Abstract
In internally fertilizing organisms, mating involves a series of highly coordinated molecular interactions between the sexes that occur within the female reproductive tract. In species where females mate multiply, traits involved in postcopulatory interactions are expected to evolve rapidly, potentially leading to postmating-prezygotic (PMPZ) reproductive isolation between diverging populations. Here, we investigate the postmating transcriptional response of the lower reproductive tract of Drosophila mojavensis females following copulation with either conspecific or heterospecific (Drosophila arizonae) males at three time points postmating. Relatively few genes (15 total) were differentially regulated in the female lower reproductive tract in response to conspecific mating. Heterospecifically mated females exhibited significant perturbations in the expression of the majority of these genes, and also down-regulated transcription of a number of others, including several involved in mitochondrial function. These striking regulatory differences indicate failed postcopulatory molecular interactions between the sexes consistent with the strong PMPZ isolation observed for this cross. We also report the transfer of male accessory-gland protein (Acp) transcripts from males to females during copulation, a finding with potentially broad implications for understanding postcopulatory molecular interactions between the sexes.
Keywords: gene expression, reproduction, sexual selection, sexual conflict, speciation
In internally fertilizing organisms, the female reproductive tract serves as the arena for a series of highly coevolved molecular interactions between the sexes that are critical for successful reproduction (1, 2). Postcopulatory interactions should further increase in complexity in species in which females mate with more than one male, as intense sexual selection propels the rapid evolution of traits mediating female choice, male competitive ability, and sexual conflict (3, 4). This, in turn, may facilitate divergence of such traits between populations following different coevolutionary trajectories, leading to postmating-prezygotic (PMPZ) reproductive isolation (5). Consistent with these expectations are the rapid evolution of morphological and molecular reproductive traits associated with postcopulatory processes (6) and the recognition that PMPZ barriers can serve as potent and rapidly evolving forms of reproductive isolation (5).
The availability of genomic resources for an increasing number of species provides a platform for elucidating the molecular basis of postcopulatory molecular interactions between males and females. For example, recent genomic studies on Drosophila melanogaster (7–14), Anopheles gambiae (15), and Apis mellifera (16, 17) have begun to characterize the female postmating response by identifying changes in the transcriptome and/or proteome of mated females. In D. melanogaster, sperm or other specific components of the seminal fluid are known to induce some of these changes, which ultimately trigger physiological responses in females (18). Male accessory-gland proteins (Acps), in particular, modulate a variety of physiological processes in D. melanogaster females including immune response, oogenesis, oviposition, sperm transfer and storage, and female receptivity (18). Although considerable progress has been made in understanding the nature and scope of postcopulatory molecular interactions between males and females, comparable studies on additional species, especially those with different mating systems, are necessary to generalize these findings. Moreover, although accumulating evidence suggests that postcopulatory incompatibilities between the sexes often result in significant PMPZ reproductive isolation between species (5), the molecular and genetic bases of such incompatibilities have yet to be identified.
Drosophila mojavensis and Drosophila arizonae are recently diverged (<1 Mya) sister species (19) with partially overlapping distributions in the arid regions of southwestern United States and northwestern Mexico. The mating systems of these two species are characterized by frequent female remating relative to D. melanogaster (20), along with extensive intersexual coevolution of postcopulatory traits (21–23), including rapid evolution of both male and female reproductive proteins (24–28). Consistent with expectations, interspecific crosses also exhibit strong PMPZ isolation, particularly those involving D. mojavensis females. Heterospecifically mated D. mojavensis females exhibit perturbations in a number of processes occurring within the female reproductive tract that result in a high incidence of failed fertilizations, a reduced rate of oviposition, and ultimately the production of few hybrid offspring (25). These problems are associated with deficiencies in the heterospecifically mated female's storage and retention of sperm, and also in degrading the insemination reaction, a temporary mass that forms in the uterus immediately after conspecific copulation (25). In contrast to conspecific matings, where the mass is typically eliminated within several hours, following heterospecific matings the mass often persists for days, interfering with oviposition and in some cases even permanently sterilizing females (21, 25). Whereas premating and postzygotic isolating barriers between D. mojavensis and D. arizonae vary in strength depending on the source population of males and females (29–31), PMPZ isolation is strong in all crosses involving D. mojavensis females, suggesting that this barrier may have been among the earliest to evolve.
