Abstract
The simple yet powerful technique of induced pluripotency may eventually supply a wide range of differentiated cells for cell therapy and drug development. However, making the appropriate cells via induced pluripotent stem cells (iPSCs) requires reprogramming of somatic cells and subsequent redifferentiation. Given how arduous and lengthy this process can be, we sought to determine whether it might be possible to convert somatic cells into lineage-specific stem/progenitor cells of another germ layer in one step, bypassing the intermediate pluripotent stage. Here we show that transient induction of the four reprogramming factors (Oct4, Sox2, Klf4, and c-Myc) can efficiently transdifferentiate fibroblasts into functional neural stem/progenitor cells (NPCs) with appropriate signaling inputs. Compared with induced neurons (or iN cells, which are directly converted from fibroblasts), transdifferentiated NPCs have the distinct advantage of being expandable in vitro and retaining the ability to give rise to multiple neuronal subtypes and glial cells. Our results provide a unique paradigm for iPSC-factor–based reprogramming by demonstrating that it can be readily modified to serve as a general platform for transdifferentiation.
Although successful transdifferentiation from one cell type to another by overexpressing lineage-specific genes in vivo (1, 2) and in vitro (3, 4) has been reported, until recently these methods were only effective for fate switching within the major lineages, i.e., ectoderm, mesoderm, and endoderm. However, the generation of iN cells (5) using neural-specific transcription factors has established that interlineage transdifferentiation is also possible in vitro. These transdifferentiation schemes entail overexpression of different sets of lineage-specific transcription factors. A more recent example reported single-factor transdifferentiation of fibroblasts into blood precursors using long-term ectopic expression of OCT4 (6); through extensive binding to the regulatory regions of key hematopoietic genes, OCT4 also appears to be participating in regulating hematopoietic programs acting as a lineage-specific transcription factor in this context. An important aspect of this study is the ability to generate a mitotically active progenitor population that can be further differentiated into a variety of blood cells—a critical feat that has yet to be accomplished in transdifferentiation to neural and endoderm lineages.
In an effort to devise a more general transdifferentiation strategy that might give rise to a broad array of unrelated cell types—including lineage-specific precursors—we attempted to direct conventional four iPSC-factor–based reprogramming (7, 8) toward alternative outcomes. Specifically, studies indicating that iPSCs are generated in a sequential and stochastic manner (9–11) led us to hypothesize that we might be able to manipulate cells at an early and epigenetically highly unstable state induced by the reprogramming factors. Different conditions could potentially give rise to a multitude of cell types (12) with more stable epigenetic profiles. In this context, induced pluripotency is only one—and perhaps among the less likely—of many possible outcomes. Indeed, studies have found partially or incompletely reprogrammed cells expressing multiple lineage-specific markers (7, 13–17), although these cells did not appear to represent physiologically relevant cell types. Accordingly, we hypothesized that it might be possible to deliberately bias the early reprogramming process toward a defined cell type by using inductive and/or permissive signaling conditions, after which the desired cells could be selected and/or expanded. On the basis of this same hypothesis and using a similar methodology, our group has recently shown that direct reprogramming into cardiomyocytes can be achieved (18).
In the present study, we have directly reprogrammed fibroblasts to functional neural stem/progenitor cells (NPCs) over an abbreviated period of four-factor induction. This direct reprogramming process is clearly distinct from conventional reprogramming to iPSCs or forward differentiation of pluripotent cells. Our findings not only represent a unique successful transdifferentiation of somatic cells into proliferating NPCs, but also form the basis of a methodology for interlineage transdifferentiation into multi- or oligopotent cells.
