Abstract
The class III phosphatidylinositol 3-kinase (PI3KC3) is crucial for autophagosome biogenesis. It has been long speculated to nucleate the autophagosome membrane, but the biochemical mechanism of such nucleation activity remains unsolved. We recently identified Barkor/Atg14(L) as the targeting factor for PI3KC3 to autophagosome membrane. Here, we show that we have characterized the region of Barkor/Atg14(L) required for autophagosome targeting and identified the BATS [Barkor/Atg14(L) autophagosome targeting sequence] domain at the carboxyl terminus of Barkor. Bioinformatics and mutagenesis analyses revealed that the BATS domain binds to autophagosome membrane via the hydrophobic surface of an intrinsic amphipathic alpha helix. BATS puncta overlap with Atg16 and LC3, and partially with DFCP1, in a stress-inducible manner. Ectopically expressed BATS accumulates on highly curved tubules that likely represent intermediate autophagic structures. PI3KC3 recruitment and autophagy stimulation by Barkor/Atg14(L) require the BATS domain. Furthermore, our biochemical analyses indicate that the BATS domain directly binds to the membrane, and it favors membrane composed of phosphatidylinositol 3-phosphate [PtdIns(3)P] and phosphatidylinositol 4,5-biphosphate [PtdIns(4,5)P2]. By binding preferentially to curved membranes incorporated with PtdIns(3)P but not PtdIns(4,5)P2, the BATS domain is capable of sensing membrane curvature. Thus, we propose a novel model of PI3KC3 autophagosome membrane nucleation in which its autophagosome-specific adaptor, Barkor, accumulates on highly curved PtdIns(3)P enriched autophagic membrane via its BATS domain to sense and maintain membrane curvature.
Keywords: omegasome, phagophore, lysosome, endocytosis, membrane trafficking
Autophagy is characterized by the formation of double-membrane vesicles called autophagosomes that engulf cytoplasm, organelles, protein aggregates, and microorganisms and deliver these components to the lysosome for degradation (1). Dysfunction of autophagy has been implicated in multiple human diseases including cancers, neurodegenerative diseases, diabetes, and infectious diseases (2, 3). The autophagy pathway is highly conserved from yeast to humans; at least 34 autophagy-related proteins have been identified (4–7). Among these autophagy proteins, the class III phosphatidylinositol 3-kinase (PI3KC3) complex composed of Vps34, p150/Vps15, and Beclin 1/Atg6 has an evolutionary conserved role in autophagosome formation (8). It has been long speculated that PI3KC3 nucleates the initiating autophagosome membrane (also called phagophore or the isolation membrane) (9). However, direct biochemical evidence to support the nucleation activity of PI3KC3 is still missing.
One major barrier to understanding the function of PI3KC3 in autophagosome biogenesis is the pleiotropy of this lipid kinase in multiple membrane trafficking pathways (8, 9). By generation of phosphatidylinositol 3-phosphate [PtdIns(3)P], an event that might ignite a cascade of signaling pathways, PI3KC3 is required for autophagy and endocytosis via assembly of distinct protein complexes (10–16). The recent identification of an autophagic specific adaptor, Barkor/Atg14(L), of PI3KC3 paved the way for its specific role on autophagosome membrane (10–15). Barkor/Atg14(L) mainly localizes to initiating autophagosome membrane, evidenced by colocalization with both the early autophagosome marker Atg16 and the omegasome marker DFCP1 (17). Omegasome is a potential precursor of autophagosome originated from endoplasmic reticulum (ER) (18). The N-terminal cysteine repeats target Barkor/Atg14(L) to ER, and such distribution is crucial for its function in autophagy activation (17). However, the ER targeting of Barkor/Atg14(L) is neither induced by stress nor inhibited by a dominant negative Ulk1/Atg1 mutant (17). The biochemical mechanism underlying the stress-inducible Barkor/Atg14(L) accumulation on autophagic membrane remains unknown.
