Abstract
Translational control of many mRNAs in developing metazoan embryos is achieved by alterations in their poly(A) tail length. A family of cytoplasmic poly(A)-binding proteins (PABPs) bind the poly(A) tail and can regulate mRNA translation and stability. However, despite the extensive biochemical characterization of one family member (PABP1), surprisingly little is known about their in vivo roles or functional relatedness. Because no information is available in vertebrates, we address their biological roles, establishing that each of the cytoplasmic PABPs conserved in Xenopus laevis [PABP1, embryonic PABP (ePABP), and PABP4] is essential for normal development. Morpholino-mediated knockdown of PABP1 or ePABP causes both anterior and posterior phenotypes and embryonic lethality. In contrast, depletion of PABP4 results mainly in anterior defects and lethality at later stages. Unexpectedly, cross-rescue experiments reveal that neither ePABP nor PABP4 can fully rescue PABP1 depletion, establishing that PABPs have distinct functions. Comparative analysis of the uncharacterized PABP4 with PABP1 and ePABP shows that it shares a mechanistically conserved core role in promoting global translation. Consistent with this analysis, each morphant displays protein synthesis defects, suggesting that their roles in mRNA-specific translational regulation and/or mRNA decay, rather than global translation, underlie the functional differences between PABPs. Domain-swap experiments reveal that the basis of the functional specificity is complex, involving multiple domains of PABPs, and is conferred, at least in part, by protein–protein interactions.
Keywords: global translational control, mRNA-specific translational control, RNA-binding protein, translation initiation, end-to-end complexes
Early vertebrate development is directed by maternally transcribed mRNAs (1) that are deadenylated upon their exit from the nucleus and are stored in a translationally inactive state (2). During specific developmental stages, subsets of mRNAs are readenylated in a highly regulated process known as cytoplasmic polyadenylation (3, 4) concomitant with their translational activation. Although they have been best studied during early development, dynamic changes in poly(A) tail length also occur in other cell types. The function of the poly(A) tail in stimulating translation is mediated by the binding of the cytoplasmic poly(A)-binding protein (PABP) family of proteins, which are structurally and functionally distinct from nuclear PABPs (5, 6). Although vertebrates express multiple cytoplasmic PABPs, which contain a conserved domain organization, most studies have focused on the prototypical member PABP1 [also known as PABP, cytoplasmic 1 (PABPC1)]. The N terminus of PABP1 contains four nonidentical RNA recognition motifs (RRMs) which mediate both RNA and protein interactions. The C terminus comprises a highly variable proline-rich region that mediates PABP–PABP interactions important for oligomerization along the poly(A) tail and the PABP C-terminal domain (PABC) [also known as the MLLE domain] responsible for several protein–protein interactions (5, 6).
Biochemical studies suggest that metazoan PABP1 stimulates translation initiation by simultaneously binding the poly(A) tail and interacting with other translation factors located at the 5′ UTR, including eukaryotic initiation factor (eIF)4G and poly(A)-binding protein interacting protein 1 (PAIP1) (7–9). These interactions mediate the circularization of the mRNA, promoting recruitment of the small ribosomal subunit (40S) and perhaps recycling of translation initiation factors and terminating ribosomes (10). Furthermore, PABP1 has a largely uncharacterized effect on large ribosomal subunit (60S) recruitment and participates in translation termination via binding to eukaryotic release factor 3 (eRF3) (5, 6, 11). In addition to this core role in translation, PABP1 also functions in mRNA turnover and nonsense-mediated decay (12, 13). Moreover, it is emerging that PABP1 has a variety of mRNA-specific roles, including translational activation and miRNA-mediated repression (5, 14). This multifunctionality is not fully understood but appears to be conferred by the ability of PABP1 to interact with key factors. The other PABPs remain largely uncharacterized, although the molecular functions of embryonic PABP (ePABP, also known as ePAB or PABP1-like) have been partially described (15–18).
Given the key role of PABP1 in regulating posttranscriptional gene expression and its extensive functional characterization, it is surprising that relatively little is known about the biological roles of PABP proteins. Deletion of the single poly(A)-binding protein (PAB1) in Saccharomyces cerevisiae, which serves both nuclear and cytoplasmic functions, is lethal (19), although in Schizosaccharomyces pombe spPABP is not essential (20). In Drosophila melanogaster, deletion/reduction of dPABP or its deregulated expression leads to embryonic lethality/male sterility or neurophysiological defects, respectively (21, 22). Caenorhabditis elegans encodes two cytoplasmic PABPs, and mutations in pab-1 cause defects in the germline and affect longevity (23, 24). However, an absence of studies precludes any conclusion regarding the essential nature of PABPs in vertebrates.