In the present study we sought to identify and compare the transcriptional changes that occur in female D. mojavensis reproductive tracts following conspecific and heterospecific matings at three postcopulatory time points. We first compared virgin and conspecifically mated females to identify genes involved in the normal female postmating response. We then compared this transcript set with the transcript differences observed in con- vs. heterospecific crosses, as genes found in both sets are candidates for involvement in PMPZ isolation. Our design thus allows us to investigate the genetic basis of PMPZ isolation in an internally fertilizing organism at its very early stages.
Results
We used D. mojavensis whole genome microarrays to compare patterns of transcript abundance in the lower reproductive tracts of virgin and mated females at 15 min, 2 h, and 6 h postcopulation. In addition to finding differences in the expression of lower reproductive tract genes in the three female treatments, we also found that a significant number of male-derived transcripts had been transferred at mating.
Few Transcripts Differ in Abundance Between Conspecifically Mated and Virgin Females.
Using the false discovery rate (FDR) to control for multiple testing (Q= 0.05), we identified 18 genes for which transcript levels differed between conspecifically mated and virgin females at one or more time points (Table 1). The number of genes in this set is small compared with the much larger set (160) that differed between heterospecifically and conspecifically mated females, reflecting a relative paucity of regulatory changes in the female reproductive tract following conspecific mating. The ratio of differentially regulated genes in the two comparisons is largely unaffected by the choice of FDR cutoff, as a more liberal cutoff (Q = 0.1) yields similar results (27 genes in the conspecific–virgin comparison; 246 genes in the conspecific–heterospecific comparison). We subsequently determined that 3 of the 18 transcripts in this set came from males (see below Males Transfer ACP mRNA Transcripts to Females During Mating). Twelve of the remaining 15 were definitively of female origin and thus represent genes differentially regulated in response to mating. We were unable to determine the sex of origin of the 3 remaining transcripts; in the absence of evidence to the contrary, we assume that they also represent genes that were differentially regulated in females.
Table 1.
Identity, predicted molecular function, and fold change in expression (relative to virgin females) of mating responsive transcripts in the lower reproductive tracts of D. mojavensis females at three time points postmating
| Fold change |
|||||
| D. mojavensis gene | D. melanogaster homolog | Molecular function | 15 min | 2 h | 6 h |
| GI10424 | None | Unknown | 2 | ||
| GI10632 | Npc2h* | Unknown | 19.9 | ||
| GI11600 | None | Unknown | 10 | ||
| GI14543 | Adam* | Translation initiation factor | −1.6 | ||
| GI14761 | ripped pocket* | Sodium ion transport | 82.3 | 64 | 25.8 |
| GI14846 | None | Unknown | 10.2 | ||
| GI14885 | CG6770 | Stress response | −2.4 | ||
| GI14996 | Thor* | Immune/stress response | −2.7 | ||
| GI15007 | Thor | Immune/stress response | −3.2 | ||
| GI16692 | CG13936 | Protein binding | −2.2 | ||
| GI17134 | CG14069 | Unknown | 10.7 | ||
| GI18586 | CG34193* | Unknown | 4.2 | ||
| GI20303 | CG30273* | Unknown | 78 | 66.8 | 33.7 |
| GI22307 | CG15515 | Protein binding | 5.9 | ||
| GI23227 | CG6972 | Protein binding | 1.6 | ||
| GI23324 | CG7685 | Hydrolase, glycosidase | 1.8 | ||
| GI23726 | Obp93A | Odorant binding | 11.3 | 6.3 | |
| GI23890 | Scpr-C* | Unknown | 5.7 | ||
*Homology determined by BLAST to the D. melanogaster genome (results in Table S1).