In general, membrane-associated proteins not only bridge the interplay between lipids and proteins; they are also involved in generating, sensing, and stabilizing local regions of membrane curvature. Many proteins or protein complexes are known to be crucial for membrane deformation, including protein coat complexes (clathrin, COPI, and COPII) or Bin-Amphiphysin-Rvs (BAR) domain containing proteins (19–26). These proteins that modulate membrane structures often associate with amphipathic alpha helices either by inserting small hydrophobic patch of amphipathic alpha helices into the membrane leaflet, or by mounting the membrane surface to the intrinsically curved protein scaffolds. In addition to the hydrophobic interactions, membrane-associated proteins also quite often bind to the lipid headgroups (26). Phosphoinositides (PtdIns) are particularly important because their headgroups are easily modified (27). For example, PtdIns(4,5)P2 is important for the budding of clathrin-coated vesicles and the fusion of vacuoles (28–31), whereas PtdIns(3)P is required for the invagination of vesicles into late endosomes and autophagosome biogenesis (8, 9, 13, 32).
Barkor/Atg14(L) function is highly specific to autophagosome formation because of its localization on the phagophore (10–14). In this study, we investigated the autophagosome targeting of Barkor/Atg14(L) through deletion mutagenesis. We identified a previously uncharacterized Barkor/Atg14(L) autophagosome targeting sequence (BATS) domain in Barkor/Atg14(L) that specifically recognizes early autophagic structures. The BATS domain is required for Beclin 1 recruitment and autophagy activation. Within this domain, the hydrophobic patch of an amphipathic alpha helix directly interacts with membrane. In addition, we found that BATS preferentially bound to more highly curved membranes in a PtdIns(3)P dependent manner. Hence, we propose a biochemical mechanism of PI3KC3 autophagosome membrane nucleation through autophagosome binding and membrane curvature sensing of Barkor/Atg14(L).
Results
Identification of the Barkor/Atg14(L) Autophagosome Targeting Sequence (BATS).
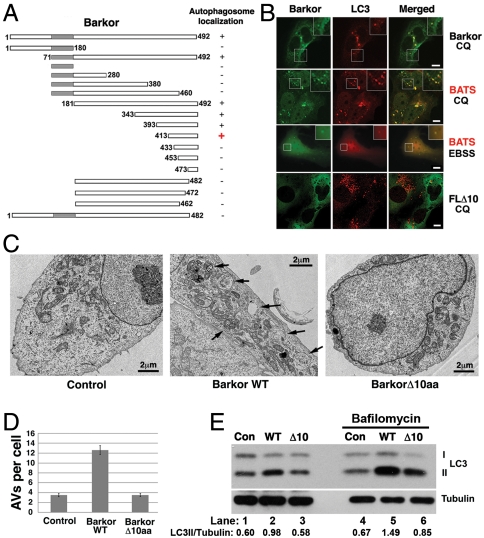
Barkor/Atg14(L) was recently identified as a stoichiometric subunit of PI3KC3 (10–12, 14) that specifically localizes to initiating autophagosomes and targets PI3KC3 to autophagosome (10). It is reported that N-terminal cysteine repeats of Barkor/Atg14(L) is essential for ER localization and autophagy function (16). However, the Barkor/Atg14(L) ER membrane association is neither induced by starvation nor blocked by Ulk1 dominant negative mutant (16), suggesting there is another stress-inducible membrane association mechanism for Barkor/Atg14(L). We performed an in-depth deletion mutagenesis analysis to search for protein sequence that is required for autophagosome targeting. To better visualize autophagic vacuoles, chloroquine (CQ), a drug that blocks autophagy flow by raising the pH of endo/lysosomes (33), was used to treat U2OS cells stably expressing Myc-tagged LC3. In these cells, we coexpressed serial deletion mutants of Barkor/Atg14(L) tagged with humanized Renilla reniformis green fluorescence protein (hrGFP) and tested their colocalization with LC3 upon CQ treatment (Fig. 1A and Fig. S1). By this way, we mapped the autophagosome membrane association domain to the C terminus of Barkor/Atg14(L). We found that the extreme C terminus, the last 80 amino acids of Barkor, is sufficient for autophagosomes targeting (Fig. 1 A and B and Fig. S1). Consequently, we determined that the final 80 amino acids in the C-terminus of Barkor/Atg14(L) define a minimum required region for autophagosome binding and named this region BATS for Barkor/Atg14(L) autophagosome targeting sequence. BATS puncta not only accumulated upon CQ treatment, but also strongly induced by Rapamycin and starvation treatments (Fig. 1B and Fig. S2). To consolidate the requirement for the carboxyl terminal of Barkor/Atg14(L) in autophagosome targeting, we generated mutants missing various lengths of amino acids from C terminus of Barkor. Deletion of last 10 amino acids from full-length Barkor/Atg14(L) is sufficient to abolish the cytosolic puncta and colocalization with LC3 (Fig. 1 A and B). This demonstrates the essential role for these amino acids in autophagosome association.