During Xenopus laevis development, in which the effects of cytoplasmic polyadenylation have been studied extensively, the expression patterns of the two identified PABPs are distinctive: ePABP protein is abundant during oogenesis and early embryogenesis and is absent later in development. PABP1 protein is reciprocally expressed, being present at low levels during oogenesis and early embryogenesis and gradually increasing after the resumption of zygotic transcription at the midblastula transition (15, 16). This apparent switching of PABPs raises interesting questions about their respective molecular and/or biological roles during different developmental stages. Here we address the roles of PABPs during vertebrate development and explore their functional redundancy. We reveal that morpholino-mediated depletion of PABP1 and ePABP in X. laevis causes severe embryonic phenotypes and lethality. In contrast, phenotypes resulting from depletion of the newly identified PABP4 [also known as inducible PABP (iPABP) or PABP, cytoplasmic 4 (PABPC4)] are not observed until later in development and comprise mainly anterior defects. We show that the observed phenotypes are caused, at least in part, by defects in global protein synthesis and that the previously uncharacterized PABP4 binds poly(A) and stimulates translation in a manner similar to PABP1 and ePABP. However, cross-rescue experiments show that neither PABP4 nor ePABP is able to rescue the PABP1-deficient phenotype completely, indicating partially differential molecular functions. Taken together our data demonstrate that although the different members of the PABP family share a role in global translation, they appear to be functionally distinct, with multiple regions of PABP contributing to specificity.
Results
PABP1 and ePABP Are Essential for X. laevis Development.
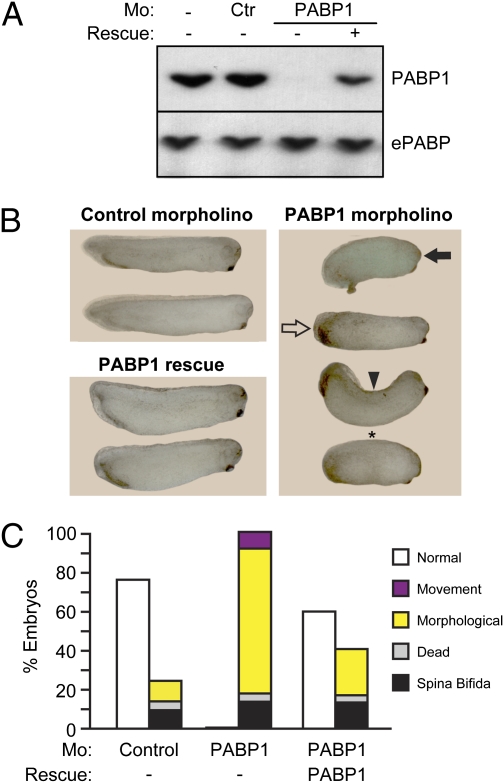
To analyze the phenotypic consequences of PABP deficiency in vertebrates, we injected X. laevis embryos with morpholino antisense oligonucleotides that blocked the translation of specific PABPs (Fig. S1). Strikingly, 100% of embryos injected with a PABP1-specific morpholino, which depleted PABP1 levels by >90% in vivo (Fig. 1A), were defective, exhibiting a wide variety of anterior and posterior defects (Fig. 1 B and C). Specific morphological defects became apparent at stage 25, and all embryos died by stage 30/31. As is typical of morpholino-mediated knockdown, a range of phenotypes was observed: Phenotypes at stage 29/30 included abnormal development of eyes, cement gland, tail, and fin, different degrees of body axis curvature, and defective posterior elongation, with single embryos often having multiple defects (Fig. 1B and Fig. S2A). In some cases development was arrested at stage 18–20. The few embryos that appeared morphologically normal had movement defects, because they did not respond when prodded (Fig. 1C). In contrast, only 10 to 25% of embryos injected with control morpholino exhibited defects, including spina bifida and early death, representing nonspecific effects. Importantly, injection of another morpholino directed against a different region of PABP1 mRNA resulted in very similar phenotypic effects (Fig. S2B). To probe further the specificity of the PABP1 phenotypes, a modified PABP1 mRNA that could not be recognized by the morpholino (morpholino-resistant) was injected at different concentrations. In the absence of morpholino, this mRNA did not cause defects (Fig. S3A). When coinjected with PABP1 morpholino, it rescued PABP1 protein expression (Fig. 1A) and the morphological and movement defects (Fig. 1 B and C and Fig. S2A), with normal embryos observed at ∼80% of the frequency seen with control morpholino. Taken together, these data establish that normal vertebrate embryogenesis depends on PABP1 expression.
Fig. 1.