At 15 min postmating, only six genes showed differences in expression (excluding male-derived genes) between mated and virgin females, with transcript levels being significantly higher in mated females for all but one of the genes (Table 1). Although most of these genes have predicted D. melanogaster orthologs (some we identified by BLAST to the D. melanogaster genome; Table S1), analysis of gene ontology terms revealed that the molecular function is known for only two: a highly up-regulated gene that codes for an odorant binding protein (Obp 93A) and a down-regulated gene that is involved in translation initiation (Adam) (Table 1). At 2 h postmating, only two additional genes showed differences in expression between mated and virgin females, including one gene involved in protein binding (CG15515). The peak number of genes exhibiting differences in expression occurred at 6 h postmating and was associated with a shift from primarily up-regulation at earlier time points to more down-regulation at 6 h (Table 1). Most notably, three genes with predicted roles in immune/stress response (a pair of Thor homologs and CG6770) were down-regulated at this time.
Regulation of Mating Responsive Genes Is Perturbed in Heterospecific Crosses.
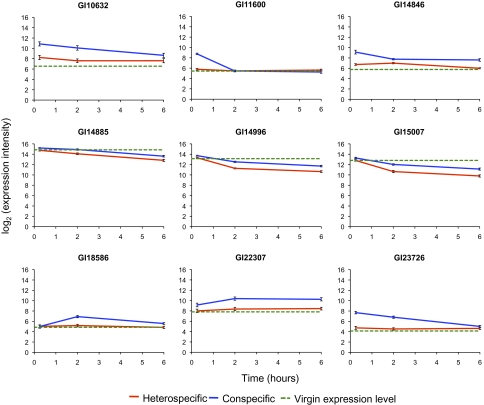
Striking differences in patterns of relative transcript abundance between conspecifically and heterospecificially mated females occurred. The majority (9/15) of mating-responsive transcripts identified from the virgin–conspecific comparison also differed between conspecifically and heterospecifically mated females, suggesting that the normal postmating transcriptional response was highly perturbed in heterospecifically mated females (Fig. 1). Moreover, transcriptional variation between the crosses was not limited to mating responsive genes identified from the conspecifically mated–virgin comparison, as heterospecifically mated transcript levels also differed for an additional 148 genes (Table S2). Analysis of gene ontology terms revealed that a number of terms associated with mitochondrial function were overrepresented in this gene set, with almost all being down-regulated in the heterospecific cross (Table S3). The contrast between the two sets of transcript differences is striking and consistent with the previous physical evidence of severe mismatch between heterospecific reproductive tracts.
Fig. 1.
Relative transcript abundance of mating-responsive genes that differed between con- and heterospecifically mated females. The line for virgins corresponds to the average of the two replicates.
Males Transfer Acp mRNA Transcripts to Females During Mating.