Results
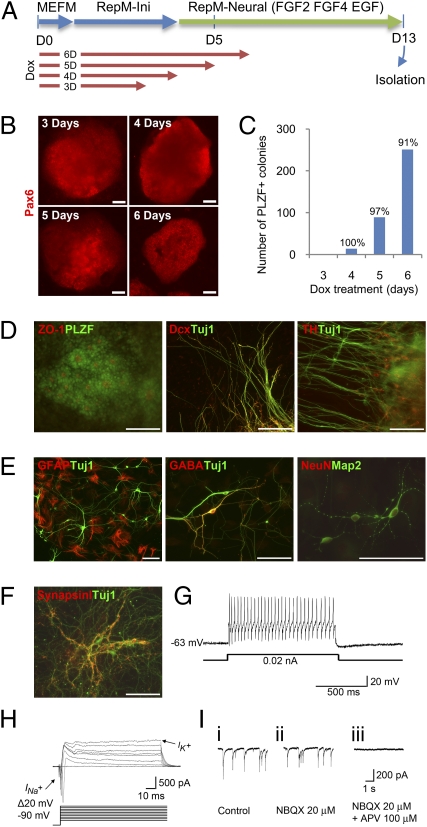
To rigorously test our hypothesis, we attempted an interlineage transdifferentiation from fibroblasts to NPCs using the doxycycline (dox)-inducible secondary mouse embryonic fibroblast (MEF) system (11, 19, 20). Inducible overexpression allows precise temporal control over the expression of the conventional iPSC-reprogramming factors, avoiding potentially detrimental effects arising from their constitutive overexpression. To ensure the survival of MEFs during the beginning of the reprogramming procedure, they were kept in MEF and reprogramming initiation medium (RepM-Ini; without leukemia inhibitory factor, LIF) for the first 3–6 d of dox treatment. Thereafter, neural reprogramming medium (RepM-neural) was applied to induce the generation and/or proliferation of nascent NPCs. RepM-neural contains FGF2, EGF, and FGF4 to support NPCs (21, 22). We tried dox treatments between 3 and 6 d to determine the optimal duration of reprogramming factor expression (Fig. 1A). We initially found that a minimum of 3 d of dox treatment was sufficient to obtain Pax6+ colonies after an additional 8–9 d in culture in RepM-neural (Fig. 1B). These colonies typically contained several hundred cells that nearly homogenously expressed promyelocytic leukemia zinc finger (PLZF), a rosette NSC marker (23), and Pax6, an early neural transcription factor (24) (Fig. 1 B–D). Extending dox treatment to 6 d increased the number of colonies (0.69 and 0.5% colony generation efficiency, n = 2). Surprisingly, we found that almost 100% of colonies showed neural transdifferentation regardless of the tested duration of dox treatment (95.9% ± 4.6 SEM, n = 3, Fig. 1C).
Fig. 1.
Transdifferentiation by transient expression of the conventional four reprogramming factors generates functional neural stem/progenitor cells. (A) Scheme for the transdifferentiation of dox-inducible secondary MEF cells into neural stem/progenitor cells (NPCs). Duration of dox (4 μg/mL) treatment is for the indicated number of days. Different media were added sequentially as described in Materials and Methods. (B) Pax6 immunostaining on day 13 of colonies arising from the indicated durations of dox treatment; 8 μg/mL dox was used for the 3-d treatment. (C) Number of PLZF-expressing colonies generated with different durations of dox treatment, as analyzed on day 13. Percentages of PLZF+ colonies over total colonies are shown. (D) Immunostaining of colonies on day 13 with various neural or neuronal markers. (E) Immunostaining of spontaneously differentiated cells from isolated colonies on day 13 with various mature neuronal or glial markers. Long-term differentiated neurons showed characteristic synapsin I expression patterns (F) and generated a full train of action potentials during injection of 20 pA current (whole-cell configuration in the current-clamp mode) (G) as well as fast Na+ currents and outward K+ currents (whole-cell configuration in the voltage-clamp mode) (H). Mature neurons also showed glutamate-mediated excitatory postsynaptic currents (EPSCs, I, i), indicating synapse formation between neurons. EPSC amplitude was partially blocked by the AMPA-type receptor antagonist NBQX (I, ii), and the remaining component was completely abrogated by the NMDA-type receptor antagonist APV (I, iii) (whole-cell configuration in the voltage-clamp mode). (Scale bars, 100 μm.)
