Abstract
Anionic lipids play a variety of key roles in membrane function, including functional and structural effects on respiratory complexes. However, little is known about the molecular basis of these lipid–protein interactions. In this study, NarGHI, an anaerobic respiratory complex of Escherichia coli, has been used to investigate the relations in between membrane-bound proteins with phospholipids. Activity of the NarGHI complex is enhanced by anionic phospholipids both in vivo and in vitro. The anionic cardiolipin tightly associates with the NarGHI complex and is the most effective phospholipid to restore functionality of a nearly inactive detergent-solubilized enzyme complex. A specific cardiolipin-binding site is identified on the basis of the available X-ray diffraction data and of site-directed mutagenesis experiment. One acyl chain of cardiolipin is in close proximity to the heme bD center and is responsible for structural adjustments of bD and of the adjacent quinol substrate binding site. Finally, cardiolipin binding tunes the interaction with the quinol substrate. Together, our results provide a molecular basis for the activation of a bacterial respiratory complex by cardiolipin.
Keywords: bioenergetics, EPR spectroscopy, metalloprotein, molybdenum
The structural and functional integrity of biological membranes is vital to life. They provide specialized permeability barriers that separate the cytoplasm of the cell from the environment and internally compartmentalize eukaryotic cells into organelles. The membrane is also a dynamic noncovalent supramolecular organization of individual lipid molecules whose combined physical and chemical properties define the matrix within which membrane proteins are organized (1). In addition, the interplay of lipids and membrane proteins is crucial to several fundamental processes ranging from respiration, photosynthesis, signal transduction, and solute transport to motility. An increasing number of membrane protein structures revealed the presence of tightly bound lipids, some of them located at specific sites (2). For instance, the list of mitochondrial proteins that bind the anionic cardiolipin lipid (CL) with high affinity is long and includes, among others, the respiratory complexes I, III, IV, and V; the ADP/ATP carrier; and even peripherical membrane proteins such as cytochrome c (for reviews, see refs. 3–8). Defined lipid species confer structural stability and control insertion and folding processes (9, 10). They are also involved in assembly or oligomerization of multisubunit complexes or supercomplexes or directly affect the function of membrane proteins (11–18). However, a major question in general biochemistry, still open, is how lipids affect, at a molecular level, respiratory complexes function.
In the bacterium Escherichia coli, the resolution of the structure of several respiratory complexes has revealed the presence of tightly bound phospholipids. In particular, cardiolipin, which consists of two phosphatidyl residues linked by a glycerol moiety, has been observed at the interface of adjacent monomers in the dimeric succinate dehydrogenase (19) or in the trimeric formate dehydrogenase N (FdnGHI) (20). Together with the latter, the nitrate reductase complex (NarGHI) forms the paradigmatic Fdn-Nar protonmotive redox loop formulated in Mitchell’s chemiosmotic theory (21). Both FdnGHI and NarGHI are membrane-bound heterotrimeric enzymes that contain a molybdenum cofactor, two hemes, and five FeS centers (22). Whereas the FdnGHI complex is oriented toward the periplasmic side of the membrane, NarGHI is cytoplasmically oriented. The catalytic subunit (FdnG or NarG) is a membrane-associated subunit composed of a [4Fe-4S] cluster in addition to the molybdenum cofactor (23, 24). The electron-transfer subunit (FdnH or NarH) is another membrane-associated subunit, which contains four FeS clusters. The membrane-anchor subunit (FdnI or NarI) incorporates two heme b groups, and NarI strongly stabilizes a semiquinone intermediate species at a QD site located in close proximity with the heme bD on the periplasmic side of the membrane (25–28). This QD site corresponds to the quinol oxidation site in NarGHI. Examination of the NarGHI crystal structure revealed the presence of a partly defined electron density attributed to a yet unidentified phospholipid (23). Accordingly, the role of the associated phospholipids remains to be established in this enzyme as well as in formate or succinate dehydrogenases.
Here, we investigate the consequences that lipids have on the structure and function of the nitrate reductase complex. Our results demonstrate that CL binds specifically in the vicinity of the QD site and that this binding activates this respiratory complex by allowing the correct interaction with the quinol substrate.
Results
Quinol:Nitrate Oxidoreductase Activity of the Nitrate Reductase Complex Critically Depends on Phospholipid Binding.
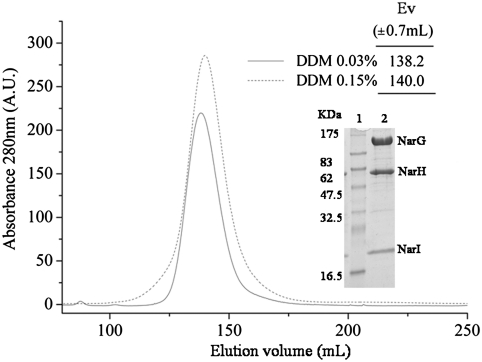
E. coli nitrate reductase complex was isolated from inner membrane vesicles (IMVs) by solubilization with 1% dodecyl maltoside (DDM) followed by chromatographic steps with either 0.03 or 0.15% of DDM. Both procedures yielded a complex in a dimeric state as shown by size-exclusion chromatography (Fig. 1). Upon SDS-PAGE, the complex is resolved into three distinct bands corresponding to the three structural subunits and shows no detectable instability (Fig. 1 Inset). EPR analysis of these preparations shows that detergent solubilization does not induce significant differences of the redox and spectroscopic properties of FeS and Mo centers when compared to NarGHI-enriched IMVs (Table S1 and Fig. S1 and Fig. S2). Maintenance of the integrity of the NarGH complex is confirmed by only slightly affected specific benzyl viologen:nitrate oxidoreductase activity values (Table 1). However, the specific quinol:nitrate oxidoreductase activity values were systematically lower for detergent-solubilized samples than for IMVs (Table 1). Increased delipidation of the enzyme complex using 0.15% DDM as compared to 0.03% leads to a further decrease in activity without affecting the dimeric organization or the stability (Table 1). Importantly, loss of activity cannot be attributable to an inhibitory action of detergents because incubation with increasing DDM concentrations in the activity assay (up to 0.08%; i.e., 8-fold the critical micellar concentration of DDM) has no effect (SI Text).