Fig. 1.
The identification of the BATS domain required for autophagosome targeting and autophagy activation. (A). Schematic representation of the deletion mutants of Barkor. All mutants are tagged with hrGFP and FLAG. Autophagosome localization is defined as cytosolic puncta overlapping with LC3 upon CQ treatment. (B). Cells stably expressing Myc-LC3 were transfected with various Barkor/Atg14(L) fragments tagged with hrGFP. Cells were treated with 200 μM CQ for 2 h and stained with anti–c-Myc antibody for LC3 and green fluorescence for Barkor/Atg14(L) mutants. (C). U2OS cells that overexpress full-length Barkor, BarkorΔ10aa and U2OS parental cells were visualized using a transmission electron microscope. AVs are marked by black arrows. Scale bar, 2 μm. (D). AVs per cross-section were counted (control, 3.45 ± 0.35; Barkor/Atg14(L) Δ10aa, 3.5 ± 0.32; Barkor/Atg14(L) WT: 12.6 ± 0.9). (E). LC3 and tubulin were detected in cells described in C; 2-h treatment with 50 nM Bafilomycin was used to block lysosomal degradation.
To test whether Barkor/Atg14(L) membrane association is necessary for its autophagy function, we expressed the targeting deficient Barkor/Atg14(L) Δ10aa mutant in U2OS cells. This mutant only lacks the final 10 amino acids on the C terminus required for autophagosome targeting; it is still capable of interacting with Beclin 1. We speculated that if Barkor/Atg14(L) membrane association is required for autophagy, BarkorΔ10aa mutant might be unable to stimulate autophagy as wild-type Barkor. Electron microscopy allows the direct analysis of autophagosome formation. Autophagic vacuoles (AVs) can be captured by a high-resolution transmission electron microscope. Barkor/Atg14(L) overexpression leads to increased number and size of AVs in U2OS cells (Fig. 1C) (10). However, in U2OS cells overexpressing the Barkor/Atg14(L) Δ10aa mutant, the promotion effect of Barkor/Atg14(L) on autophagosome formation is abolished (Fig. 1 C and D). The autophagy flux was also determined in Barkor/Atg14(L) wild-type and Δ10aa mutant expressing cells. LC3-II conversion is compromised in both normal or Bafilomycin-treated cells expressing Δ10aa mutant compared to the cells expressing Barkor/Atg14(L) wild-type cells (Fig. 1E). These combined results suggest that Barkor/Atg14(L) autophagosome association via its carboxyl terminus is required for autophagosome formation.
BATS Associates with Autophagosome Membrane via the Hydrophobic Side of an Amphipathic Alpha Helix.
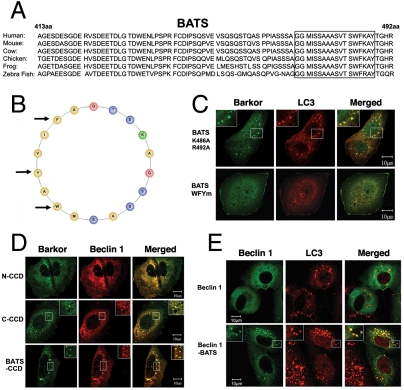
The final 80 amino acids of the C terminus of Barkor/Atg14(L) define a minimum required region for autophagosome binding (Fig. 1A). Through bioinformatics analysis, we determined specific characteristics of BATS that aid in its ability to function as a targeting sequence. The BATS domain is highly conserved in vertebrates (Fig. 2A). Secondary structural analysis revealed that 19 amino acids of BATS form a classical amphipathic alpha helical wheel with hydrophobic and hydrophilic residues that align on opposite sides of the helix (Fig. 2B). This arrangement may allow the hydrophobic patch of the BATS domain to shallowly embed into the lipid bilayer. To investigate the hydrophobic interaction between BATS amphipathic alpha helix and the lipid bilayer, we replaced three hydrophobic residues, tryptophan (W), phenylalanine (F), and tyrosine (Y), with arginine (R) residues. As expected, the expression of this WFYm mutant lacked BATS puncta (Fig. 2C). Alternatively, the mutation of two hydrophilic residues (K486A, R492A) on the polar side has no effect on BATS puncta formation (Fig. 2C), suggesting that the hydrophobic interaction between BATS and lipid bilayer is critical for membrane association.