We found typical neural rosette structures, PLZF expression, and luminal expression of ZO-1 (23) in the transdifferentiated colonies (Fig. 1D). In many colonies, we also found Tuj1+ neurons, some of which coexpress the early cortical neuronal marker Dcx, and even the dopaminergic neuronal marker tyrosine-hydroxylase (Fig. 1D). Importantly, these expression profiles are not observed in iN cells (5). Flow cytometry analysis of the transdifferentiated cells revealed that a population of cells expressing Prominin-1, PSA-NCAM, and A2B5 began to emerge after day 7 (Fig. S1 B and C). This population, in which various neural progenitors (25) appear to coexist, likely represents the colony-forming cells (subset 2 in Fig. S1A). Notably, this population did not contain a significant number of cells expressing SSEA-1, a pluripotent stem cell marker (Fig. S1B). The heterogeneity of the reprogrammed cells (Fig. 1D) probably reflects parallel paths of reprogramming, some of which result in a more direct transdifferentiation to mature neuronal fates—e.g., because mature neuronal cells coexist with transdifferentiated NPCs (Fig. 1D) and PSA-NCAM–expressing cells were generated as early as day 7 in the newly emerging population (Fig. S1C).
Following transdifferentiation, mitotically active neural colonies—which have typically been found to express Pax6 (23)—were isolated by manual picking and subcultured en bloc in conventional NPC medium containing FGF2 and EGF for expansion of NPCs (Fig. 1B). Each small cluster could form a colony within several days after isolation; however, despite mild enzymatic passaging and the avoidance of single cell dissociation, the NPCs appeared to lose their ability to form colonies within 3–5 passages. At this point, the NPCs were dissociated into single cells then cultured in N2 medium without any cytokines for further differentiation. After 1–2 wk of spontaneous differentiation, we observed NeuN and Map2-expressing mature neurons, GABAergic neurons, and GFAP-expressing astrocytes (Fig. 1E). The fully differentiated neurons that had formed by day 20 showed characteristic expression of synapsin I (Fig. 1F). To more rigorously characterize these apparently mature neurons, we examined their electrophysiological properties by patch clamp technique. The neurons showed evoked action potentials and fast Na+ currents (INa+) (n = 3 out of 4 cells recorded, Fig. 1 G and H). We also detected established synaptic connections and spontaneous excitatory postsynaptic currents (EPSCs, Fig. 1I), confirming that they are functional neurons capable of forming synapses. These results collectively suggest that the Pax6- and PLZF-expressing NPCs are functional and can be differentiated into mature neurons and glial cells. Compared with the circuitous process of reprogramming to iPSCs and subsequent redifferentiation to NPCs, our induced transdifferentiation method—requiring only a single step that is complete within 13 d and showing almost 100% newly generated colonies that are mostly composed of NPCs—is a highly efficient, direct, and rapid process for generating NPCs.
Importantly, the Nanog-GFP marker harbored by the secondary MEF cells was never expressed during transdifferentiation (Fig. S2). Indeed, its activation was found to require at least 9 d of dox treatment followed by a week of subsequent culture (19). Thus, we concluded that no fully reprogrammed iPSCs were generated within the time span of our experiment, and that the Pax6 and PLZF-expressing cells were directly reprogrammed from fibroblasts instead of redifferentiation from intermediate pluripotent cells. To better understand and characterize this transdifferentiation process, we performed the following additional analyses.