Fig. 1.
Gel permeation chromatography of delipidated NarGHI-DDM isolated complex. Elution profile of NarGHI isolated with 0.03% n-dodecyl-β-D-maltoside (DDM 0.03%) or 0.15% (DDM 0.15%) on a Sephacryl HR 300 column. Purified NarGHI elution volume (Inset table) was determined by calibration with molecular weight standards as indicated in Materials and Methods. Inset figure: Molecular weight standard is presented in lane 1 (Protein Marker Broad Range, Biolabs); profile of 20 μg of NarGHI DDM 0.15%-isolated protein loaded in a 12% SDS-PAGE (lane 2). A.U., arbitrary units; Ev,:elution volume.
Table 1.
Enzymatic activities and phospholipid content of DDM-isolated NarGHI and of the NarGHIW162A variant
| mol lipid/mol NarGHI | ||||||
| BV activity | Quinol activity | CL | PG | PE | ||
| NarGHI | membrane | 121 ± 10 | 13.3 ± 1.3 | — | — | — |
| DDM 0.03% | 70 ± 6 | 8.3 ± 0.5 | 3.5 | 1.3 | 2.6 | |
| DDM 0.15% | 65 ± 5 | 1.2 ± 0.1 | 1 | 0.2 | 0.2 | |
| NarGHIW162A | membrane | 125 ± 13 | 13.3 ± 1.6 | — | — | — |
| DDM 0.03% | 68 ± 6 | 4.9 ± 0.4 | 2 | 1.1 | 2.8 | |
| DDM 0.15% | 66 ± 4 | 0.4 ± 0.1 | 0.9 | ND | ND | |
Specific benzyl viologen (BV) and menadiol (quinol) activities have been measured and expressed in units/mg of NarGHI. The phospholipid content was calculated from TLC analysis of the lipid extract. The TLC data are the average of at least three independent experiments that vary by no more than 10% from the mean. ND, not detected.
Cardiolipin Binds Specifically to the Nitrate Reductase Complex.
The phospholipid content of the enzyme preparations was estimated by thin-layer chromatography (TLC) and ranged between 7 and 8 mol/mol of nitrate reductase. Interestingly, TLC analysis shows a specific enrichment in CL during enzyme purification as it accounts for approximately 50% of the total phospholipids content when using 0.03% DDM (Table 1). It is worth mentioning that CL represents a minor phospholipid in the inner membrane of E. coli with a percentage never exceeding 5–10% (29).
Incubation of the enzyme with a large excess of DDM (0.15%) followed by ion-exchange chromatography to remove the resulting phospholipid-detergent mixed micelles did not produce phospholipid-free nitrate reductase. Indeed, the total amount of phospholipids bound to the enzyme was reduced from about 7.4 to 1.4. Using this detergent concentration, the nearly inactive enzyme is still organized as a dimer, and we found that the only lipid bound in stoichiometric amount is CL. The presence of this remaining CL is thus not sufficient to promote quinol oxidase activity.
Quinol Oxidase Activity Is Most Effectively Recovered upon Reconstitution in Proteoliposomes Enriched in Anionic Phospholipids.
Purified NarGHI complex was used to further examine the phospholipid requirement of quinol oxidase activity. The phospholipid composition of the E. coli inner membrane varies between 70–80% of phosphatidyletanolamine (PE), 20–25% of phosphatidylglycerol (PG), and 5–10% of CL. To mimic the native situation, the NarGHI complex was reconstituted into liposomes having the above-mentioned lipid composition. The specific activity of NarGHI reconstituted into liposomes was significantly higher than that measured with purified enzyme preparation and comparable to that measured in NarGHI-enriched IMVs (Tables 1 and 2). The loss of quinol oxidase activity associated with delipidation is thus fully reversible upon reconstitution into liposomes.
Table 2.
Quinol oxidase activity of NarGHI in proteoliposomes
| Activity, U/mg | |
| EcL | 14.6 ± 1.4 |
| EcL + PE | 14.3 ± 1.4 |
| EcL + PG | 17.9 ± 0.3 |
| EcL + CL | 21.4 ± 1.3 |
| Control | 1.4 ± 0.1 |
Pure lipids and extracts are from E. coli. Mix composition in molecules of PE∶PG∶CL are as follows: EcL (13∶4∶1), EcL + PE (20∶4∶1), EcL + PG (13∶8∶1), EcL + CL (13∶4∶2). Proteoliposomes were prepared using a 0.15% DDM purified NarGHI complex. In control experiments, purified NarGHI was treated identically with the exception of liposomes. EcL, total lipid extract.