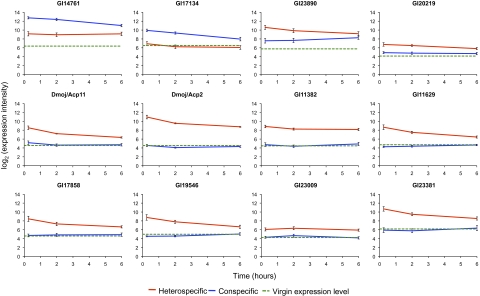
Because Acps are typically expressed only in male tissues, we were surprised that conspecifically and heterospecifically mated females showed large differences in relative transcript abundance for several previously identified Acps and/or male reproductive transcripts. Given that most transcripts contain fixed sequence differences between D. mojavensis and D. arizonae, we used a combination of RT-PCR and sequencing to confirm that at least 12 transcripts in the female reproductive tract were of male origin (Table 2), including 10 that were in higher abundance in the heterospecific vs. conspecific cross and 3 that were in higher abundance in the conspecific cross vs. virgins (one gene was in both sets) (Fig. 2). We verified these differences among conspecific, heterospecific, and virgin treatments for three of the genes (DmojAcp2, GI17858, and GI23890) using quantitative PCR (Table S4). We also independently verified the transfer of male transcripts by repeating the heterospecific mating experiments followed by RT-PCR and sequencing of 10 of the male-derived transcripts. We were able to confirm the transfer of 7 of these transcripts (Table 2). The number of male transcripts we identified likely underestimates the total number transferred due to difficulties identifying the origin of some transcripts, and the fact that we examined only a subset of all those that differed in abundance between the conspecific and heterospecific crosses.
Table 2.
Transcripts that differed between con- and heterospecifically mated females that were (i) candidate D. mojavensis/D. arizonae Acps (ii), genes from D. melanogaster male reproductive tracts, or (iii) proteins that were transferred in the ejaculate of D. melanogaster
| D. moj ID | Gene | Source of transcript | Confirm§ | D. moj Acp | D. mel male RT | Protein transferred in D. mel | D. mel ortholog | Conspecific mating effect | Reference |
| GI17858 | Male | No† | Yes | Yes | CG14034‖ | 45, 47 | |||
| GI20219 | Male | Yes | Yes | Yes | Yes | CG30488 | 45–47 | ||
| GI19546 | Male | No† | Yes | Yes | CG34002 | 45, 46 | |||
| GI20999 | Dmoj/Acp11 | Male | Yes | Yes | — | 27 | |||
| GI20988 | Dmoj/Acp2 | Male | Yes | Yes | — | 27, 45 | |||
| GI11629 | Adk3 | Male | Yes | Yes | CG6612 | 47 | |||
| GI23890 | ScprC | Male | Yes | Yes | CG5106 | Yes | 47 | ||
| GI14761 | rpk | Male | No¶ | CG1058‖ | Yes | ||||
| GI11382 | Male | Yes | — | ||||||
| GI17134 | Male | Yes | CG14069 | Yes | |||||
| GI23381 | Male | Yes | Yes | Yes | CG17097 | 45–47 | |||
| GI23009 | Male | Yes | Yes | CG10284‖ | 45, 47 | ||||
| GI16594 | Female | Yes | CG1318 | 45 | |||||
| GI22128 | Female | Yes | CG34215‖ | 45 | |||||
| GI13447 | Cam | Female | Yes | CG8472 | 47 | ||||
| GI10528 | Unknown† | Yes | Yes | Yes | CG14061 | 45–47 | |||
| GI18622 | Dmoj/Acp2b* | Unknown | Yes | — | 45 | ||||
| GI10529 | Unknown† | Yes | Yes | CG9997 | 45, 46 | ||||
| GI10530 | Unknown† | Yes | — | 45 | |||||
| GI13594 | Unknown‡ | Yes | CG18233‖ | 45 | |||||
| GI13596 | Unknown‡ | Yes | CG18233‖ | 45 | |||||
| GI20607 | Unknown‡ | Yes | CG4812 | 45 | |||||
| GI21941 | capt | Unknown‡ | Yes | CG33979 | 47 |
Transcripts were screened by RT-PCR and sequencing to determine whether they were of male or female origin.
*Previously unannotated D. mojavensis gene.
†Did not amplify.
‡Amplified, but problems sequencing.
§Independent replicate experiment.
¶Female origin in replicate experiment.
‖Orthologous call made by BLAST to D. melanogaster genome; see Table S1.
Fig. 2.
Relative transcript abundance of male-donated transcripts in conspecifically mated females, heterospecifically mated females, and virgins over the time course of the experiment. The line for virgins corresponds to the average of the two replicates.