Fig. 2.
The amphipathic alpha helix of BATS is responsible for autophagosome membrane binding. (A). The last 80 amino acids (BATS) of Barkor/Atg14(L) homologues in vertebrates were aligned, and a highly conserved membrane interaction helix was predicted and noted by a box. (B). The predicted membrane interaction helix is presented as an amphipathic helical wheel. Hydrophobic residues are yellow, the hydrophilic residues are blue, and positively charged residues are green. Three hydrophobic residues mutated in C are marked by arrows. (C). Cells stably expressing Myc-LC3 were coexpressed with different BATS mutants, which were tagged with hrGFP (BATS K486A, R492A and BATS WFYm-BATS mutant W484R, F485R, Y488R). Cells were treated with 200 μM CQ for 2 h and stained with c-Myc antibody for LC3 and green fluorescence for BATS mutants. (D). U2OS cells were coexpressed with Myc-Beclin 1 and different Barkor/Atg14(L) mutants tagged by hrGFP and treated with 200 μM CQ. (E). Cells stably expressing Myc-LC3 were coexpressed with either GFP-Beclin 1 or GFP-Beclin 1-BATS, and treated with 200 μM CQ for 2 h.
We have previously shown that recruitment of Beclin 1 to autophagosomes by Barkor/Atg14(L) requires its coiled-coil domain (CCD) (10). If BATS is sufficient for membrane targeting, we anticipate that CCD tethered to BATS might be sufficient to direct Beclin 1 to autophagosomes. To test this hypothesis, we generated a chimeric protein consisting only of a CCD and BATS fusion. This cognate Barkor/Atg14(L) is sufficient to localize to the cytosolic puncta decorated with LC3 (Fig. S3) and to recruit Beclin 1 to autophagosomes (Fig. 2D). To bypass the Barkor–Beclin 1 interaction via the CCD, we generated another chimera protein consisting of Beclin 1 directly fused to BATS (Beclin 1–BATS). We hypothesized that BATS alone would be sufficient to guide Beclin 1 to the autophagosome. We expressed this Beclin 1–BATS fusion construct and determined its subcellular localization. Beclin 1–BATS, but not Beclin 1 alone, localized to autophagosomes and overlapped with LC3 (Fig. 2E), suggesting that BATS is adequate for Beclin 1 recruitment to autophagosomes. Moreover, Beclin 1–BATS as well as BATS puncta that overlapped with LC3 were dramatically induced upon treatments of Rapamycin and CQ (Fig. S2). Thus, BATS is sufficient to direct Barkor/Atg14(L) and Beclin 1 to autophagosomes in a stress-inducible manner and trigger autophagy activation.
BATS Localizes to Highly Curved Early Autophagic Structures.
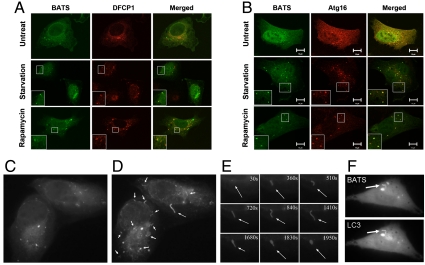
It has been reported that Barkor/ATG14L localizes to ER and overlaps with or adjacent to DFCP1 positive omegasomes, which serves as a platform for autophagosome formation (16, 34). We speculate that stress-induced BATS puncta might overlap with early autophagosome markers. We examined the colocalization of BATS with DFCP1 and Atg16. BATS partially colocalizes with DFCP1, a marker for ER-associated omegasome (17), upon starvation or CQ treatment (Fig. 3A). Remarkably, BATS puncta significantly colocalize with Atg16, a phagophore or the isolation membrane marker, upon starvation or Rapamycin treatment (Fig. 3B). These data indicate that BATS locates on early autophagic structures.
Fig. 3.