To examine if there is a lipid specificity in stimulating NarGHI activity, we prepared liposomes with varying phospholipids composition. NarGHI activity was the lowest when reconstituted with PE-enriched liposomes, increased with PG-enriched, and was the highest with CL-enriched liposomes (Table 2). These results demonstrate that anionic phospholipids, in particular CL, were the most effective phospholipids to restore functionality of a nearly inactive DDM-solubilized enzyme complex.
In Vivo Modification of the Phospholipid Composition of the Membrane Affects Quinol Oxidase Activity.
To further corroborate the importance of anionic phospholipids on quinol oxidase activity of the NarGHI complex, the influence of the phospholipid composition of the E. coli membrane was assessed in vivo. Basically, NarGHI was overproduced in both a wild-type strain and in its pssA variant. For the latter, the resulting lack of PE is accompanied by a strong increase in anionic phospholipids, which can represent up to 95% of the total phospholipid content (30, 31). Although the benzyl viologen activity of both NarGHI-enriched membranes was identical (70 vs. 76 units/mg of NarGHI), we observed a 2.5-fold increase of the quinol oxidase activity in the pssA mutant strain having a higher level of anionic phospholipids (12 vs. 5 units/mg of NarGHI). Thus, the quinol oxidase activity of the NarGHI complex is sensitive to the lipid composition of the inner membrane.
Tight Binding of Cardiolipin Occurs at a Specific Binding Site at the Interface of the Three Subunits.
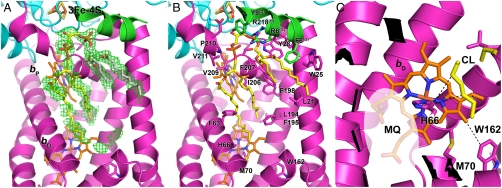
X-ray crystal structure of E. coli NarGHI was described at a resolution of 1.9 Å (23). Although not fully attributed, a large region of extended electron density is clearly visible around both hemes bD and bP [Protein Data Bank (PDB) ID code 1Q16]. A fraction of this density was attributed to a single molecule of PG (Fig. 2A). Noteworthy is the fact that this electron density feature is observed in all the electron density maps associated with the NarGHI mutants (PDB ID codes 1Y4Z, 1Y5I, 1Y5L, IY5N, 3EGW, 3IR5, 3IR6, and 3IR7). Based on the deposited X-ray data and on our knowledge that NarGHI copurifies with CL molecules, we reinterpreted this extended electron density map and assigned it to a CL (Fig. 2A). Interestingly, in the model so obtained, the stabilizing ligands of the CL phosphodiester headgroups are NarG-R6, NarH-R218, and NarI-Y28, while a large number of nonpolar interactions between the hydrophobic lipid tails and the NarI subunit are made. A high conservation of the hydrophobic character of the residues present in the cavity housing the acyl chains of the CL is observed in the NarI family PFAM PF02665. One of the acyl chains of CL is located approximately 4 Å away from heme bD and its ligand His66. Among the residues present in the immediate surrounding of this acyl chain, only the NarI-Trp162 residue is strictly conserved (Fig. 2 B and C). Although substitution of this residue into Ala had no measurable effect on NarGHI activity in IMVs, purification of the NarGHI-W162A complex yielded a stable and mature enzyme with a nearly twofold reduced quinol oxidase activity (Table 1). TLC analysis showed that this pronounced effect was due to a lower affinity of the variant enzyme for CL (Table 1).
Fig. 2.
Cardiolipin binding cavity within the NarI subunit of the E. coli nitrate reductase. (A) Electron density map (Fo-Fc contoured at 1.5σ) around a hydrophobic cavity of the NarI subunit and modelling of a CL molecule. The carbon atoms of the PG moiety of CL, as it was assigned by Bertero et al. (23), are colored in white. The ribbon is colored according to the subunit content with magenta (NarI), blue (NarH), and green (NarG). (B) View of the interaction network of the CL (yellow) molecule with NarGHI residues (all the side chains of residues located around 4 Å of CL as well as the conserved Trp162 residue are depicted in stick, and residue name and number is indicated for the NarI subunit; the subunit name is indicated in superscript otherwise). (C) Close-up view of the heme bD showing the van der Waals contact between NarI His66 Cδ and the CL molecule. It may help to maintain the Nδ1 in the right position for quinol substrate (MQ, menaquinol) binding (indicated as a translucent area).
Lipid Binding Is Necessary for Quinol Substrate Fixation at the QD Site.