PABP1 depletion causes multiple developmental defects and embryonic lethality. (A) Western blot analysis of whole-cell extracts from stage 14–16 embryos injected with control (Ctr) or PABP1-A (PABP1) morpholino (Mo) ± 10 ng of PABP1 rescue mRNA, using antibodies specific for PABP1 and ePABP. (B) Representative photographs of stage 29/30 embryos injected with control or PABP1-A morpholino ± 10 ng of PABP1 rescue mRNA. PABP1 morphants show abnormal development of anterior (closed arrow) and posterior (open arrow) structures, spinal curvature (arrowhead), and developmental arrest at stage 18–20 (asterisk). (Additional photographs are shown in Fig. S2.) (C) Percentages of control, PABP1 morpholino, and PABP1 rescue embryos (stage 29/30) displaying the indicated phenotypes. Morphological defects include spinal curvature; abnormal development of eye, cement gland, tail, and fin; ventral edema; absence of posterior development; and developmental arrest at stage 18–20. Data represent the average of nine (PABP1 morpholino) or six (PABP1 rescue) independent experiments, with ∼1,200 or 900 embryos per experimental point, respectively.
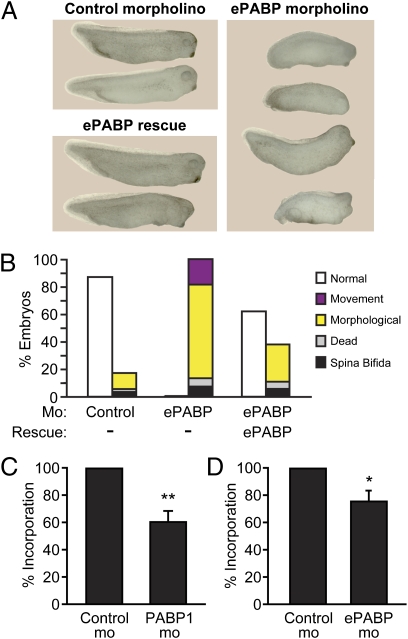
Embryos injected with a morpholino that blocks ePABP translation in vitro and in vivo (Figs. S1B and S4A) showed a similar range of morphological and movement defects, although death occurred later, by stage 35. This finding was perhaps surprising, because ePABP is the predominant PABP in early embryos, but may result from the higher levels of maternal ePABP (15, 16). ePABP defects were present in both anterior and posterior structures and also were fully penetrant (Fig. 2 A and B and Fig. S4B). Efficient phenotypic rescue was achieved by coinjecting a morpholino-resistant ePABP mRNA (Fig. 2 A and B and Fig. S4B), excluding off-target effects. Embryos injected with the ePABP mRNA in the absence of morpholino did not display developmental defects (Fig. S3A). Importantly, an alternative ePABP-specific morpholino generated similar phenotypes (Fig. S4C), further showing the morphological and movement defects to be specific. These data provide insight into the biological role of ePABP, establishing that, like PABP1, it is essential for vertebrate development.
Fig. 2.
ePABP is essential for development. (A) Representative photographs of stage 30–32 embryos injected with control or ePABP-A (ePABP) morpholino ± 10 ng of ePABP rescue mRNA. (Additional photographs are shown in Fig. S4.) (B) Percentages of control, ePABP morpholino, and ePABP rescue embryos (stage 30–32) displaying the indicated phenotypes. Legend is as in Fig. 1C. Data represent the average of seven (ePABP morpholino) or three (ePABP rescue) independent experiments, with ∼900 or 300 embryos per experimental point, respectively. Control and (C) PABP1 or (D) ePABP morphants were labeled metabolically with [35S]methionine, and newly synthesized proteins were TCA precipitated. Data are shown as percentage of [35S]methionine incorporation relative to control (set to 100%) and represent the average of two independent experiments, with ∼60 embryos per experimental point. Error bars indicate SEM. *P < 0.05 and **P < 0.01 versus control. P values were determined by t test.
To gain insight into the mechanism underlying the PABP1 and ePABP phenotypes, we analyzed the effect of their depletion on global protein synthesis in embryos. PABP1 and ePABP morphants both showed a significant reduction in global protein synthesis, of 40% and 25%, respectively (Fig. 2 C and D); with the latter perhaps reflecting the higher levels of maternal ePABP (15, 16).
PABP4 Depletion Causes Anterior Defects and Embryonic Lethality.
Bioinformatic analysis identified a PABP mRNA in X. laevis encoding a protein that is most closely related to mammalian PABP4 (Fig. S5 A and B). This newly identified PABP4 maintains the same domain organization as PABP1 (5) and exhibits high homology to human PABP4 (81% identity) and X. laevis PABP1 (75% identity). PABP4 mRNA, like PABP1, was detected in a wide variety of adult tissues (Fig. S5C), consistent with studies in mammals (25). PABP4 mRNA is expressed in early oocytes, decreases during later oogenesis, and reappears from stage 7/8 embryos (Fig. S5D) at the onset of zygotic transcription, when ePABP is still the predominant PABP protein.