BATS localizes to highly curved early autophagic structures. (A). U2OS cell were transfected with hrGFP-BATS together with Myc-DFCP1. Forty-eight hours after transfection, cells were either treated by EBSS for 1 h or by 200 μM CQ for 2 h; Myc-DFCP1 was stained with Rhodamine Red antibody and detected by laser confocal microscope. (B) U2OS cells were transfected with hrGFP-BATS together with Myc-Atg16. Forty-eight hours after transfection, cells were either treated with EBSS for 1 h or with 2 μM rapamycin for 4 h; Myc-Atg16 was stained with Rhodamine Red antibody and detected by laser confocal microscope. (C and D) U2OS cells expressing GFP-tagged BATS (2 d after transfection) were treated with EBSS medium and observed under the fluorescence microscope. Images were captured immediately after the addition of EBSS medium (C) or at 30-min time points (D) (also see Movie S1). Arrows mark tubules. (E). Arrows along the time course (0–30 min) mark one representative tubule after starvation. (F). U2OS cells expressing GFP-BATS and Tomato-LC3 were captured after addition of EBSS medium; both channels were captured every 2 s. Two representative images were presented (also see Movie S2). An arrow marks a tubule.
Autophagosome undergoes dynamic changes progressing from a small crescent-shaped compartment to a cup-shaped membrane and finally to closed ring structure (35), which could be captured with time-lapse video microscopy. We observed a clear overlap of BATS with Atg16 and LC3 in the steady state, which urged us to study the dynamic interaction of BATS with autophagic membrane upon starvation. GFP-tagged BATS was ectopically expressed in U2OS cells and observed in a time-lapse video microscopy analysis following starvation. GFP-BATS expressing cells cultured in nutrient rich medium were switched to Earle’s balanced salt solution (EBSS) medium and observed under fluorescence microscope lively. BATS signal was initially presented as cytosolic puncta. Upon starvation, BATS was enriched on many growing narrow tubules. These BATS positive tubules are often highly curved but have rather undirected curvatures (switching between concave and convex) (Fig. 3 C–E and Movie S1). Time-lapse video taken in cells expressing GFP-BATS and Tomato-LC3 proved that the BATS tubule is also LC3 positive (Fig. 3F and Movie S2), indicating that BATS decorates on the highly curved autophagic structures. The half-life of BATS tubules (about 30 min) is much longer than a forming autophagosome (about 5–10 min) (35). This observation, along with the tubules with unstable curvatures, suggests that BATS is likely involved in membrane curvature sensing and stabilization, a process that probably requires additional factors (PtdIns production, etc.).
To explore the relationship between these BATS tubules and ER, we observed GFP-BATS and DsRed2-ER signals together upon starvation in a time-lapse experiment. The majority of BATS puncta colocalized with ER network. We found that BATS tubules are extended from ER-associated BATS puncta (Movie S3), suggesting that BATS tubules might represent the newly formed highly curved membrane sprouting from omegasomes.
Recombinant BATS Domain Directly Bound to PtdIns(3)P Enriched Membrane.
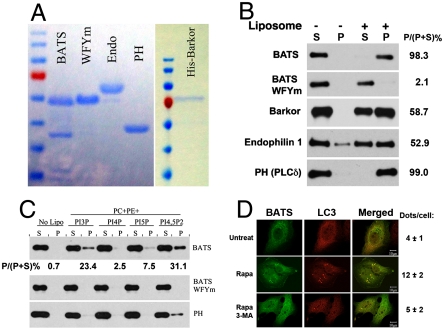
The autophagosome membrane association of the BATS domain led us to speculate that BATS could directly bind liposomes. Recombinant GST-tagged BATS wild type and the WFYm mutant were expressed and purified from Escherichia coli, and full-length His-tagged Barkor/Atg14(L) was expressed and purified from baculovirus-infected insect cells (Fig. 4A). Recombinant endophilin 1 and PH domain of PLCδ were also purified from E. coli. These proteins were incubated with liposome prepared from Folch fractions of brain lipid extracts in a cosedimentation assay (Fig. 4B). The BATS domain alone does not precipitate. Instead, the BATS domain coprecipitated with liposome (Fig. 4B), indicating that BATS directly binds to the lipid bilayer. In contrast, the BATS WFYm mutant with the hydrophobic residues changed to hydrophilic amino acids failed to precipitate with liposome (Fig. 4B). Full-length Barkor/Atg14(L) purified from insect cells is also capable of liposome binding (Fig. 4B). We failed to purify Barkor/Atg14(L) with C terminal 10 amino acids deletion from insect cells because of its instability. However, we speculate that multiple membrane binding domains may exist along Barkor/Atg14(L). For example, its N terminus is able to associate with ER membrane (16). As positive controls, both endophilin 1 and the pH domain of PLCδ cosedimented with liposomes (Fig. 4B). These data show that BATS liposome binding is mediated by the hydrophobic residues in the amphipathic alpha helix.