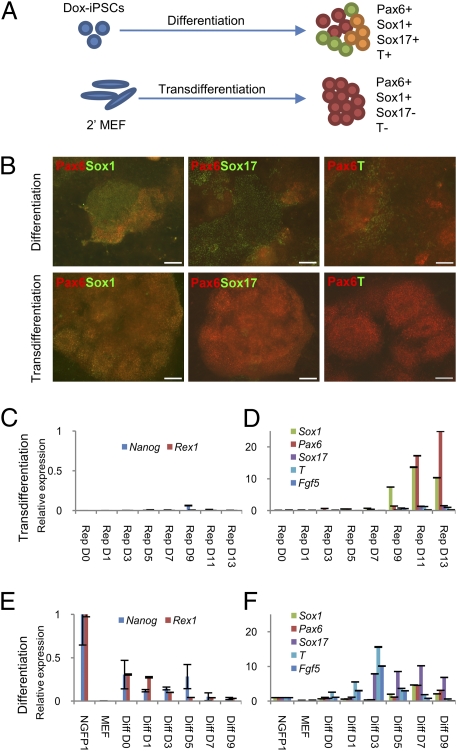
First, we sought to more definitively rule out the possibility that the generation of NPCs might first require the formation of transient pluripotent intermediates. To this end, we tested RepM-neural treatment directly on the iPSCs (NGFP1), which were used in blastocyst injections to make the secondary MEF cells, hypothesizing that we would get results similar to MEFs if the generation of NPCs relied on the generation of a small number of pluripotent cells early in the transdifferentiation process. However, we found that using iPSCs as a starting population resulted in a highly complex mixture of neuroectodermal (Sox1+ and Pax6+), endodermal (Sox17+), and mesodermal (T+) cells (Fig. 2 A and B). On the contrary, most colonies transdifferentiated from MEFs were almost entirely composed of cells expressing Sox1, the earliest neuroectodermal marker (21), and Pax6 (Fig. 2 A and B). RT-PCR analysis corroborated these results, as Sox1 and Pax6 expression was exclusive in transdifferentiation (Fig. 2D). As above, cells generated from iPSCs displayed a much more arbitrary profile of lineage-specific marker gene expression (Fig. 2 E and F). Although conventional neural differentiation does not rely on NPC-supporting cytokines, we could not do a comparison in their absence because no cell growth of transdifferentiated cells was ever observed without these cytokines. These results show that transdifferentiated NPCs arise directly from fibroblasts without any dependence on the generation of pluripotent intermediates.
Fig. 2.
Direct reprogramming is a highly efficient method of deriving pure neural progenitor cells. (A) Experimental overview and cartoon representation of results from panels B, D, and F. Cells after 9 d of differentiation from iPSCs, or after 13 d of transdifferentiation show the indicated marker expression profiles. (B) Day 9 immunostaining of cells differentiated from iPSCs, or transdifferentiated NPCs on day 13, with antibodies against the indicated markers. Pax6 and Sox1 demarcate early neuroectoderm. Sox17 is indicative of endoderm. T expression is early mesodermal. (Scale bars, 100 μm.) (C–F) qRT-PCR analysis of the indicated markers’ expression in cells harvested at multiple time points during the direct reprogramming process (C and D) and differentiation from iPSCs (E and F). Pluripotency genes (C and E) and lineage-specific genes (D and F) are shown in separate graphs. All values are relative to expression in iPSCs.
Second, we analyzed neural marker expression over time to pinpoint the onset of neural specification during transdifferentiation. As early as day 3, Pax6 expression was increased by 5.6-fold compared with day 0 (D0) MEFs. Sox1 expression started on day 5 and increased dramatically thereafter (Fig. 2D). Critically, during iPSC differentiation, Sox1 expression also begins on day 5 (Fig. 2F). This latter finding implies that if transdifferentiation relied on the generation of a pluripotent intermediate, this intermediate would have to arise at least 5 d before initial Sox1 expression. Because Sox1 expression commences on day 5 during transdifferentiation, this leaves no time for the reprogramming of MEFs to pluripotency. Further, even minute amounts of Nanog expression are not detected until much later in the transdifferentiation process (Fig. 2C). In short, Sox1+ NPCs cannot be arising from putative pluripotent intermediates generated during transdifferentiation.
We also investigated the possible generation of epiblast stem cells (EpiSCs) during the transdifferentiation because FGF2 supports self-renewal of EpiSCs, but could not detect any FGF5 expression (26) at any point in the process (Fig. 2D). In addition, Sox17 and Brachyury (T), which are not only lineage-specific markers but are also highly expressed in EpiSCs (27), were not detected either (Fig. 2D). Collectively, our results strongly support the notion that transdifferentiated NPCs are derived from nonpluripotent intermediate cells arising during the early phase of the process.