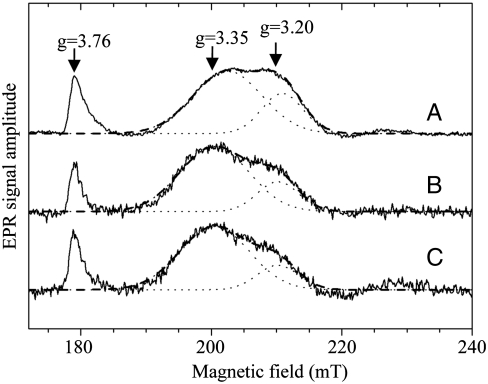
The location of a CL molecule immersed in a large cavity exposing both hemes and the loss of quinol oxidase activity associated to the disturbance of CL binding lead to question the structure–function relationships between these two observations. Interestingly, examination of the EPR signals of both hemes revealed a significant perturbation of the heme bD signature upon delipidation of the enzyme complex (Fig. 3). Indeed, whereas the signal associated to heme bD appears heterogeneous in NarGHI-enriched IMVs (gz values are approximately 3.35 and approximately 3.20), progressive delipidation of the enzyme leads to a significant decrease of the high-field feature with a gz ∼ 3.20 and a concomitant increase of the gz ∼ 3.35 component as confirmed by quantitative analysis of the EPR spectra (SI Text). Overall, our results indicate that loss of lipids associated with the NarGHI complex is responsible for structural perturbations of the heme bD center.
Fig. 3.
X-band EPR spectra of b-type hemes in NarGHI-enriched IMVs and NarGHI-DDM isolated. NarGHI in IMVs (A), NarGHI solubilized with 0.03% DDM (B), and NarGHI solubilized with 0.15% DDM (C). NarGHI concentration: (A) 82 μM, (B) 93 μM, and (C) 80 μM. IMVs and purified NarGHI samples were poised at +238 mV (A), +213 mV (B), and +195 mV (C) as described in Materials and Methods. Proximal heme (bP) gz value is approximately 3.76 and distal heme (bD) gz values are approximately 3.35 and approximately 3.20. EPR spectra were recorded under the following conditions: temperature, 12.5 K; microwave power, 20 mW at 9.4 GHz; modulation amplitude, 1 mT; modulation frequency, 100 kHz; and number of accumulations, 9. A polynomial baseline correction was applied to the experimental spectrum. The heme bD EPR signal was resolved into two components with Gaussian line shapes as described in SI Text. They are depicted as dotted lines together with their sum (dashed lines). The ratio of the area A of the two components Ag∼3.35/Ag∼3.20 is 2.3 (A), 3.6 (B), and 4.3 (C).
To address the origin of the loss of quinol oxidase activity, substrate binding was first assayed using fluorescence quenching titration (FQT) of a substrate analogue, HOQNO. HOQNO binds to the NarGHI complex present in IMVs with an affinity of approximately 0.25 μM, which drops down 14-fold to a value of approximately 3.4 μM after solubilization with 0.03% DDM (Table 3). Further delipidation using 0.15% DDM completely prevents HOQNO binding. Therefore, delipidation consecutive to purification of the enzyme complex must be associated to structural perturbation of the QD site in the vicinity of heme bD. To further substantiate this observation, we took advantage of the ability of the NarGHI complex to highly stabilize a semiquinone intermediate at the QD site (25–28). Interestingly, progressive delipidation using 0.03 and 0.15% DDM is associated to reduced amount of semiquinone bound in the enzyme complex (Table 3). These data demonstrate that lipids are necessary for quinol binding to NarGHI and generation of the SQ intermediate at the QD site.
Table 3.
HOQNO binding determined by FQ titration and EPR analysis of semiquinone (SQ) associated with NarGHI-enriched IMVs or DDM-isolated enzyme
| HQNO | SQ | ||
| Kd, μM | Em | SQ/FS4 | |
| Membrane | 0.25 ± 0.04 | −95 ± 10 | 10% |
| DDM 0.03% | 3.4 ± 0.1 | −90 ± 20 | 4% |
| DDM 0.15% | NQD | −90 ± 20 | 2% |
Em is the redox potential of the two-electron Q/QH2 couple at which the menasemiquinone concentration is maximal. Quantitation of the semiquinone content was achieved by double-integrating its EPR signal recorded under nonsaturating conditions (temperature 60 K; microwave power 0.1 mW) and is given as a percentage relative to the FS4 content determined using the same procedure in a sample fully oxidized by potassium ferricyanide (10 mM). NQD, no quench detected.
Discussion
Evidence is accumulating that specific lipids play important roles in membrane proteins, but how specifically lipids interact with and enable membrane proteins to achieve their full functionality remains unclear. X-ray structures of purified membrane proteins have revealed tight and specific binding of lipids. General features of lipid binding have been deduced. However, only in a few has the causal link between lipid binding and enzyme activity been understood. For instance, CL has been found associated to a number of bacterial and eukaryotic respiratory complexes, even though the functions associated to lipid binding often differ considerably. Our work has now identified a tight and specific interaction between CL and the nitrate reductase A (NarGHI), a bacterial respiratory complex. Delipidation of the complex leads to a loss of catalytic activity, whereas CL is the most effective phospholipid to restore functionality of a nearly inactive detergent-solubilized enzyme complex. More importantly, our study describes at a molecular level the activation of a bacterial respiratory complex by CL.