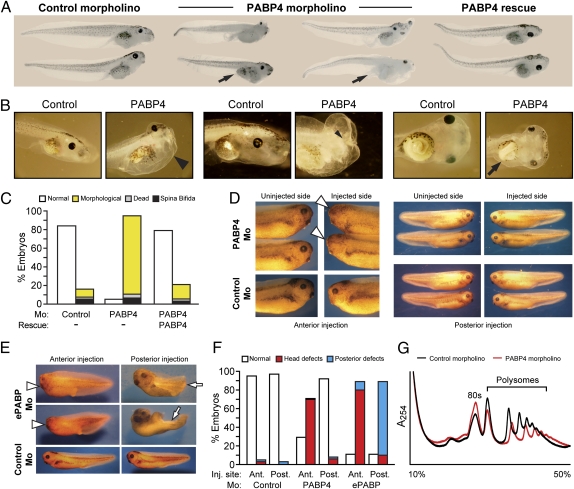
We investigated the developmental role of this newly identified PABP by injecting a PABP4-specific morpholino (Fig. S1C). Interestingly, morphological defects were observed only after stage 29/30, later than in PABP1 and ePABP morphants, and embryos did not die until stage 50. A number of specific phenotypes were observed, but, in sharp contrast to PABP1 and ePABP morphants, the formation of anterior structures appeared more sensitive to PABP4 depletion than posterior structures (Fig. 3A). Frequent defects included cephalic and ventral edema, malformation of the head, and poor eye development (Fig. 3B). Severe deformities of the digestive tract also were frequently observed. In normal stage 41/42 embryos the intestine begins to twist, with ∼360° torsion by stage 44 and 2.5–3.5 revolutions by stage 47. Many PABP4-deficient embryos failed to exhibit normal intestinal coiling (Fig. 3B). Development of the proctodeum was often impaired or even absent (Fig. 3A). Remarkably, the incidence of morphologically defective embryos was >90% at stages 47–50 (Fig. 3C), and the embryos also displayed abnormal swimming motions. Because attempts to generate PABP4-specific antibodies proved unsuccessful, we used a FLAG-tagged morpholino-resistant PABP4 mRNA for rescue experiments at a dose that did not cause phenotypic defects although the protein was easily detectable (Fig. S3 B and C). Coinjection of this mRNA with the PABP4 morpholino efficiently rescued the morphological phenotypes (Fig. 3 A and C), confirming their specificity.
Fig. 3.
PABP4 depletion causes anterior defects and embryonic lethality. (A) Representative photographs of stage 45–47 embryos injected with control or PABP4 morpholino ± 1 ng of PABP4-FLAG rescue mRNA. Arrows indicate abnormal or absent proctodeum. (B) Control (stage 45/46) and PABP4 morphants, which show ventral and cephalic edema (large arrowhead), absence of eye (small arrowhead), and reduced intestinal torsion (arrow). (C) Percentages of control, PABP4 morpholino, and PABP4 rescue embryos (stage 47–50) displaying the indicated phenotypes. Morphological defects include ventral and cephalic edema, abnormal head and eye development, and digestive tract defects. Data represent the average of three independent experiments, with ∼400 embryos per experimental point. (D and E) Representative photographs of stage 35–37 control embryos and embryos injected anteriorly (Ant) or posteriorly (Post) with (D) PABP4 or (E) ePABP-A (ePABP) morpholino. Arrowheads indicate abnormal head/eye development; and arrows indicate abnormal posterior development. (F) Percentages of control, PABP4, and ePABP-A morpholino embryos from D and E displaying the indicated phenotypes. Data represent the average of five (PABP4) or three (ePABP) independent experiments, with ∼120 or 90 embryos, respectively, per experimental point. (G) Polysomal profiling of stage 40–42 control and PABP4 morphants over a 10–50% sucrose gradient with a 60% cushion. A representative experiment is shown, and the positions of the 80S ribosome and polysomes are indicated.
In contrast to PABP1 and ePABP, PABP4 depletion appeared to be associated mainly with anterior defects, leading us to inject the morpholino into a single presumptive anterior or posterior cell at the four- to eight-cell stage, targeting only one side of the embryo. At stage 35–37 PABP4 depletion only caused defects following injection into an anterior blastomere; 70% of embryos showed abnormal eye development on the injected side (Fig. 3 D and F), indicating that formation of anterior structures is more sensitive to PABP4 depletion. In contrast, ePABP knockdown caused defective phenotypes after either anterior or posterior injection (Fig. 3 E and F), consistent with the results shown in Fig. 2A.