Third, we analyzed changes in histone modification marks in the Sox1 and Oct4 promoters by chromatin immunoprecipitation (Fig. 3). It was found that Sox1 promoter was characterized by the bivalent chromatin domains enriched in both trimethyl histone H3 Lys-4 (H3-K4me3) and trimethyl histone H3 Lys-27 (H3-K27me3) (28). Our analysis showed that the Sox1 promoter region underwent a burst of activating H3K4 trimethylation on day 8, the level of which reached that of control adult NPCs by day 12. Repressive H3K27me3 marks showed a concomitant down-regulation from day 4 onward. Slightly higher amounts of H3K27me3 than adult NPCs on day 12 may be the result of some unreprogrammed cells from the whole culture harvest. Importantly, the Oct4 promoter was persistently marked by H3K27me3 and showed a level of H3K4me3 similar to adult NPCs. These results indicate that epigenetic commitment to the formation of NPCs starts as early as day 8 in our direct reprogramming, without a parallel commitment to pluripotency.
Fig. 3.
Histone modification during direct reprogramming. Sox1 and Oct4 promoter loci were analyzed by chromatin immunoprecipitation with antibodies against the epigenetic marks of H3K4me3 and H3K27me3 during direct reprogramming. Samples were obtained on day 0 (D0), day 4 (D4), day 8 (D8), and day 12 (D12). Adult NPCs (aNPC) were used as a control. Blue and red bars indicate the extent of immuoprecipitation from normal IgG and each of the antimodified histone antibodies, respectively. Data are percentage of input chromatin amount, analyzed by qPCR (mean ± SEM, n = 3).
Fourth, we surmised that the fate choice between NPCs and iPSCs is determined by exposure to signaling specific to each cell type. To test this hypothesis, we examined the effects of using LIF-containing medium (Fig. 4A). Strikingly, even brief exposure (as little as 1 d) to LIF clearly decreased the number of PLZF-expressing colonies on day 9 (Fig. 4B). Conversely, flow cytometric analysis showed that LIF exposure increases the number of SSEA1-expressing cells (Fig. S3 B and C). Gene expression analysis also showed down-regulation of Sox1 and Pax6 with concomitant up-regulation of Nanog and Rex1 under the same conditions (Fig. 4 C and D). The correlated expression between SSEA1 and Nanog as well as Rex1 implies an overall increase in pluripotent character by extended LIF treatment. Accordingly, we observed that small clusters of Nanog-GFP expressing cells were generated when we used RepM-Pluri (essentially ESC maintenance medium) throughout, instead of RepM-neural. Interestingly, Sox17, T, and FGF5 expression on days 9, 11, and 13 was also increased by LIF exposure (Fig. S4B). This increase in expression of genes specific to other lineages might result from the spontaneous redifferentiation of Nanog-GFP–expressing cells generated by LIF exposure or perhaps from an induction of EpiSC-like cells. These results imply that intermediate cells may embark on the path toward becoming iPSCs or NPCs by responding to LIF or NPC-supporting medium, respectively. This cytokine-dependent cell-type specification during direct reprogramming has also been shown in other recent studies (6, 29). Thus, it is clear that the choice between conversion to NPCs and complete reprogramming to iPSCs could be influenced at a very early point during the transdifferentiation process.
Fig. 4.
Fate choice is dictated early during the transdifferentiation process by different environmental cues. (A) Schematic of experiments involving differential exposure to LIF- containing medium (RepM-Pluri) from day 4 onward for the indicated number of days. Dox treatment was for 5 d, beginning on D0. (B) PLZF+ colony number expressed as a percentage of the total from each indicated sample on day 9. (C and D) Quantitative analysis of mRNA levels of indicated marker genes in each sample (harvested on day 9). All values are relative to expression in iPSCs. Pax6 immunostaining (E) and total colony number (F) of TTFs transdifferentiated in the presence or absence of JAK inhibitor. Immunostaining of reprogrammed NPCs from TTFs; neural rosette formation (G) and their long-term differentiation into various neuronal subtypes (H–L). (Scale bars, 100 μm.)