We have provided evidence that CL binding tunes the interaction with the quinol substrate and the generation of the semiquinone catalytic intermediate at the QD site. One tentative model that is consistent with our experimental observations is that CL binding via one of its acyl chains promotes a correct positioning of the heme bD and of its ligand His66 involved in the interaction with the semiquinone intermediate (Fig. 2C; CL is in van der Waals contact with the Cδ of His66 and heme bD). It is worth mentioning that such particular positioning of the acyl chain is likely due to unsaturation of the fatty acid chain as can be deduced from the observed kink in the modeled CL. Unsaturation of the acyl chains of lipids is known to dramatically affect their structure and reactivity and thus has a direct bearing on biological function (32–36). The close proximity of one of the acyl chains of CL with heme bD and of NarI-His66 ligand, together with the modification in the EPR line shape of heme bD elicited by this lipid molecule, are highly suggestive of structural modifications at the heme center. A similar situation has recently been observed in the cytochrome bc1 from yeast where the presence of lipids has a strong influence on the spectral properties of the complex (16, 17). Given the absence of a X-ray structure of NarGHI with bound quinone, EPR studies allowed us to identify and characterize semiquinone intermediates of both natural substrates, ubiquinol and menaquinol (25–28). Importantly, in both cases, the semiquinone species interact at the QD site via a H-bond to the His66 residue. The gz parameter of b-type cytochromes with histidine axial ligands is known to be highly sensitive to their environment and in particular to the distortion at the heme center controlled by the orientation of the imidazole rings (37, 38). In support of this, we observed a decrease of heterogeneity of the heme bD EPR signature upon delipidation of the enzyme complex. Our work now provides a unique facet to the function played by these heme-ligand His residues by controlling interaction with the substrate through contacts with lipid molecules.
The NarGHI heterotrimer is likely stabilized by the ordered CL molecule that is bound in a pocket lined with residues coming from all three subunits and stabilized through multiple electrostatic and hydrophobic interactions on the cytoplasmic side of the inner membrane. The CL is mainly stabilized by residues NarG-R6, NarH-R218, and NarI-Y28 for the phosphodiester headgroups and by nonpolar aromatic residues for the lipid tails of NarI subunit. Tightly bound CL has been identified in crystals of mitochondrial bc1 complex (12, 13), cytochrome c oxidase (39), ADP-ATP carrier (40), as well as in crystallized prokaryotic proteins, such as the photoreaction center (41–43), the formate dehydrogenase N (20), and in succinate dehydrogenase (19). In this regard, it is important to notice the ubiquitous nature of CL–protein interactions and the general trend here followed in the case of the nitrate reductase where CL binds preferentially at monomer interfaces of oligomeric assemblies and at subunit interfaces of multisubunit complexes.
During our study, it appeared that a nearly inactive but stable NarGHI complex was obtained after purification with large excess of DDM. The resulting enzyme is still dimeric as judged by size-exclusion chromatography and binds a single CL per NarGHI complex. Binding of this CL, although not involved in nitrate reductase functioning, may be responsible for the dimeric organization of the complex. Careful examination of all available NarGHI X-ray data has revealed the presence of an additional electron density partly attributed to a phosphatidic acid molecule at the interface between the symmetrically related NarI subunits. This lipid molecule may correspond to the stoichiometric CL found associated with the dimeric and inactive solubilized NarGHI complex.
Reversible binding of CL may provide a convenient way to finely adjust the activity of this respiratory complex depending on the metabolic demand. Because quinol oxidase activity is shown here to be strongly dependent on the presence of bound CL, any process that modulates the concentration of this lipid should greatly affect the extent of nitrate reductase functioning. In support of this assumption, we observed that chromosomal expression of the NarGHI complex yields a threefold increase in quinol oxidase activity compared to the overexpressed situation under identical growth conditions (Table S2). The nearly unchanged CL content of the membrane cannot overcome the overexpression of a fully mature and stable NarGHI complex and allows functionality of only a fraction of it. Such heterogeneity is directly observed in NarGHI-enriched IMVs on the EPR signal associated to heme bD center. Moreover, using a PE-deficient mutant strain enriched in anionic phospholipids, we observed that the NarGHI complex displays a 2.5-fold increase in quinol oxidase activity. Interestingly, bacteria contain variable amounts of CL depending on their physiological state. Although CL is only a trace component during exponential growth, its amount has been shown to increase in the stationary phase (44), in response to energy deprivation (45) or to osmotic stress (46). In E. coli, the expression level of cardiolipin synthase (cls gene) is also influenced by respiratory conditions, the highest level being obtained under anaerobiosis (47). Furthermore, cls expression depends upon the nature of the terminal electron acceptor. An additional interesting observation is that CL localizes to the polar and septal regions of the cytoplasmic membrane (29, 48). This localization of CL may help to maintain the proper spatial segregation of proteins, including the osmosensory transporter that was also shown to localize to the poles (29). Thus, we hypothesize that varying the amount of CL in the E. coli cells temporally and spatially provides an elegant, mechanistically simple way of turning on or off the activity of respiratory complexes depending on the energy demand. Other anionic phospholipids such as PG may also work as seen in our studies but at a much reduced level. Finally, as mentioned above, unsaturation of the fatty acid chains of CL is also an important parameter to consider in modulating the activity of respiratory complexes (36, 49).
Concluding Remarks
Our results provide a comprehensive description of an active role for CL in quinol substrate binding to a respiratory complex. This special dependency may also hold true in other CL-associated respiratory complexes described so far in energy-conserving membranes, e.g,. in mitochondria.
Materials and Methods
Bacterial Strains and Plasmids.