Fig. 4.
Membrane binding activity of the BATS domain. (A). GST-tagged constructs of wild-type BATS, BATS WFYm mutant, endophilin 1, and PH domain of PLCδ were recombinantly expressed and purified from E.coli. Full-length His-tagged Barkor/Atg14(L) was expressed and purified from baculovirus-infected insect cells. (B). Purified recombinant proteins described in A were tested for their liposome binding activity in a cosedimentation assay. Liposomes generated from Folch fraction I of brain extract from bovine brains were used in this assay. S denotes supernatant and P denotes pellet. The binding efficiency is calculated as bound proteins (P) versus input (P + S). (C). Recombinant wild-type BATS, BATS WFYm mutant and the PH domain of PLCδ were incubated with liposomes incorporated with different forms of phosphoinositides and tested for their liposome binding in a cosedimentation assay. S denotes supernatant and P denotes pellet. (D). Cells expressing GFP-BATS and Myc-LC3 were left untreated, treated with Rapamycin alone or in combination with 3-Methyladenine (3-MA), and observed under the fluorescence microscope. Dots per cells were calculated in at least 50 cells from three independent experiments.
Because the early autophagosome structures are decorated by PtdIns(3)P, we investigated whether the BATS domain binds to phosphoinositides. We tested BATS binding to liposomes composed of neutral lipids, phosphatidylcholine (PC) and phosphatidylethanolamine (PE), as well as different phosphoinositides. Interestingly, BATS preferentially binds to liposomes incorporated with either PtdIns(3)P or PtdIns(4,5)P2, but not PtdIns(4)P or PtdIns(5)P (Fig. 4C). This suggests that PtdIns(3)P and PtdIns(4,5)P2 are likely involved in BATS-mediated membrane association. PtdIns(3)P production has consistently been considered an essential event during autophagosome formation (8, 9). To investigate if the PtdIns(3)P binding is required for BATS autophagosome association, we treated BATS-expressing cells with a PI3KC3 inhibitor, 3-methyleadenine (3-MA). BATS puncta are induced upon rapamycin treatment but diminished upon 3-MA coadministration (Fig. 4D), indicating that BATS autophagosome association is dependent on PtdIns(3)P production.
Membrane curvature sensing by BATS.
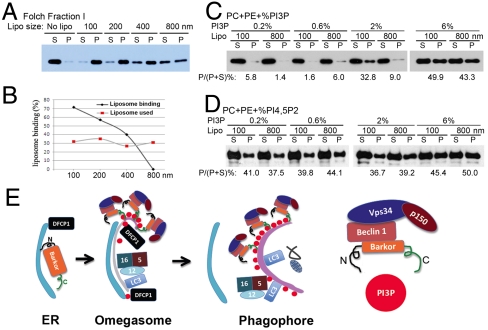
The time-lapse video microscopy analysis suggests that BATS might bind to the curved membrane. The amphipathic alpha helix of the BATS domain is not highly charged but serine/threonine enriched at its polar side (Fig. 2B), resembling the BAR domains that serve as sensors of membrane curvature (26, 36–38). We speculated that the BATS domain might serve as a sensor of curvature, binding more tightly to curved, as opposed to flat, membranes. We tested this hypothesis using liposomes of different intrinsic curvatures. For this purpose, we generated liposomes with different sizes made from the Folch fractions of the brain lipid extracts. Recombinant BATS domain binds preferentially to the highly curved membranes of small liposomes rather than to the relatively flat membranes of large liposomes (Fig. 5A). The quantity of liposomes was normalized by Texas-Red–labeled PE incorporated into liposomes (Fig. 5B). The sensitivity of the BATS domain to the vesicle size and therefore to the extent of curvature confirms that it is a membrane curvature sensor.
Fig. 5.