Finally, we did not observe any expression of pluripotency (Oct4), neural (Sox1 and Pax6), or neuronal (Tuj1) markers in the starting cells or in the cells cultured in RepM-neural media without dox induction (Fig. S5), strongly suggesting an absence of contaminating neural cells in the starting MEF population. Nonetheless, to rigorously rule out any contribution from contaminating neural crest cells and mesenchymal stem cells that might exist in rather heterogeneous MEF populations (30), we reprogrammed adult tail-tip fibroblasts (TTFs) using the dox-inducible STEMCCA system (31) following the same method described above. We successfully transdifferentiated TTFs into Pax6-expressing NPCs in 13 d (Fig. 4E) with an efficiency of 0.07% (±0.006%, n = 2) using just 6 d of induction—shorter than the 8 d required for the generation of pluripotent cells from TTF with this system (31). These reprogrammed NPCs from adult TTFs showed the same characteristics as reprogrammed cells obtained from secondary MEFs: both populations expressed neural rosette markers (PLZF and ZO-1, Fig. 4G) and were able to differentiate into Dcx-, TH-, GABA-, Map2-, NeuN-, and synapsin I-expressing neurons (Fig. 4 H–L). These results confirm that exogenous genes delivered lentivirally were able to successfully reprogram adult TTFs. In parallel with four-factor reprogramming, we also tried three factors (i.e., omitting c-Myc). However, otherwise identical conditions failed to produce any neural cells in the latter case.
Because short-term LIF treatment seems to increase the pluripotent character of the reprogrammed cells—although levels of Nanog and Rex1 expression are still quite low compared with iPSCs (Fig. 4D)—a small number of LIF-responsive cells might exist in our cultures. To strictly eliminate the possibility of these cells, we inhibited Janus kinase (JAK)/Stat3 signaling, which is directly involved in pluripotent reprogramming (32) by treating cultures with a small-molecule inhibitor of JAK. Although this inhibition decreased the number of colonies by around 30% (Fig. 4F), almost all remaining colonies homogeneously expressed Pax6 (Fig. 4E). These results imply that our transdifferentiation takes a strictly independent path from reprogramming to pluripotency, which can be blocked by JAK inhibitor 1 treatment. When we analyzed the properties of neurons differentiated from the NPCs derived using JAK inhibitor treatment with the secondary MEF system, they showed very similar results to cells not treated with JAK inhibitor (Fig. S6). Thus, JAK inhibitor treatment does not adversely affect direct transdifferentiation into functional NPCs.
In summary, we have successfully transdifferentiated fibroblasts into functional and proliferating NPCs. Such four iPSC-factor–based transdifferentiation can be guided by modulating transgene expression time and the culture environment (such as specific cytokines) and will likely prove useful for transdifferentiation to other lineages as well.
Discussion
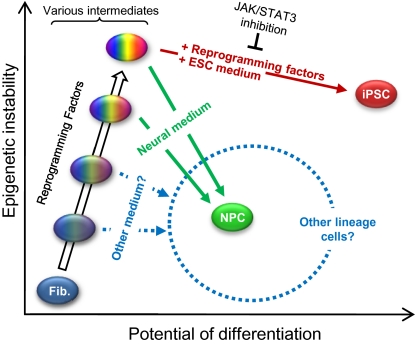
We have shown a direct cell type switch from fibroblasts to functional NPCs by transient expression of the four reprogramming factors, whereby the process is clearly distinct from and does not depend on the generation of iPSCs. This transdifferentiation process is highly specific and efficient, reaching completion within 9–13 d. Our method achieves an interlineage cell type change much like that of iN cells (5), with one critical advantage: the resulting cells are expandable progenitor cells. Another advantage of our method is the use of general reprogramming factors instead of lineage-specific transcription factors. Because the four Yamanaka factors were chosen for pluripotent cell generation (7), they are generally only considered useful for the derivation of iPSCs. However, our results suggest a different paradigm in which various developmentally plastic intermediate cells may be generated in the process and that iPSCs are perhaps only one of many possible outcomes of the four-factor reprogramming. The ultimate result may depend largely on extrinsic signaling inputs (Fig. 5). Interestingly, in the newt, Sox2, Klf4, and c-Myc are temporarily up-regulated after lens removal or limb amputation to initiate the regeneration process (33). This observation lends support to our hypothesis that the four conventional iPSC factors not only induce reprogramming to iPSCs, but may also be capable of mediating direct fate switching between differentiated cells. Indeed, we have also found that fibroblasts can be transdifferentiated into spontaneously contracting cardiac cells by temporary expression of the same four reprogramming factors under different culture conditions within 11 d (18). These results collectively imply a generally nonspecific/undirected reprogramming process induced by the four iPSC factors (i.e., not specifically directed toward the pluripotent state as it has been regarded) and suggest a unique strategy/paradigm to significantly expand and exploit iPSC-factor–based reprogramming. Changing the duration of transgene expression and culture conditions may allow a transient, plastic developmental state established early to effectively serve as a cellular platform for transdifferentiation toward various lineages.