The E. coli strains and plasmids are described in Table S3. E. coli nar-deficient strain JCB4023 was used for overexpression of NarGHI or NarGHIW162 mutant from the plasmid pVA700. Utilization of the PE-deficient strain (AD93) is described in SI Text. Plasmid pRAC was constructed by cloning a narJI (SacII/SalI) fragment from a pVA700 plasmid into pGEM-T (Promega). Site-directed mutagenesis was performed as described previously (50) using pRAC plasmid as template and primers listed in Table S4, with mutation verified by sequence analysis. Subsequently, the mutated SacII/SalI fragment was cloned into pVA700.
Overexpression and NarGHI Purification.
NarGHI expressing cells were grown in terrific broth under semianaerobic conditions at 37 °C, and NarGHI-enriched IMVs were isolated in 100 mM MOPS, 5 mM EDTA at pH 7.5 as described before (50). NarGHI was purified as described in SI Text.
Lipids Extraction and TLC Analysis.
Lipids were extracted from purified NarGHI using a modified Bligh and Dyer extraction method (51), analyzed by TLC, and are described in SI Text.
Enzyme Activity and Protein Quantification.
Nitrate reductase activity was measured with standard assays using reduced benzyl viologen or menadiol as electron donors (52, 53). The NarGHI protein concentration in IMVs or solubilized preparations was estimated by the method of Lowry or using rocket immunoelectrophoresis as described in ref. 50.
Gel Filtration Chromatography.
Samples were analyzed in a Sephacryl HR 300 column (322 mL in HK 75/225, GE Healthcare) at a flow rate of 1 mL min-1 at 4 °C. To assess molecular weights, a calibration curve was calculated based on separation of standard proteins (molecular weight markers 29–669 kDa, Sigma Aldrich). The running buffer contained 20 mM Tris-HCl pH 7.5, 0.03% DDM, 100 mM NaCl, 5 mM EDTA, 5% (vol/vol) glycerol.
Proteoliposomes Preparation.
Lipid total extract, lipid polar extract, and pure phospholipids from E. coli were purchased from Avanti Lipids. First, liposomes were elaborated by mixing 10 mg of lipids with 17 μg of menadione (Sigma Aldrich) in chloroform. This mix was dried with argon and dispersed in 1 mL of reconstitution buffer (20 mM Mes, pH 6.5, 2 mM MgSO4). Three freeze/thawing cycles with liquid nitrogen were performed, and sonication was applied for 15 s with 45-s interval until turbidity vanished completely (pure PE was not sonicated). Vesicules were extruded with Millex HV (Millipore) 0.45 μM filters and incubated with purified NarGHI in a ratio of 85 μg of protein per milligram of liposome in presence of 0.14% (vol/vol) of Thesit (Sigma Aldrich) in a total volume of 1 mL. Suspension was gently agitated, and detergent was removed with 240 mg of SM2 Bio-Beads (Biorad Laboratories). Samples were centrifuged at 100,000 × g for 30 min at 4 °C. The pellet was finally resuspended in 400 μL of reconstitution buffer. In control experiments, the purified NarGHI was treated identically with the exception of liposomes. No effect on the stability and activity of the complex has been noticed.
Fitting of Cardiolipin into the NarGHI Complex.
Coordinates and structure factors of the NarGHI complex were obtained from the Protein Data Bank in Europe (PDBe) server (http://www.ebi.ac.uk/pdbe/; PDB code 1Q16). The omit map was generated using Refmac5 (54) from the CCP4 suite and by omitting the PG, phosphatidic acid, and all the cofactors from the map calculation. A cardiolipin molecule was then fitted into this omit map using Coot (55) and the real space refinement option (Fig. 2). The Coot’s stereochemical parameters (bonds, angles, planes, chiral, and nonbonded) were satisfying, and the electron density map allows us to model the fatty acid moieties of the CL up to 9, 5, 11, and 15 carbon atoms. The longer one extends in contact with heme bD while the remaining parts of the fatty acid moieties are disordered.
Redox Titration Experiments and EPR Spectroscopy.
NarGHI-enriched IMVs or purified NarGHI were studied by redox titrations performed as mentioned in 28. Redox potentials are given in the text with respect to the standard hydrogen electrode. EPR measurements were performed on a Bruker Elexsys E500 spectrometer. X-band EPR spectra were recorded using a standard rectangular Bruker cavity (ST 4102) fitted to an Oxford Instruments Helium flow cryostat (ESR900).
FQT with 2-n-Heptyl-4-hydroxyquinoline-N-oxide (HOQNO).
The affinity of NarGHI preparations for HOQNO was determined by performing FQT experiments as described in ref. 56. HOQNO concentrations were quantified by UV-visible absorption using ε350 = 8.80 mM-1 cm-1. Fluorescence was followed with a PerkinElmer Life Sciences LS-50B luminescence spectrometer or a Tecan Infinite 200. The observed fluorescence (Fobs) was fitted to an equation describing ligand binding to a single site as described in ref. 56.
Supplementary Material
Acknowledgments.
We thank Prof. D. Jahn and Dr. A. Walburger for critical reading, Drs. L.F. Wu and E. Bouveret for discussions, Drs. S. Canaan and R. Dhoubib for providing advice concerning TLC, and L. Sylvi for technical support. This work was supported by the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche, and the Université de Provence. R.A.C. was supported by a fellowship from the Agence Nationale de la Recherche.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010427108/-/DCSupplemental.