Because PABP4 depletion led to the most distinct phenotypes, and this vertebrate-specific PABP remains largely uncharacterized, we investigated whether differences in its putative core function in translation contribute to its developmental requirement. To assess PABP4 function in vivo, the polysome profiles of PABP4-deficient embryos were analyzed, revealing a clear reduction in polysome peaks compared with control embryos (Fig. 3G), which reflects a significant decrease in global protein synthesis. Thus, defects in translation appear to be an important facet of the PABP4 phenotype. Consistent with this result, PABP4 was found to associate with polysomal as well as nonpolysomal fractions when expressed in oocytes, reminiscent of ePABP (Fig. S6A). To explore the extent of the mechanistic similarity between PABP4 and PABP1/ePABP, we directly examined its quantitative ability to stimulate translation and its interaction with protein partners. Tethered function analysis in intact oocytes (9, 17) revealed that PABP4 stimulates the translation of mRNAs to a level equivalent to PABP1 and ePABP (Fig. S6 B–D). Moreover, we found that PABP4 binds poly(A) RNA (Fig S7A) consistent with observations in mammals (25, 26), interacts with translation factors known to be protein partners of X. laevis PABP1 and ePABP (9, 17, 27), and engages in PABP–PABP interactions (Fig. S7 C–E), suggesting that PABP4 may bind mRNAs cooperatively with other PABPs.
Thus, both the ability of PABP4 to stimulate translation and its mode of action seem to be conserved, revealing that differences in this core function do not appear to underlie the unique developmental requirement for this PABP.
PABP1, ePABP, and PABP4 Have Distinct Functions.
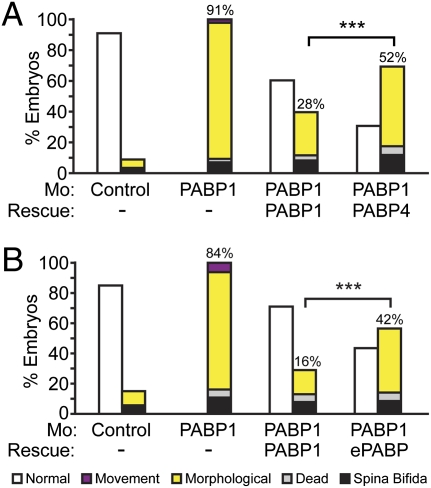
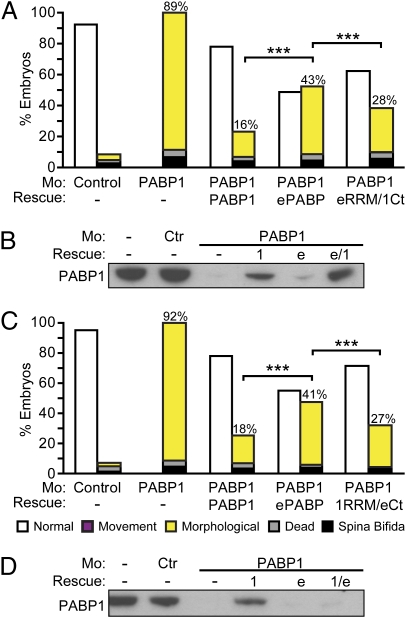
Conservation of their core role in translation does not preclude the possibility that other distinct molecular functions and/or differences in their temporo-spatial expression contribute to the different phenotypes associated with PABP1, ePABP, or PABP4 depletion. Thus, cross-rescue experiments were undertaken with mRNAs containing identical generic 5′ and 3′ UTRs to negate differences in their expression. Injection of mRNAs encoding either PABP1-FLAG or PABP4-FLAG into PABP1 morphants resulted in similar levels of ectopic PABP proteins (Fig. S3C), as expected. PABP1-FLAG mediated efficient rescue, reducing the PABP1-specific phenotypes (morphological and movement) from 91% to 28% of embryos (Fig. 4A and Fig. S8A). PABP4-FLAG expression did not rescue the specific phenotypes to the same extent, because they were reduced only from 91% to 52% (Fig. 4A). Strikingly, cross-rescue of PABP1 with an ePABP mRNA only reduced the percentage of specific phenotypes from 84% to 42%, whereas PABP1 self-rescue reduced these specific phenotypes to 16% (Fig. 4B and Fig. S8B). Thus, these data establish that neither PABP4 nor ePABP can completely substitute for PABP1 during development, providing evidence that metazoan members of this family have nonredundant as well as overlapping molecular functions.
Fig. 4.
PABP4 and ePABP cannot rescue PABP1-depleted embryos efficiently. Embryos were injected with control or PABP1-A (PABP1) morpholino ± (A) 1 ng of PABP1-FLAG or PABP4-FLAG rescue mRNA or (B) 10 ng of PABP1 or ePABP rescue mRNA. Percentages of stage 29/30 embryos displaying the indicated phenotypes are shown; numbers indicate the percentage of PABP1-specific phenotypes in each case (morphological plus movement defects). Data represent the average of approximately (A) 350 embryos or (B) 500 embryos per experimental point, in three or four independent experiments, respectively. ***P < 0.0001, as determined by Fisher's exact test. (Photographs are shown in Fig. S8.)
Multiple Determinants Underlie PABP Specificity.