Fig. 5.
Model for direct reprogramming of fibroblasts (Fib.) to neural stem/progenitor cells. By adding neural medium to four factor-induced intermediate cells comprising various epigenetic states, a fate switch to neural stem/progenitor cells (NPCs) can be achieved. Alternatively, iPSCs can be generated by prolonged expression of the four factors with concomitant incubation in ESC medium. Cells belonging to other lineages could likely also be isolated, depending on the type of medium used.
Although iN cells are functional neurons (5), they lack the potential to generate the diverse neuronal subtypes that can be derived from iPSCs. Furthermore, these postmitotic neurons may not be very suitable to the study of certain neurological diseases, due not only to their nonproliferative state (which severely limits their numbers), but also to their inability to recapitulate disease phenotypes occurring at the neural progenitor stage (34). Currently, iPSCs derived from patients with late-onset neurological diseases (e.g., those with Parkinson disease and amyotrophic lateral sclerosis) do not readily recapitulate disease phenotypes when redifferentiated (35, 36), although some patients’ iPSCs with genetic defects show their symptoms (37, 38). These findings imply that complete reprogramming to a pluripotent state may reset certain epigenetic hallmarks of the disease state, thereby necessitating long-term aging under conditions of stress for its repeated manifestation. Considering this negative correlation between the degree of reprogramming and the manifestation of a particular disease phenotype, we think that the transdifferentiated NPCs—which are derived with limited reprogramming, thus avoiding the establishment of a pluripotent state—could eventually prove to be a better-suited model system than iPSCs when studying such late-onset diseases.
For secondary MEF cells, the transdifferentiation process may at first glance seem inefficient compared with iPSC reprogramming using the same cells (19). However, considering the abbreviated induction period, the efficiency is reasonable, i.e., comparable to initial reprogramming as confirmed by retrospective analyses (39).
We observed no significant up-regulation of EpiSC markers such as Sox17, Brachyury (27), and FGF5 (26) during our direct reprogramming. Nonetheless, a recent study by the Schöler group showed direct reprogramming to EpiSCs can be achieved in 3–5 wk (29), which is substantially slower than our method. Thus, a temporary emergence of EpiSCs during our direct reprogramming—which can be achieved within 12 d—is highly unlikely. Interestingly, 5 d of induction followed by long-term LIF treatment caused increased expression of Sox17 and FGF5 with extremely low levels of Brachyury and Nanog (Fig. S4). These cells may partially resemble EpiSCs; however, the absence of LIF in our transdifferentiation media makes the generation of such cells nearly impossible. As expression of exogenous factors is tightly regulated in the inducible system (ref. 20 and Fig. S4A), to achieve fully reprogrammed EpiSCs or iPSCs status, a much longer induction of reprogramming factors may be required.
In a recent development, lentiviral expression of OCT4 alone was shown to mediate direct reprogramming of human fibroblasts to blood progenitors (6). Although OCT4 expression is down-regulated after long-term differentiation, long-term induction is required and there is always a risk of lentiviral reactivation of OCT4 expression, which may induce dysplastic lesions (40). Our study shows that temporary expression of four reprogramming factors is sufficient to induce lineage-specific transdifferentiation. If our transdifferentiation process can be replicated using nonviral and temporary expression methods such as transient transfection (41, 42), protein transduction (43, 44), mRNA transfection (45), or small molecules (46), it could form the basis of safe and convenient transdifferentiation across a broad lineage spectrum.
