditions (Left) and peak amplitude of response to Tg (Right) for the two cell populations. The number of cells analyzed for each condition is indicated within the columns. A.U., arbitrary units.
GFP-b(5)tail Expression Selectively Activates the ATF6 Arm of the UPR.
Because the UPR elicits ER expansion by stimulating phospholipid synthesis (2–4), we systematically investigated the possible activation of its three arms, Ire1, PERK, and ATF6, in our system.
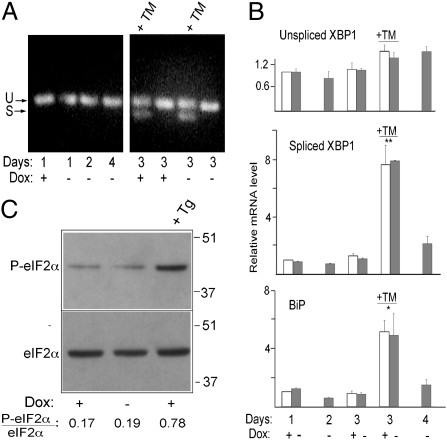
When activated, Ire1 promotes unconventional cytoplasmic splicing of the mRNA coding for the bZIP transcription factor XBP1 (1). We first assessed possible XBP1 mRNA splicing by RT-PCR, using forward and reverse primers lying outside of the spliced intron. As shown in Fig. 3A, we could detect no generation of spliced XBP1 mRNA at any time after Dox removal, whereas splicing was clearly detected when the cells were exposed to tunicamycin (TM), which induces ER stress by inhibiting N-glycosylation. We considered the possibility that small differences in XBP1 mRNA splicing not detectable by conventional RT-PCR might underlie stimulation of PtdCho synthesis, and therefore evaluated generation of the spliced form by RT-quantitative PCR (qPCR; the design of this assay is shown in Fig. S2). This assay also failed to reveal any enhancement in XBP1 mRNA splicing in the induced cells (Fig. 3B), although it did reveal a highly significant increase in cells exposed to TM. RT-qPCR also revealed no increase in the transcript coding for one of the major stress-sensitive ER chaperones, BiP, but did reveal a statistically significant increase of this mRNA when cells were exposed to TM (Fig. 3B, Lower). Likewise, no induction of UPR-responsive genes was revealed by RT-qPCR of another mRNA specifying a stress-responsive membrane protein (RAMP4) (19) or by immunoblotting of well-characterized UPR up-regulated components of the folding machinery (Fig. S3).
Fig. 3.
Absence of XBP1 splicing or PERK activation in GFP-b(5)tail–expressing cells. (A) Evaluation of XBP1 mRNA splicing by RT-PCR on cells maintained in the presence or absence of Dox for the indicated times. The +TM lanes contained reactions from cells treated for 4 h with the drug (6 μg/mL). U and S indicate the products of unspliced and spliced XBP1 mRNA, respectively. (B) Quantification of XBP1 and BiP mRNAs by RT-qPCR. Values are normalized to those of the +Dox cells at 1 d after replating. Shown are mean values ± SEM from three independent experiments. Statistical probability, indicated by the asterisks, refers to TM-treated cells vs. all untreated cells (*P < 0.05; **P < 0.01 by one-way ANOVA followed by Tukey's posttest). (C) Total and P-eIF2α detected by immunoblotting. Lysates were from GFP-b(5)tail–HeLa-TetOff cells grown in the presence or absence of Dox for 48 h. Cells were treated with 300 nM Tg for 30 min where indicated. The ratio of P-eIF2α to total eIF2α is indicated below the lanes.
We next investigated possible activation of PERK by analyzing the phosphorylation status of its target, eukaryotic initiation factor 2α (eIF2α). Immunoblotting revealed no increase in phosphorylated eIF2α (P-eIF2α) after Dox removal but did reveal a fourfold increase in cells treated with Tg (which induces stress by depleting ER luminal Ca2+) (Fig. 3C), suggesting that this arm of the UPR is also not involved in the ER proliferation response.