Our data reveal the existence of functional differences between PABP family members (Fig. 4), which are not linked to their function in global translation (Figs. 2 C and D and 3G and Figs. S6 and S7). The C-terminal region is the most diverse among PABPs, leading us to hypothesize that it may be responsible for these differences. To address this hypothesis, domain-swap experiments were undertaken between the RRM and C-terminal regions of PABP1 and ePABP, which share 83% and 56% identity, respectively. First, the C terminus of PABP1 was appended to the RRM region of ePABP (eRRM/1Ct) and was coinjected with the PABP1 morpholino. Interestingly, rescue with eRRM/1Ct reduced PABP1-specific phenotypes from 89% to 28%, compared with a reduction from 89% to 43% with wild-type ePABP (Fig. 5A), showing that substitution of the PABP1 C terminus results in a more efficient rescue. However, this rescue was less efficient than wild-type PABP1, although Western blotting showed that the hybrid protein was expressed similarly in embryos (Fig. 5B). Importantly, the reciprocal rescue of PABP1 phenotypes using a construct in which the ePABP C terminus was appended to the RRMs of PABP1 (1RRM/eCt), also gave an intermediate level of cross rescue (Fig. 5 C and D). Thus, our data reveal that multiple regions underlie the unique requirement for PABP1.
Fig. 5.
Multiple regions of PABP1 determine specificity. Embryos were injected with control or PABP1-A (PABP1) morpholino ± 10 ng of PABP1, ePABP, or either (A) eRRM/1Ct or (C) 1RRM/eCt rescue mRNA. Percentages of stage 26–31 embryos displaying the indicated phenotypes are given. Numbers indicate the percentage of PABP1-specific phenotypes in each case (morphological plus movement defects). Data represent the average of approximately (A) 160 embryos or (C) 135 embryos per experimental point, in four or three independent experiments, respectively. ***P < 0.0001, as determined by Fisher's exact test. (B and D) Western blots of embryos from A and C using an anti-PABP1 antibody, which does not recognize 1RRMs/eCt (1/e) because it is raised against a C-terminal PABP1 peptide. 1, PABP1; e, ePABP; e/1, eRRM/1Ct.
Discussion
By providing phenotypic studies of PABPs in vertebrate development, we have uncovered essential but distinct functional roles for all three members of the X. laevis PABP family. These results significantly extend the previous analysis of PABP1 in yeast and invertebrates (19–24). In each case, developmental defects were accompanied by a significant loss of protein synthesis, establishing the role of metazoan PABPs in protein synthesis in vivo and showing that a decrease in overall translation contributes to the phenotypes. However, given the pleiotropic nature of PABP1, it is likely that defects in other aspects of posttranscriptional regulation (5, 12–14) also contribute to the phenotypes. Interestingly, injection of PABP mRNAs in nondepleted embryos did not result in phenotypic consequences, suggesting that overexpression is tolerated.
Altered PABP levels or differences in their temporo-spatial expression may contribute to the observed phenotypes. However, cross-rescue experiments revealed that neither PABP4 nor ePABP, even when similarly expressed, can substitute completely for PABP1, because the percentage of defective phenotypes obtained is approximately two and three times greater, respectively, than with the PABP1 self-rescue. These results strongly indicate that differential molecular functions also must contribute to the requirement for multiple PABPs during development, which is intriguing, given the high degree of amino acid similarity, especially between PABP1 and PABP4. Thus, our results identify the existence of biologically relevant differences between the functions of individual PABP family members in metazoa.
Importantly, mechanistic analysis of PABP function provides insight into the basis of these differences. We establish that PABP1, ePABP, and PABP4 each has an important role in determining overall translation rates in vivo, and SDS/PAGE analysis of [35S]methionine-labeled extracts from embryos suggest that synthesis of a wide range of proteins is affected (Fig. S9), confirming an effect on global translation. These global defects are consistent with our finding that PABP4 can bind poly(A) and stimulate translation similarly to PABP1 and ePABP, sharing interactions with basal translation factors. The evolutionary conservation of PABP4 suggests that this ability may be maintained in other species. Thus, all three PABPs appear to play indistinguishable rather than distinct roles in global translation, excluding this as a mechanism to explain the specificity. This situation differs from the eIF4A family in which one member (eIF4AIII) appears to have an alternative function in posttranscriptional regulation (28, 29).
Since their ability to promote global translation does not vary significantly, their distinct functions must be related to other aspects of posttranscriptional control in which their roles are less well characterized. Cross-rescue experiments swapping the less conserved C-terminal region of PABP1 and ePABP, although unable to formally exclude subtle or cell type-specific differences in PABP stability, revealed that multiple regions contribute to PABP specificity. Because the C-terminal region mediates interactions with proteins but not with RNA (Fig. S10A), our results strongly suggest that PABP-specific protein interactions with partners other than basal translation factors (which interact with all three) play a role in the requirement for individual PABPs. The high conservation of the PABC domain suggests that differential protein interactions may be mediated by the proline-rich region, which is highly variable and predicted to be alternatively spliced in mammals, providing great scope for PABP-specific protein–protein interactions. It is tempting to speculate that PABP-specific protein partners may be involved in mRNA-specific regulation, as misregulation of even a single essential mRNA could be sufficient to cause lethality. In this regard it is interesting to note that translational activation by mRNA-specific binding proteins that directly recruit PABP through C-terminal interactions has been described in X. laevis (30). However, the few characterized interactions mediated by this region do not appear to be PABP specific (Fig. S10 A and C); thus exhaustive identification of the PABP interactome is required.