Membrane curvature sensing by the BATS domain. (A). Folch fraction derived liposomes with different sizes (100–800 nm) were incubated with recombinant BATS in a cosedimentation assay. S denotes supernatant and P denotes pellet. (B). Liposome binding was evaluated as the percentage of BATS bound to liposomes compared to the binding of BATS to 800 nm liposomes. Liposome loading was evaluated by the measurement of fluorescence-labeled PE in the liposomes. (C). Three hundred fifty micromolar lipid with equal amount of PC, PE combining a gradient increased ratio of PtdIns(3)P (0.2, 0.6, 2, and 6 mol%) were made into liposomes with two different sizes (100 and 800 nm), and their binding to recombinant BATS were tested by cosedimentation assays. S denotes supernatant and P denotes pellet. The binding efficiency (bound protein versus input) is denoted as P/(P + S). (D). The similar liposome binding assay as described in (C) was performed using PtdIns(4,5)P2. (E). Proposed functional model of Barkor/Atg14(L) BATS domain in the membrane targeting of Barkor/Atg14(L) on highly curved PtdIns(3)P enriched membrane. N, N terminus; C, C terminus. Please see text for more explanation.
To investigate the role that PtdIns(3)P plays in BATS membrane curvature sensing, we incubated recombinant BATS with two different sizes of liposomes (100 and 800 nm) composed of different amounts of PtdIns(3)P (0.2, 0.6, 2, and 6%). The membrane binding of BATS to liposomes with 0.2 and 0.6% PtdIns(3)P incorporated is barely detectable, regardless of the size of liposomes. When the concentration of PtdIns(3)P of liposomes is increased to 2%, there is a clear preference for BATS to interact with 100-nm liposomes rather than 800-nm liposomes (Fig. 5C). Further increasing PtdIns(3)P to 6% in liposomes induced strong interaction between BATS and liposomes, and no obvious difference of BATS binding to 100-nm or 800-nm liposomes was observed (Fig. 5C). These data indicate that BATS senses membrane curvature in a PtdIns(3)P-dependent manner, and local high concentration of PtdIns(3)P is enough to override the curvature sensing activity of BATS. Interestingly, BATS could only sense the membrane curvature of liposomes incorporated with PtdIns(3)P but not PtdIns(4,5)P2 (Fig. 5D).
Discussion
In this study, we identified a membrane association BATS domain of Barkor/Atg14(L) that specifically targets Barkor/Atg14(L) to highly curved PtdIns(3)P enriched early autophagic membrane. The BATS domain is essential for the function of Barkor/Atg14(L) in autophagy activation. We have also determined that the BATS domain utilizes the hydrophobic residues within the amphipathic alpha helix to interact with membrane. BATS localizes to Atg16 and LC3 positive autophagic membrane, and also partially colocalizes with DFCP-1 marked omegasome. In vitro, the BATS domain preferentially binds to the highly curved membrane in a PtdIns(3)P-dependent manner. These data indicate that Barkor/Atg14(L) is concentrated on the curved autophagic membrane enriched in PtdIns(3)P via the BATS domain in a stress-inducible manner.
It is reported that Barkor/Atg14(L) targets to ER membrane through its N-terminus cysteine repeats. Upon autophagic stress, Barkor/Atg14(L) recruits Beclin 1-Vps34-p150 complex to ER to produce PtdIns(3)P on local ER membrane and likely cradles the precursor membrane of autophagosome. Stress-inducing high membrane curvature attracts Barkor/Atg14(L) because of the high binding affinity of the C-terminal BATS domain with curved membrane, which leads to more PtdIns(3)P production and further enhanced Barkor/Atg14(L) membrane binding via BATS. The concentration of Barkor/Atg14(L) on curved membrane might be crucial to stabilize the membrane curvature, and also to recruit downstream effectors Atg5/Atg12/Atg16 and LC3 to the curved membrane and extend membrane curvature to engulf cargoes and complete autophagosome (Fig. 5E). When fusion starts, it is likely that PtdIns(3)P is dephosphorylated by phosphatases, which leads to the dissociation of PI3KC3 complex and Atg5/Atg12/Atg16 from autophagosome. Barkor/Atg14(L)and PI3KC3 dissociation from autophagosome might cause membrane curvature instability and prime the membrane for fusion with endo/lysosomes to form autolysosomes.