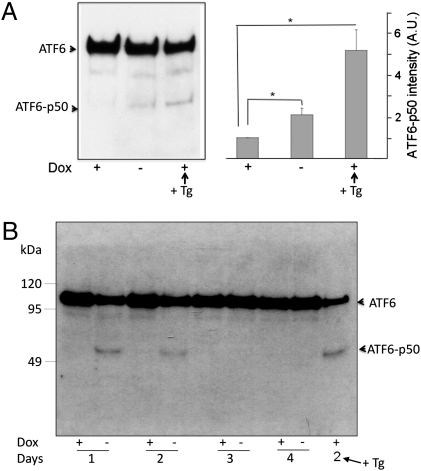
To investigate a possible involvement of the transcription factor ATF6α, we probed for the generation of the active 50-kDa form, which is generated from the constitutively expressed inactive membrane integrated form by proteolytic processing (20, 21). We first analyzed cleavage of a transiently transfected FLAG-tagged version of full-length ATF6α by immunoblotting with anti-FLAG antibodies (Fig. 4A). At 2 d of induction, a statistically significant twofold increase in cleaved ATF6α was observed. We then confirmed the GFP-b(5)tail–induced ATF6α cleavage by probing the endogenous transcription factor with specific antibodies. As shown in Fig. 4B, ATF6α cleavage was apparent already at 1 d after Dox removal, before the peak PtdCho synthesis (Fig. 1B); it remained detectable at 2 d but was no longer observable at later times. Cleavage of the transcription factor was insensitive to Dox removal in HeLa-TetOff cells not transfected with GFP-b(5)tail (Fig. S4).
Fig. 4.
GFP-b(5)tail expression activates ATF6α. (A) Immunoblotting analysis of FLAG-tagged ATF6α. GFP-b(5)tail–HeLa-TetOff cells were transiently transfected with a plasmid coding for FLAG-tagged ATF6α and maintained in culture for 48 h in the presence or absence of Dox. (Left) Representative blot. (Right) Densitometric analysis of blots from three independent experiments. Mean values ± SEM are shown (*P = 0.04 and *P = 0.02 for −Dox and for +Dox + Tg cells, respectively, vs. +Dox cells by Student's two-tailed t test after logarithmic transformation of the densitometric readings). A.U., arbitrary units. (B) Time course of endogenous ATF6α cleavage in GFP-b(5)tail–HeLa-TetOff cells assessed by immunoblotting. In A and B, cells were treated with 300 nM Tg for 1 h where indicated.
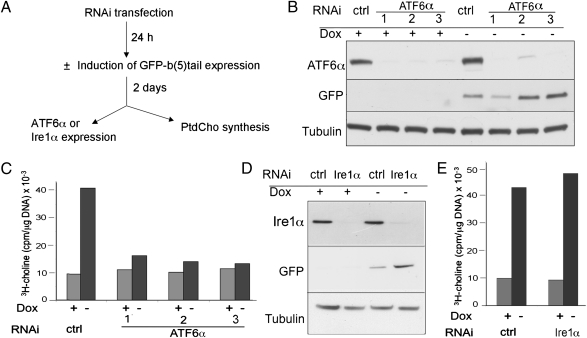
ATF6α but Not Ire1α Is Required for the ER Proliferation Response.
To establish whether the ER proliferation response requires ATF6, silencing experiments were carried out according to the scheme of Fig. 5A. Three days after RNAi transfection, ATF6α protein levels were severely depleted by each of three different RNAi duplexes (Fig. 5B). As expected, removal of Dox caused a robust increase in PtdCho synthesis in the cells transfected with control RNAi, but this response was nearly completely abrogated in cells transfected with each of the three silencing duplexes (Fig. 5C).
Fig. 5.