References
- 1.Dowhan W. Molecular basis for membrane phospholipid diversity: Why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 2.Hunte C, Richers S. Lipids and membrane protein structures. Curr Opin Struct Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Fry M, Green DE. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. J Biol Chem. 1981;256:1874–1880. [PubMed] [Google Scholar]
- 4.Drose S, Zwicker K, Brandt U. Full recovery of the NADH∶ubiquinone activity of complex I (NADH∶ubiquinone oxidoreductase) from Yarrowia lipolytica by the addition of phospholipids. Biochim Biophys Acta. 2002;1556:65–72. doi: 10.1016/s0005-2728(02)00307-9. [DOI] [PubMed] [Google Scholar]
- 5.Sharpley MS, Shannon RJ, Draghi F, Hirst J. Interactions between phospholipids and NADH∶ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry. 2006;45:241–248. doi: 10.1021/bi051809x. [DOI] [PubMed] [Google Scholar]
- 6.Schlame M, Ren M. The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim Biophys Acta. 2009;1788:2080–2083. doi: 10.1016/j.bbamem.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi AS, Zhou J, Gohil VM, Chen S, Greenberg ML. Cellular functions of cardiolipin in yeast. Biochim Biophys Acta. 2009;1793:212–218. doi: 10.1016/j.bbamcr.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mileykovskaya E, Zhang M, Dowhan W. Cardiolipin in energy transducing membranes. Biochemistry (Mosc) 2005;70:154–158. doi: 10.1007/s10541-005-0095-2. [DOI] [PubMed] [Google Scholar]
- 9.Booth PJ, Curran AR. Membrane protein folding. Curr Opin Struct Biol. 1999;9:115–121. doi: 10.1016/s0959-440x(99)80015-3. [DOI] [PubMed] [Google Scholar]
- 10.Dowhan W, Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu Rev Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schagger H, et al. Phospholipid specificity of bovine heart bc1 complex. Eur J Biochem. 1990;190:123–130. doi: 10.1111/j.1432-1033.1990.tb15554.x. [DOI] [PubMed] [Google Scholar]
- 12.Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 14.van Dalen A, de Kruijff B. The role of lipids in membrane insertion and translocation of bacterial proteins. Biochim Biophys Acta. 2004;1694:97–109. doi: 10.1016/j.bbamcr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the assembly of membrane proteins and organization of protein supercomplexes: Implications for lipid-linked disorders. Subcell Biochem. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wenz T, et al. Role of phospholipids in respiratory cytochrome bc(1) complex catalysis and supercomplex formation. Biochim Biophys Acta. 2009;1787:609–616. doi: 10.1016/j.bbabio.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Hielscher R, Wenz T, Hunte C, Hellwig P. Monitoring the redox and protonation dependent contributions of cardiolipin in electrochemically induced FTIR difference spectra of the cytochrome bc(1) complex from yeast. Biochim Biophys Acta. 2009;1787:617–625. doi: 10.1016/j.bbabio.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Gold VA, et al. The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci USA. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yankovskaya V, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 20.Jormakka M, Tornroth S, Byrne B, Iwata S. Molecular basis of proton motive force generation: Structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 21.Richardson D, Sawers G. Structural biology. PMF through the redox loop. Science. 2002;295:1842–1843. doi: 10.1126/science.1070366. [DOI] [PubMed] [Google Scholar]
- 22.Blasco F, et al. The coordination and function of the redox centres of the membrane-bound nitrate reductases. Cell Mol Life Sci. 2001;58:179–193. doi: 10.1007/PL00000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertero MG, et al. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol. 2003;10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 24.Lanciano P, et al. New method for the spin quantitation of [4Fe-4S](+) clusters with S = (3)/(2). Application to the FS0 center of the NarGHI nitrate reductase from Escherichia coli. J Phys Chem B. 2007;111:13632–13637. doi: 10.1021/jp075243t. [DOI] [PubMed] [Google Scholar]
- 25.Grimaldi S, et al. Direct evidence for nitrogen ligation to the high stability semiquinone intermediate in Escherichia coli nitrate reductase A. J Biol Chem. 2010;285:179–187. doi: 10.1074/jbc.M109.060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arias-Cartin R, et al. HYSCORE evidence that endogenous mena- and ubisemiquinone bind at the same Q site (Q(D)) of Escherichia coli nitrate reductase A. J Am Chem Soc. 2010;132:5942–5943. doi: 10.1021/ja1009234. [DOI] [PubMed] [Google Scholar]
- 27.Grimaldi S, Lanciano P, Bertrand P, Blasco F, Guigliarelli B. Evidence for an EPR-detectable semiquinone intermediate stabilized in the membrane-bound subunit NarI of nitrate reductase A (NarGHI) from Escherichia coli. Biochemistry. 2005;44:1300–1308. doi: 10.1021/bi048009r. [DOI] [PubMed] [Google Scholar]
- 28.Lanciano P, Magalon A, Bertrand P, Guigliarelli B, Grimaldi S. High-stability semiquinone intermediate in nitrate reductase A (NarGHI) from Escherichia coli is located in a quinol oxidation site close to heme bD. Biochemistry. 2007;46:5323–5329. doi: 10.1021/bi700074y. [DOI] [PubMed] [Google Scholar]