Proteins that strongly tubulate membranes, such as epsin1 ENTH domain, are often highly charged at the polar side of their amphipathic helix and bind membranes independent of liposome curvature (29). In contrast, weakly charged amphipathic helices tend to serve as membrane curvature sensor. In this study, we demonstrate that Barkor/Atg14(L) functions as a membrane curvature sensor. Whether the BATS domain and Barkor/Atg14(L) can deform membrane by itself or in cooperation with other PI3KC3 subunits has yet to be investigated. The driving and sensing of autophagosome membrane curvature could be coordinated with protein scaffolds and lipid modification. One possible protein scaffold is the Atg5/Atg12/Atg16 complex, which recruits LC3/Atg8 and its conjugation enzymes to the nucleating autophagosome membrane. Both Atg5/Atg12/Atg16 and LC3 recruitment and conjugation are critical for the elongation of the initiating membrane. PtdIns(3)P generation by the PI3K kinase is also indispensable for membrane formation, which is required for the orientation of the Atg5/Atg12/Atg16 complex to autophagosome membrane rather than other types of membrane (39). Identification of the BATS domain as a sensor of initial membrane curvature should provide a biochemical platform to test the necessity of different proteins or nonprotein factors for reconstitution of autophagosome biogenesis in the near future.
Material and Methods
All Barkor/Atg14(L) mutants were cloned into the Bgl II and EcoR I sites on hrGFP-N1 and a 3xFlag tag was inserted between the Sal I and BamH I sites. hrGFP-BATS triple R mutation (W484R, F485R, Y488R) was cloned from full-length Barkor using the following primers: P1: GCGAATTCGgctggagaatcagatgagagcg and P2: GCGAATTCGAacggtgtccagtgcgagcttt acgccgggaggtcaccga. See SI Materials and Methods for other details.
Supplementary Material
Acknowledgments.
We thank Qiming Sun and Tsz Mei Jesmine Cheung for technical assistance. We thank Pietro De Camilli for the expression constructs of Endophilin A1 and the PH domain of PLCδ. We thank Randy Schekman, Jeremy Thorner, and Harvey McMahon for scientific discussion, and Livy Wilz for the critical reading of the manuscript. The work is supported by a New Investigator Award for Aging from the Ellison Medical Foundation, Hellman Family Foundation and National Institutes of Health Grant R01 (CA133228) to Q.Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.J.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016472108/-/DCSupplemental.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie Z, Klionsky DJ. Autophagosome formation: Core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 5.Ohsumi Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: Identification of a novel receptor, Atg34p. J Biol Chem. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backer JM. The regulation and function of class III PI3Ks: Novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 9.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex—At the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5:713–716. doi: 10.4161/auto.5.5.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 15.Itakura E, Mizushima N. Atg14 and UVRAG: Mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5:534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 16.Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci USA. 2010;107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsunaga K, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson JC, Legg JA, Machesky LM. Bar domain proteins: A role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–498. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 20.McMahon HT, Kozlov MM, Martens S. Membrane curvature in synaptic vesicle fusion and beyond. Cell. 2010;140:601–605. doi: 10.1016/j.cell.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Martens S, McMahon HT. Mechanisms of membrane fusion: Disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 22.Lee MC, et al. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 23.Lippincott-Schwartz J, Liu W. Membrane trafficking: Coat control by curvature. Nature. 2003;426:507–508. doi: 10.1038/426507a. [DOI] [PubMed] [Google Scholar]
- 24.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 25.Sudhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 27.Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 28.Ford MG, et al. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 29.Ford MG, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 30.Wickner W. Membrane fusion: Five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 31.Mima J, Wickner W. Phosphoinositides and SNARE chaperones synergistically assemble and remodel SNARE complexes for membrane fusion. Proc Natl Acad Sci USA. 2009;106:16191–16196. doi: 10.1073/pnas.0908694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Noriega A, Grubb JH, Talkad V, Sly WS. Chloroquine inhibits lysosomal enzyme pinocytosis and enhances lysosomal enzyme secretion by impairing receptor recycling. J Cell Biol. 1980;85:839–852. doi: 10.1083/jcb.85.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peter BJ, et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 37.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Madsen KL, Bhatia VK, Gether U, Stamou D. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 2010;584:1848–1855. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 39.Fujita N, Itoh T, Fukuda M, Noda T, Yoshimori T. The Atg16L cmplex secifies the ste of LC3 lpidation for mmbrane bogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.