Silencing of ATF6α but not of Ire1α deletes the increase in PtdCho synthesis driven by GFP-b(5)tail. (A) Design of the experiments. (B) Silencing of ATF6α by three different siRNAs in induced and noninduced cells, shown by immunoblotting. (C) Stimulation of PtdCho synthesis by GFP-b(5)tail expression is strongly reduced after transfection with the three different ATF6α siRNAs. (D) Immunoblotting shows silencing of Ire1α by a pool of three siRNAs, both in induced and noninduced cells. (E) PtdCho synthesis stimulated by GFP-b(5)tail expression is unaffected by transfection with the Ire1α siRNA pool. ctrl, control.
Because XBP1 is known to induce phospholipid synthesis (3), we considered the possibility that early activation of Ire1 not detected at 1 d of induction (Fig. 3) might be required in addition to ATF6α. To test this possibility, we silenced Ire1α with a pool of three RNAi duplexes. Although Ire1α was efficiently knocked down by this treatment, the silencing had no effect on the ER proliferation response to GFP-b(5)tail expression (Fig. 5 D and E).
Discussion
Although the proliferation response of the ER to increased levels of membrane proteins has been known for decades (5), the underlying mechanisms have been unclear. The failure to clearly define the pathways involved in this response may have been attributable to the experimental systems that were used in previous studies, which were generally conducted on cultured cells heavily overexpressing ER membrane proteins. Under these stressful conditions, the cells likely activated signaling pathways that are not normally involved in the homeostatic adaptation of the ER to physiological variations in membrane protein expression (2, 7, 9–13). In the TetOff cells that we have used here, the levels of expression of the transfected protein remain at physiological levels. Furthermore, the few translocated polar residues of GFP-b(5)tail are unlikely to create a folding problem in the ER lumen. In agreement, GFP-b(5)tail expression did not appear to cause any ER stress, as judged by undetectable ROS production, absence of gross alterations in Ca2+ storage, and unchanged levels of chaperones/folding enzymes normally up-regulated by the UPR.
Although both spliced XBP1 and cleaved ΑΤF6α are known to induce phospholipid synthesis (2–4), our results demonstrate that the ATF6 arm of the UPR alone is selectively activated and triggers membrane expansion in response to moderate expression of a membrane protein. ATF6α cleavage preceded the peak of PtdCho synthesis, and silencing of the transcription factor nearly completely abrogated the proliferation response. In contrast, we found no evidence for activation of either Ire1 or PERK, and silencing of Ire1α had no effect on the response of the cell to GFP-b(5)tail expression. Our work demonstrates that ATF6α, in addition to its well-known response to lumenal unfolded proteins, links membrane protein expression to ER expansion under conditions in which there is no indication of stress. Rather, activation of this pathway may serve a preemptive function in avoiding the problems that would otherwise be caused by membrane protein overload.
It is noteworthy that the early up-regulation of PtdCho synthesis we observed was much larger than would have been required to maintain a constant protein-to-phospholipid ratio in the face of the modest load of GFP-b(5)tail. A similar early overshoot in phospholipid synthesis also occurs in tissues that increase the expression of ER membrane proteins (5). Thus, both in cultured cells and in tissues, the ER appears to overreact to minute changes in the protein-to-phospholipid ratio. The exaggerated up-regulation of synthesis is presumably partly counterbalanced by enhanced degradation, because the three- to fourfold increase in PtdCho synthesis we observed was not matched by the degree of ER expansion assessed morphometrically in our system (15).
Our work uncovers a previously unrecognized function for ATF6α and opens a number of interesting questions that merit investigation. The first concerns the pathways linking the activated transcription factor to increased phospholipid synthesis. Analysis of cells transfected with cleaved ATF6α has implicated both transcriptional and posttranscriptional mechanisms (4). It will be interesting to determine whether cleaved ATF6α generated in response to the expression of ER membrane proteins operates by the same mechanisms and to identify the molecular players linking the transcription factor to phospholipid synthesis.