- 29.Romantsov T, et al. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol. 2007;64:1455–1465. doi: 10.1111/j.1365-2958.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- 30.Mikhaleva NI, Santini CL, Giordano G, Nesmeyanova MA, Wu LF. Requirement for phospholipids of the translocation of the trimethylamine N-oxide reductase through the Tat pathway in Escherichia coli. FEBS Lett. 1999;463:331–335. doi: 10.1016/s0014-5793(99)01661-0. [DOI] [PubMed] [Google Scholar]
- 31.DeChavigny A, Heacock PN, Dowhan W. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J Biol Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 32.Yung BY, Kornberg A. Membrane attachment activates dnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:7202–7205. doi: 10.1073/pnas.85.19.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mileykovskaya E, et al. Effects of phospholipid composition on MinD-membrane interactions in vitro and in vivo. J Biol Chem. 2003;278:22193–22198. doi: 10.1074/jbc.M302603200. [DOI] [PubMed] [Google Scholar]
- 34.Brandner K, et al. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: Implications for Barth Syndrome. Mol Biol Cell. 2005;16:5202–5214. doi: 10.1091/mbc.E05-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 36.van Gestel RA, et al. The influence of the acyl chain composition of cardiolipin on the stability of mitochondrial complexes; an unexpected effect of cardiolipin in alpha-ketoglutarate dehydrogenase and prohibitin complexes. J Proteomics. 2010;73:806–814. doi: 10.1016/j.jprot.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 37.More C, et al. EPR spectroscopy: A powerful technique for the structural and functional investigation of metalloproteins. Biospectroscopy. 1999;5:S3–18. doi: 10.1002/(SICI)1520-6343(1999)5:5+<S3::AID-BSPY2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 38.Berry EA, Walker FA. Bis-histidine-coordinated hemes in four-helix bundles: How the geometry of the bundle controls the axial imidazole plane orientations in transmembrane cytochromes of mitochondrial complexes II and III and related proteins. J Biol Inorg Chem. 2008;13:481–498. doi: 10.1007/s00775-008-0372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinzawa-Itoh K, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pebay-Peyroula E, et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 41.Wakeham MC, Sessions RB, Jones MR, Fyfe PK. Is there a conserved interaction between cardiolipin and the type II bacterial reaction center? Biophys J. 2001;80:1395–1405. doi: 10.1016/S0006-3495(01)76112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McAuley KE, et al. Structural details of an interaction between cardiolipin and an integral membrane protein. Proc Natl Acad Sci USA. 1999;96:14706–14711. doi: 10.1073/pnas.96.26.14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roszak AW, Gardiner AT, Isaacs NW, Cogdell RJ. Brominated lipids identify lipid binding sites on the surface of the reaction center from Rhodobacter sphaeroides. Biochemistry. 2007;46:2909–2916. doi: 10.1021/bi062154i. [DOI] [PubMed] [Google Scholar]
- 44.Short SA, White DC. Metabolism of phosphatidylglycerol, lysylphosphatidylglycerol, and cardiolipin of Staphylococcus aureus. J Bacteriol. 1971;108:219–226. doi: 10.1128/jb.108.1.219-226.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koch HU, Haas R, Fischer W. The role of lipoteichoic acid biosynthesis in membrane lipid metabolism of growing Staphylococcus aureus. Eur J Biochem. 1984;138:357–363. doi: 10.1111/j.1432-1033.1984.tb07923.x. [DOI] [PubMed] [Google Scholar]
- 46.Catucci L, Depalo N, Lattanzio VM, Agostiano A, Corcelli A. Neosynthesis of cardiolipin in Rhodobacter sphaeroides under osmotic stress. Biochemistry. 2004;43:15066–15072. doi: 10.1021/bi048802k. [DOI] [PubMed] [Google Scholar]
- 47.Heber S, Tropp BE. Genetic regulation of cardiolipin synthase in Escherichia coli. Biochim Biophys Acta. 1991;1129:1–12. doi: 10.1016/0167-4781(91)90206-2. [DOI] [PubMed] [Google Scholar]
- 48.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cronan JE., Jr Phospholipid modifications in bacteria. Curr Opin Microbiol. 2002;5:202–205. doi: 10.1016/s1369-5274(02)00297-7. [DOI] [PubMed] [Google Scholar]
- 50.Lanciano P, Vergnes A, Grimaldi S, Guigliarelli B, Magalon A. Biogenesis of a respiratory complex is orchestrated by a single accessory protein. J Biol Chem. 2007;282:17468–17474. doi: 10.1074/jbc.M700994200. [DOI] [PubMed] [Google Scholar]
- 51.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 52.Giordani R, Buc J, Cornish-Bowden A, Cardenas ML. Kinetic studies of membrane-bound nitrate reductase A from Escherichia coli with menadiol and duroquinol, analogues of physiological electron donors. Eur J Biochem. 1997;250:567–577. doi: 10.1111/j.1432-1033.1997.0567a.x. [DOI] [PubMed] [Google Scholar]
- 53.Jones RW, Garland PB. Sites and specificity of the reaction of bipyridylium compounds with anaerobic respiratory enzymes of Escherichia coli. Effects of permeability barriers imposed by the cytoplasmic membrane. Biochem J. 1977;164:199–211. doi: 10.1042/bj1640199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Bertero MG, et al. Structural and biochemical characterization of a quinol binding site of Escherichia coli nitrate reductase A. J Biol Chem. 2005;280:14836–14843. doi: 10.1074/jbc.M410457200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.