Materials and Methods
For direct reprogramming, secondary MEFs were sequentially cultured in MEF medium, reprogramming initiation medium, and neural reprogramming medium with 3–6 d of induction by doxycycline treatment beginning on day 0. Lentivirally transduced adult TTFs (using an inducible STEMMCCA system) were reprogrammed as secondary MEFs. The detailed methods, cells, and reagents concerning direct reprogramming with secondary MEFs and TTFs, as well as all analysis such as quantitative PCR, immunocytochemistry, flow cytometry, chromatin immunoprecipitation, and electrophysiology are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Wenlin Li, Per Stehmeier, and Tongxiang Lin for critical reading of the manuscript. We appreciate the assistance of Ella Kothari and Jun Zhao at the University of California at San Diego, La Jolla, CA Stem Cell Core in blastocyst injections and Chao Zhao in caring for mice. We thank Debbie Watry for flow cytometry and general assistance. The pHAGE2-TetOminiCMV-STEMCCA plasmid was a generous gift from Dr. Gustavo Mostoslavsky. S.D. and K.Z. are supported by the National Institutes of Health (NIH) Director's Transformative R01 Program (R01 EY021374). S.D. is supported by funding from the National Institute of Child Health and Human Development, National Heart, Lung, and Blood Institute; the National Institute of Mental Health/NIH; the California Institute for Regenerative Medicine; the Prostate Cancer Foundation, Fate Therapeutics; the Esther B. O'Keeffe Foundation; and The Scripps Research Institute. K.Z. is supported by grants from the Chinese National 985 Project to Sichuan University and West China Hospital, the National Eye Institute/NIH, Veterans Administration Merit Award, the Macula Vision Research Foundation, Research to Prevent Blindness, a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research, and the Dick and Carol Hertzberg Fund. S.A.L. and M.T. were supported in part by NIH Grants P01 HD29587, P01 ES016738, P30 NS057096, and R01 EY05477 and by the California Institute for Regenerative Medicine.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103113108/-/DCSupplemental.
References
- 1.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 4.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szabo E, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artyomov MN, Meissner A, Chakraborty AK. A model for genetic and epigenetic regulatory networks identifies rare pathways for transcription factor induced pluripotency. PLOS Comput Biol. 2010;6:e1000785. doi: 10.1371/journal.pcbi.1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 15.Silva J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Sridharan R, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Efe JA, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 19.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 22.Hitoshi S, et al. Primitive neural stem cells from the mammalian epiblast differentiate to definitive neural stem cells under the control of Notch signaling. Genes Dev. 2004;18:1806–1811. doi: 10.1101/gad.1208404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkabetz Y, et al. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 25.Alcock J, Sottile V. Dynamic distribution and stem cell characteristics of Sox1-expressing cells in the cerebellar cortex. Cell Res. 2009;19:1324–1333. doi: 10.1038/cr.2009.119. [DOI] [PubMed] [Google Scholar]
- 26.Greber B, et al. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 29.Han DW, et al. Direct reprogramming of fibroblasts into epiblast stem cells. Nat Cell Biol. 2011;13:66–71. doi: 10.1038/ncb2136. [DOI] [PubMed] [Google Scholar]
- 30.Weston JA, et al. Neural crest and the origin of ectomesenchyme: Neural fold heterogeneity suggests an alternative hypothesis. Dev Dyn. 2004;229:118–130. doi: 10.1002/dvdy.10478. [DOI] [PubMed] [Google Scholar]
- 31.Sommer CA, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, et al. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maki N, et al. Expression of stem cell pluripotency factors during regeneration in newts. Dev Dyn. 2009;238:1613–1616. doi: 10.1002/dvdy.21959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchetto MC, Winner B, Gage FH. Pluripotent stem cells in neurodegenerative and neurodevelopmental diseases. Hum Mol Genet. 2010;19:R71–R76. doi: 10.1093/hmg/ddq159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 37.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 42.Jia F, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu S, et al. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.