A second very intriguing problem raised by our work is the absence of up-regulation of known target genes of ATF6α (e.g., BiP, GRP94, XBP1) under conditions in which the transcription factor was cleaved and caused increased phospholipid synthesis. Several studies have demonstrated that forced expression of cleaved ATF6α is alone sufficient to up-regulate ER chaperones (4, 20, 22); hence, an increase in classic UPR targets might have been expected also under our condition. It is, however, well known that ATF6α dimerizes with other transcription factors that modulate its activity (23–25). Thus, ATF6α's failure to activate its usual target genes in our cells is probably attributable to the presence of a particular combination of positive or negative coregulators that affect its activity and specificity. Although these coregulators remain to be identified, our results highlight the context dependency of the output of ER-initiated signaling pathways.
The third, and to us perhaps the most interesting, question concerns the mechanism by which GFP-b(5)tail expression activates ATF6α cleavage. This cleavage normally occurs in the Golgi complex by the action of two sequentially acting proteolytic enzymes, Site-1 and Site-2 proteases (21). Under basal conditions, ATF6 is an ER resident and its lumenal domain is associated with BiP (26, 27). Under stress, ATF6 is released from BiP and is thereby allowed to reach the Golgi complex (26). The molecular mechanism by which stress induces dissociation of the ATF6–BiP complex is still unclear; however, it is generally believed that unfolded/misfolded proteins within the lumen play a causal role (26, 27). How then could expression of a tail-anchored protein affect the association of ATF6 to BiP?
BiP binding is known to directly or indirectly regulate not only ATF6 but also the other two stress transducers, PERK and Ire1 (28, 29). However, the three branches of the UPR display distinct sensitivities toward different forms of stress (30), suggesting that additional sensor-specific regulatory factors are involved. The exquisite selectivity of ATF6α activation that we observe in our system argues in favor of alternative mechanisms to decreased BiP binding that would allow export of the transcription factor in response to slightly increased membrane protein expression.
Although the ATF6–BiP complex is remarkably stable under basal conditions (27), its kinetic “on” and “off” constants within the ER lumen are not known. Hence, it is conceivable that some dissociation occurs in the absence of stress, with consequent constitutive recruitment of the transcription factor to ER exit sites (31). Interactions of the transmembrane domain within the bilayer could slow this constitutive transport, and increased insertion of membrane proteins could interfere with this second retention mechanism, favoring export. This type of regulation would recall the cholesterol-dependent regulation of sterol regulatory element binding proteins (SREBPs), which, like ATF6, are activated by transport to the Golgi complex followed by proteolytic cleavage (32). Notably, within the Golgi, the SREBPs and ATF6 are processed by the same Site-1 and Site-2 proteases (21).
In conclusion, this study demonstrates a unique role for ATF6α in regulating the size of the ER in the face of changing membrane protein load. The selective activation of this transcription factor and the apparent lack of its activity on its classic target genes illustrate the intricacies and context dependency of the UPR in basal cellular physiology. Our future work will be aimed at elucidating the pathways of ATF6 activation, possible shared features of ATF6 and SREBP regulation, and the roles of ATF6 in maintaining ER homeostasis.
Materials and Methods
Cell Culture, Plasmid Transfection, and Recombinant Protein Production.
Nontransfected HeLa-TetOff cells and HeLa-TetOff cells stably transfected with GFP-b(5)tail under the Tet-responsive element were grown as previously described (15). Additional information on the induction process is given in SI Materials and Methods. GFP was imaged on paraformaldehyde (4% wt/vol)-fixed cells with a Leica TC-SP2 confocal microscope (63× oil immersion lens with an N.A. of 1.32).