Discussion
Our study provides new insights into the female postmating transcriptional response in a species that remates frequently and exhibits major differences in female postmating physiology compared with D. melanogaster. Moreover, our approach provides evidence, at the genetic level, of PMPZ incompatibilities between D. mojavensis and D. arizonae, a major source of reproductive isolation between these species. We also report the transfer of male accessory-gland transcripts (Acps) (and transcripts of other genes) from males to females during copulation. This unanticipated finding adds an interesting dimension to the assumed nature of postcopulatory interactions between males and females.
Response of the Female Reproductive Tract to Conspecific Mating.
The data indicate that only a small number of genes (15) are differentially regulated in D. mojavensis female reproductive tracts following mating, despite the fact that major physiological changes, including the formation and degradation of the insemination reaction, occur during this time. Similarly, previous studies on D. melanogaster have shown that although many genes are differentially regulated in response to mating, a relatively small subset of these genes exhibit large fold changes (more than twofold) in expression in the first 3 h after mating (13). This observation has led to the suggestion that females are “poised” for an initial response to mating that primarily involves the modification of transcripts or proteins already present in the female reproductive tract rather than large-scale changes in transcriptional regulation (13). Our results are also consistent with this hypothesis, at least to the extent that few genes appear to be regulated at the transcriptional level by mating in D. mojavensis.
Whereas the number of genes regulated by mating was small, almost all exhibited large (more than twofold) differences in expression (Table 1), as expected of fundamental components of the initial postmating response. Moreover, although D. mojavensis and D. melanogaster diverged over 40 million years ago and exhibit marked differences in female reproductive physiology, 9 of the 12 D. mojavensis mating-responsive genes with predicted D. melanogaster orthologs also were regulated by mating in previous studies in D. melanogaster (7–14) (Table S5). The remarkable concordance in the identity of mating-responsive genes in D. mojavensis and D. melanogaster supports the functional conservation of genes involved in mating, which, in some cases, may even extend across insect orders (17).
Previous studies on D. melanogaster have demonstrated that genes involved in immunity, particularly antimicrobial peptides (AMPs), are highly up-regulated after mating (8–10, 13, 32, 33). Curiously, we did not detect such a response in D. mojavensis. Although this may indicate an interesting contrast in the female response to mating in these species, it also could reflect incomplete annotation of AMPs in the D. mojavensis genome. We verified that orthologs of most D. melanogaster AMPs were on the microarray and were not differentially regulated in response to mating. However, orthologs of some others have not been identified, and BLAST searches did not reveal any likely candidates in the D. mojavensis genome. Thus, although these genes may have been included on the microarray, we would not be able to identify them as AMPs if they were differentially regulated. Nevertheless, this potential is limited because there were only three genes that were up-regulated after mating that do not have orthologous calls in D. melanogaster. Although the production of AMPs in response to mating is clouded by these issues, three other genes involved in immunity or stress response (two Thor homologs and CG6770) were down-regulated in mated females at 6 h postmating (Table 1). These same genes were also down-regulated after mating in several D. melanogaster studies, with the only exception being that Thor was up-regulated in one study that used whole body flies (Table S5).
Response of the Female Reproductive Tract to Heterospecific Mating.
This study clearly demonstrates that transcriptional regulation of mating-responsive genes is highly disrupted in heterospecifically mated D. mojavensis female reproductive tracts. Transcript levels of many of the mating-responsive genes were considerably higher in the conspecific cross (most more than twofold) and in most cases these differences were detectable by 15 min postmating and persisted throughout the time course that we examined. The fact that most of the mating-responsive genes were up-regulated in conspecifically mated females relative to virgins indicates that heterospecifically mated females fail to ramp up transcription of key genes involved in the normal female postmating response. In some cases the pattern of regulation was similar between the crosses (i.e., the gene was up-regulated relative to virgins in both crosses), but transcript levels were higher in the conspecific cross, whereas in other cases heterospecifically mated females completely failed to up-regulate key mating genes (Fig. 1). An interesting exception to this general pattern is seen for the three genes predicted to function in immune and/or stress response (two homologs of Thor and CG6770), which were down-regulated in both crosses, but more so in heterospecifically mated females.