Our data also, surprisingly, show that the relatively conserved RRM1–4 region is important for PABP specificity. This region interacts with both proteins and RNA, including, in other species, proteins that are involved in mRNA-specific regulation (31–33). Any differential RNA binding probably involves elements other than the poly(A) tail, because each of the PABPs binds poly(A) efficiently (Fig. S7 A and B). Thus, poly(A)-independent recruitment of individual PABPs to specific mRNAs, via RNA or proteins, may provide additional mechanisms for regulating mRNAs that are stored during development with short poly(A) tails.
In conclusion, our investigation provides insight into the distinct roles of vertebrate PABPs and in so doing argues against the hypothesis that members of this family are functionally redundant. These unexpected differences provide an explanation for developmental switching between PABPs (15, 16) and the presence of multiple PABPs within many mammalian cell types (34) and emphasize the importance of dissecting the roles of individual family members in whole organisms. The approach described here provides a powerful in vivo tool for future work aimed at understanding the challenging and intriguing issues of PABP function and specificity.
Methods
Plasmid construction and supporting procedures are described in SI Methods.
Animal experiments were performed under license and in accordance with the Animals in Scientific Procedures Act (1986).
The following translation-blocking morpholino antisense oligonucleotides, modified with 3′ carboxyfluorescein, were used (Gene Tools): PABP1-A: (5′ UTR) CTTCACTCTT CTCTTTACC GGATTT; PABP1-B: (AUG) GGTAGCTGG GAGCACTGGG ATTCAT; ePABP-A: (5′ UTR) GGATTCGCCC TCAGCCTGAT AACTC; ePABP-B: (AUG) CGGCTCCGGT TGCATTCATG TTTGC; PABP4: (5′ UTR) CACTGAGCAA TAATGGGACG GCTAA; Control: CCTCTTACCT CAGTTACAATT TATA.
Approximately 7 fmoles of morpholino and/or 1–10 ng of in vitro transcribed, capped, and polyadenylated mRNA were injected into both blastomeres in X. laevis embryos at the two-cell stage. Injections into single anterior or posterior blastomeres were performed with 3 fmoles of morpholino at the four- to eight-cell stage. Injected embryos were selected by fluorescence and allowed to progress for phenotypic analysis. Embryos were staged according to Nieuwkoop and Faber (35). For photography, embryos were fixed in 0.1 M 3-(N-Morpholino)propanesulfonic acid (pH 7.4), 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde and were washed and stored in 100% ethanol.
Western Blot Analysis.
Embryos were homogenized mechanically in TE buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, pH 8). One embryo per lane was separated by SDS-PAGE, transferred to PVDF membranes (Millipore), and probed with mouse monoclonal anti-PABP1 10E10 (1:2,000; Abcam), anti-FLAG M2 (1:6,000; Sigma), rabbit anti-PABP1 (1:500) (15), or anti-ePABP (1:2,000) (17). HRP-conjugated anti-mouse (Pierce Biotechnology) and anti-rabbit (Sigma) IgGs were used as secondary antibodies, and signals were detected by enhanced chemiluminescence (GE Healthcare).
Metabolic Labeling.
Embryos were coinjected with morpholino, as above, and with 35 nCi of [35S]methionine. Embryos were collected at stage 14–16, pooled into groups of five, and homogenized in TE. Trichloroacetic acid (TCA) precipitates (10%) were counted by liquid scintillation to determine [35S]methionine incorporation.
Sucrose Gradient Analysis.
Eighty stage VI oocytes or 30 stage 40–42 embryos were mechanically lysed in basic lysis buffer (300 mM KCl, 10 mM MgCl2, 20 mM Tris-HCl, pH 7.4) supplemented with 0.5% sodium deoxycholate and 150 μg/mL cycloheximide or 20 mM EDTA. Cleared supernatants were layered onto a 10–50% sucrose gradient over a 60% cushion in basic lysis buffer and centrifuged for 2 h at 247,000 × g (38,000 rpm) using a TH-641 rotor (Sorvall). Fractions were collected using a density gradient fractionation system (Teledyne Isco). Proteins were extracted from fractions by 10% TCA precipitation before Western analysis.
Supplementary Material
Acknowledgments
We thank Bertrand Cosson for the gift of pGBT9-eRF3 plasmid and PABP1 antibody; Tom Van Agtmael for statistical analysis; Richard Smith, Nick Hastie, and other colleagues for critical reading of the manuscript; and Ted Pinner for figure preparation. Work in the N.K.G. laboratory was supported by Medical Research Council core funding, a Medical Research Council Senior Non-Clinical Fellowship, and by project grants from the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017664108/-/DCSupplemental.