A plasmid coding for FLAG-tagged ATF6 (26, 27), kindly provided by Ron Prywes (Columbia University, New York, NY), was transiently transfected into GFP-b(5)tail–HeLa-TetOff cells by the calcium phosphate method. Transfections were performed on noninduced cells or on induced cells 1 d after removal of Dox. The transfection medium was replaced with fresh medium after 24 h, and the cells were lysed after a further 2 h.
A recombinant GST-GFP-b(5)tail fusion protein was expressed in Escherichia coli bl21 and purified and quantified as described previously (33).
Metabolic Labeling with 3H-Choline and Lipid Extraction.
At the times indicated in the figures, cells grown in the absence or presence of Dox were incubated with 3H-choline (2 μCi/mL, specific activity of 85.5 Ci/mmol; Perkin–Elmer) for 3 h. The cells were detached by trypsinization, centrifuged, and resuspended in PBS. Equal cell numbers were broken by repeated passages through a 26-gauge needle.
Lipids were extracted from the homogenates by the method of Bligh and Dyer (34). Nitro-benzoxadiazole–PtdCho was included in the organic solvent at 10 μg/mL to monitor phospholipid recovery. The extracted and dried-down lipids were redissolved in chloroform/methanol (2:1), and radioactivity was determined by liquid scintillation counting (Wallac 1414 Win Spectral; Perkin–Elmer Life Sciences). TLC analysis demonstrated that over 90% of the label recovered in the organic phase was in PtdCho. Radioactivity recovered in the organic phase was normalized to the DNA content of the samples. The latter was assayed by Hoechst binding (Fluorescent DNA quantification kit; BioRad) and evaluated with a Victor2 multilabel counter (Perkin–Elmer).
Immunoblotting.
Cell monolayers or cell homogenates were solubilized in preheated lysis buffer containing 2% (wt/vol) SDS, 50 mM Tris-HCl (pH 6.8), a protease inhibitor mixture (Complete; Roche), and, in the case of phosphoprotein analysis, 10 mM NaF. The protein content of the lysates was determined with the bicinchoninic acid reagent (Pierce) before SDS/PAGE and blotting. The sources of primary antibodies and details of the procedure are given in the SI Materials and Methods.
ROS Measurements.
ROS accumulation was evaluated by DCF fluorescence (35). Cells were seeded in 96-well plates and maintained in growth medium in the absence or presence of Dox. At the times indicated in the figure legend, cells were incubated for 30 min at 37 °C with 20 μM 2′,7′-dichlorodihydrofluorescein (H2DCF) diacetate (Invitrogen) and 2.5 mM probenecid (Sigma) in complete medium. The cells were washed and bathed in PBS supplemented with 1.2 mM CaCl2 and 10 mM glucose. ROS-driven conversion of H2DCF to DCF was monitored every 5 min for 60 min, using the Victor2 microplate fluorometer. The fluorescence in control samples exposed to 250 μM H2O2 increased linearly during the entire period of measurement.
Intracellular Calcium Measurements.
Cultures were loaded with the ratiometric calcium dye FURA-2 (2 μM in Krebs–Ringer solution buffered with Hepes and containing 2 mM CaCl2), as previously described (36). Cells were imaged with a 40× objective (N.A. of 1.3), with the use of Polychrome IV (TILL Photonics) as a light source. The temporal analysis of FURA-2 340:380 ratio values in a cytoplasmic area of all cells in a field was carried out under basal conditions, after addition of Tg (1 μM), and after the subsequent addition of EGTA (3 mM). At least two different coverslips were analyzed in each experiment.
RT-PCR and qPCR.
Gene expression of spliced and unspliced XBP1, of BiP, and of RAMP4 was quantitatively analyzed by RT-qPCR, with SYBR Green or Taqman technology, using the ANI Prism 7000 Sequence Detection System (Applied Biosystems). GAPDH mRNA was used as an internal control for mRNA level normalization, and the 2−ΔΔCT (cycle threshold) method was used to calculate the results. A full description of the conditions used, as well as the primers and their validation, is given in SI Materials and Methods and Fig. S2.
ATF6α and Ire1α Silencing.
Cells at ∼40% confluence were transfected with RNAi duplexes with the use of RNAiMAX Lipofectamine reagent (Invitrogen). ATF6α was silenced in separate cultures with three different duplex siRNAs, each used at a concentration of 10 nM (Stealth RNAi HSS177036, HSS177037, and HSS117915; Invitrogen). Ire1α was silenced with a pool of three siRNAs, each used at a concentration of 10 nM (Stealth RNAi HSS140846, HSS140847, and HSS176615; Invitrogen). In both cases, parallel cultures were transfected with equal concentrations (10 or 30 nM) of Stealth Negative Universal Control Medium (Invitrogen). Six hours after transfection, the medium was replaced with complete growth medium, and the cultures were maintained overnight in the presence of Dox. The following day, the cells were trypsinized, replated with or without Dox, and maintained in culture for another 48 h before analysis.
Supplementary Material
Acknowledgments
In addition to the colleagues who kindly provided reagents (listed in the text), we thank Prof. Domenicantonio Rotiroti for his encouragement and support and Marco Pozzi for helpful advice on the RNAi experiments. This work was supported, in part, by Grant IG9040 from the Italian Association for Cancer Research (AIRC) (to N.B.) and by Institutional Consiglio Nazionale delle Ricerche Grant ME.P02.007. J.M. was supported by a research contract from the University of Catanzaro.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101379108/-/DCSupplemental.
References
- 1.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: A link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bommiasamy H, et al. ATF6alpha induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci. 2009;122:1626–1636. doi: 10.1242/jcs.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orrenius S, Ericsson JL, Ernster L. Phenobarbital-induced synthesis of the microsomal drug-metabolizing enzyme system and its relationship to the proliferation of endoplasmic membranes. A morphological and biochemical study. J Cell Biol. 1965;25:627–639. doi: 10.1083/jcb.25.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schunck WH, et al. Comparison of two cytochromes P-450 from Candida maltosa: Primary structures, substrate specificities and effects of their expression in Saccharomyces cerevisiae on the proliferation of the endoplasmic reticulum. Eur J Cell Biol. 1991;55:336–345. [PubMed] [Google Scholar]
- 7.Szczesna-Skorupa E, Chen CD, Liu H, Kemper B. Gene expression changes associated with the endoplasmic reticulum stress response induced by microsomal cytochrome p450 overproduction. J Biol Chem. 2004;279:13953–13961. doi: 10.1074/jbc.M312170200. [DOI] [PubMed] [Google Scholar]
- 8.Vergères G, Yen TS, Aggeler J, Lausier J, Waskell L. A model system for studying membrane biogenesis. Overexpression of cytochrome b5 in yeast results in marked proliferation of the intracellular membrane. J Cell Sci. 1993;106:249–259. doi: 10.1242/jcs.106.1.249. [DOI] [PubMed] [Google Scholar]
- 9.Menzel R, Vogel F, Kärgel E, Schunck WH. Inducible membranes in yeast: Relation to the unfolded-protein-response pathway. Yeast. 1997;13:1211–1229. doi: 10.1002/(SICI)1097-0061(199710)13:13<1211::AID-YEA168>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 10.Takewaka T, Zimmer T, Hirata A, Ohta A, Takagi M. Null mutation in IRE1 gene inhibits overproduction of microsomal cytochrome P450Alk1 (CYP 52A3) and proliferation of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biochem. 1999;125:507–514. doi: 10.1093/oxfordjournals.jbchem.a022314. [DOI] [PubMed] [Google Scholar]
- 11.Larson LL, Parrish ML, Koning AJ, Wright RL. Proliferation of the endoplasmic reticulum occurs normally in cells that lack a functional unfolded protein response. Yeast. 2002;19:373–392. doi: 10.1002/yea.839. [DOI] [PubMed] [Google Scholar]