References
- 1.Farley BM, Ryder SP. Regulation of maternal mRNAs in early development. Crit Rev Biochem Mol Biol. 2008;43:135–162. doi: 10.1080/10409230801921338. [DOI] [PubMed] [Google Scholar]
- 2.Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 3.Belloc E, Piqué M, Méndez R. Sequential waves of polyadenylation and deadenylation define a translation circuit that drives meiotic progression. Biochem Soc Trans. 2008;36:665–670. doi: 10.1042/BST0360665. [DOI] [PubMed] [Google Scholar]
- 4.Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorgoni B, Gray NK. The role of cytoplasmic poly(A)-binding proteins in regulating gene expression: A developmental perspective. Brief Funct Genomics Proteomics. 2004;3:125–141. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- 6.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223.1–223.14. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig AW, Haghighat A, Yu AT, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 8.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derry MC, Yanagiya A, Martineau Y, Sonenberg N. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb Symp Quant Biol. 2006;71:537–543. doi: 10.1101/sqb.2006.71.061. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez CI, Wilusz CJ, Wilusz J. The interface between mRNA turnover and translational control. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2007. pp. 719–745. [Google Scholar]
- 13.Mühlemann O. Recognition of nonsense mRNA: Towards a unified model. Biochem Soc Trans. 2008;36:497–501. doi: 10.1042/BST0360497. [DOI] [PubMed] [Google Scholar]
- 14.Burgess HM, Gray NK. mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem Soc Trans. 2010;38:1517–1522. doi: 10.1042/BST0381517. [DOI] [PubMed] [Google Scholar]
- 15.Cosson B, et al. Characterization of the poly(A) binding proteins expressed during oogenesis and early development of Xenopus laevis. Biol Cell. 2002;94:217–231. doi: 10.1016/s0248-4900(02)01195-4. [DOI] [PubMed] [Google Scholar]
- 16.Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkie GS, Gautier P, Lawson D, Gray NK. Embryonic poly(A)-binding protein stimulates translation in germ cells. Mol Cell Biol. 2005;25:2060–2071. doi: 10.1128/MCB.25.5.2060-2071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thakurta AG, Ho Yoon J, Dhar R. Schizosaccharomyces pombe spPABP, a homologue of Saccharomyces cerevisiae Pab1p, is a non-essential, shuttling protein that facilitates mRNA export. Yeast. 2002;19:803–810. doi: 10.1002/yea.876. [DOI] [PubMed] [Google Scholar]
- 21.Blagden SP, et al. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev Biol. 2009;334:186–197. doi: 10.1016/j.ydbio.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Sigrist SJ, et al. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature. 2000;405:1062–1065. doi: 10.1038/35016598. [DOI] [PubMed] [Google Scholar]
- 23.Maciejowski J, et al. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics. 2005;169:1997–2011. doi: 10.1534/genetics.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko S, et al. Two mutations in pab-1 encoding poly(A)-binding protein show similar defects in germline stem cell proliferation but different longevity in C. elegans. Mol Cells. 2010;30:167–172. doi: 10.1007/s10059-010-0103-2. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Duckett CS, Lindsten T. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol Cell Biol. 1995;15:6770–6776. doi: 10.1128/mcb.15.12.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sladic RT, Lagnado CA, Bagley CJ, Goodall GJ. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur J Biochem. 2004;271:450–457. doi: 10.1046/j.1432-1033.2003.03945.x. [DOI] [PubMed] [Google Scholar]
- 27.Cosson B, et al. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol Cell. 2002;94:205–216. doi: 10.1016/s0248-4900(02)01194-2. [DOI] [PubMed] [Google Scholar]
- 28.Budiman ME, et al. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol Cell. 2009;35:479–489. doi: 10.1016/j.molcel.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 30.Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel GP, Ma S, Bag J. The autoregulatory translational control element of poly(A)-binding protein mRNA forms a heteromeric ribonucleoprotein complex. Nucleic Acids Res. 2005;33:7074–7089. doi: 10.1093/nar/gki1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duncan KE, Strein C, Hentze MW. The SXL-UNR corepressor complex uses a PABP-mediated mechanism to inhibit ribosome recruitment to msl-2 mRNA. Mol Cell. 2009;36:571–582. doi: 10.1016/j.molcel.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 33.Chang TC, et al. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katzenellenbogen RA, Vliet-Gregg P, Xu M, Galloway DA. Cytoplasmic poly(A) binding proteins regulate telomerase activity and cell growth in human papillomavirus type 16 E6-expressing keratinocytes. J Virol. 2010;84:12934–12944. doi: 10.1128/JVI.01377-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis. New York: Garland Publishing, Inc.; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.