- 12.Li S, et al. Hepatitis C virus NS4B induces unfolded protein response and endoplasmic reticulum overload response-dependent NF-kappaB activation. Virology. 2009;391:257–264. doi: 10.1016/j.virol.2009.06.039. [DOI] [PubMed] [Google Scholar]
- 13.Pahl HL, Baeuerle PA. The ER-overload response: Activation of NF-kappa B. Trends Biochem Sci. 1997;22:63–67. doi: 10.1016/s0968-0004(96)10073-6. [DOI] [PubMed] [Google Scholar]
- 14.Borgese N, Francolini M, Snapp E. Endoplasmic reticulum architecture: Structures in flux. Curr Opin Cell Biol. 2006;18:358–364. doi: 10.1016/j.ceb.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprocati T, Ronchi P, Raimondi A, Francolini M, Borgese N. Dynamic and reversible restructuring of the ER induced by PDMP in cultured cells. J Cell Sci. 2006;119:3249–3260. doi: 10.1242/jcs.03058. [DOI] [PubMed] [Google Scholar]
- 16.Stäubli W, Hess R, Weibel ER. Correlated morphometric and biochemical studies on the liver cell. II. Effects of phenobarbital on rat hepatocytes. J Cell Biol. 1969;42:92–112. doi: 10.1083/jcb.42.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omura T, Takesue S. A new method for simultaneous purification of cytochrome b5 and NADPH-cytochrome c reductase from rat liver microsomes. J Biochem. 1970;67:249–257. doi: 10.1093/oxfordjournals.jbchem.a129248. [DOI] [PubMed] [Google Scholar]
- 18.Cheng KC, Schenkman JB. Purification and characterization of two constitutive forms of rat liver microsomal cytochrome P-450. J Biol Chem. 1982;257:2378–2385. [PubMed] [Google Scholar]
- 19.Yamaguchi A, et al. Stress-associated endoplasmic reticulum protein 1 (SERP1)/Ribosome-associated membrane protein 4 (RAMP4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. J Cell Biol. 1999;147:1195–1204. doi: 10.1083/jcb.147.6.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 22.Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Li M, et al. ATF6 as a transcription activator of the endoplasmic reticulum stress element: Thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20:5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker R, et al. Identification of TFII-I as the endoplasmic reticulum stress response element binding factor ERSF: Its autoregulation by stress and interaction with ATF6. Mol Cell Biol. 2001;21:3220–3233. doi: 10.1128/MCB.21.9.3220-3233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 27.Shen J, Snapp EL, Lippincott-Schwartz J, Prywes R. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol Cell Biol. 2005;25:921–932. doi: 10.1128/MCB.25.3.921-932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 29.Pincus D, et al. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DuRose JB, Tam AB, Niwa M. Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol Biol Cell. 2006;17:3095–3107. doi: 10.1091/mbc.E06-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindler AJ, Schekman R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc Natl Acad Sci USA. 2009;106:17775–17780. doi: 10.1073/pnas.0910342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Ceppi P, et al. Two tail-anchored protein variants, differing in transmembrane domain length and intracellular sorting, interact differently with lipids. Proc Natl Acad Sci USA. 2005;102:16269–16274. doi: 10.1073/pnas.0508157102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 35.Mattson MP, Barger SW, Begley JG, Mark RJ. Calcium, free radicals, and excitotoxic neuronal death in primary cell culture. Methods Cell Biol. 1995;46:187–216. doi: 10.1016/s0091-679x(08)61930-5. [DOI] [PubMed] [Google Scholar]
- 36.Verderio C, et al. SNAP-25 modulation of calcium dynamics underlies differences in GABAergic and glutamatergic responsiveness to depolarization. Neuron. 2004;41:599–610. doi: 10.1016/s0896-6273(04)00077-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.