The overall pattern of misexpression of mating-responsive genes in heterospecifically mated females presumably reflects failed or suboptimal interactions between components of the ejaculate of heterospecific males and the female reproductive tract. In addition, the fact that transcript abundance also differed for a relatively large number of nonmating-responsive genes (148) points to a complex transcriptional response to the ejaculate of heterospecific males that is not necessarily limited only to genes directly involved in mating. Transcript abundance for the majority of these genes was lower in the heterospecific cross, with genes involved in mitochondrial function, in particular, showing lower transcript levels. Although the reason for this is unclear, the difference was detectable at all time points, and thus could have important long-term metabolic consequences for heterospecifically mated females.
Transfer and Persistence of Male Acp Transcripts in the Female Reproductive Tract.
Our study clearly demonstrates that males transfer mRNA transcripts to females during mating. At least 7 of the 12 male-donated transcripts are previously identified candidate D. mojavensis Acps, and two others are orthologous to genes expressed in D. melanogaster male reproductive tracts (Table 2). Although the role of these transcripts within the female is unclear, the fact that they represent a small, repeatable subset of transcripts from the male reproductive tract suggests that their inclusion in the ejaculate is not random. Moreover, all were still present at elevated levels in females after 6 h (Fig. 2), indicating that they are not rapidly degraded, as might be expected if they served no functional purpose.
We do not know whether male transcripts that are passed to females are contained within cells or other vessels or whether they are extracellular, but in either case they could play an important functional role. For example, recent evidence in humans suggests that sperm carry mRNA transcripts, some of which are translated de novo by mitochondrial-like ribosomal proteins and appear to play an important role in sperm motility, capacitation, and fertilization (34, 35). Moreover, other transcripts are delivered to the oocyte where they may influence early embryonic development (36). Given that at least some of the male-donated transcripts are orthologous to genes in the D. melanogaster testes and seminal vesicle, the possibility that sperm carry these transcripts is plausible. In addition to this possibility, we also note that D. mojavensis females are unusual among Drosophila in that they incorporate significant levels of peptide components of the male ejaculate into their somatic tissues (37). The mechanism for this is unknown, but could conceivably provide a route for the uptake of male-donated mRNA transcripts into female cells where they could be subsequently translated. Given that Acps play a critical role in many postcopulatory processes (2, 18), the translation of these transcripts into functional proteins within the female, if it occurs, would have important implications for understanding fertilization success, postcopulatory sexual selection, and the evolution of PMPZ isolating barriers.
Even if transferred male transcripts are extracellular and not translated, they still may be functionally important. An increasing number of studies have demonstrated that extracellular nucleic acids play pivotal roles in processes such as blood clotting and immune response (38, 39). For example, recent evidence from a study on the mechanisms of mammalian blood clotting demonstrated that extracellular RNA serves as a procoagulant, providing a template for adhesion of blood coagulation factors in response to vascular injury (38). Similarly, following bacterial infection in the insect, Galleria mellonella, extracellular RNA released from damaged cells forms a net-like coagulation structure that efficiently entraps bacteria and triggers important immune defenses (39). In light of this, it seems plausible that male-derived mRNA transcripts could be involved in the formation of the insemination reaction, which at least superficially resembles a coagulatory response in the uterus of D. mojavensis females. Although speculative, this intriguing possibility warrants further investigation.
Our study represents a step forward in understanding the genetic basis of sexual conflict and reproductive compensation (40), postcopulatory molecular interactions between the sexes, and how disruptions in these interactions contribute to the evolution of reproductive isolation between diverging populations. In addition, our discovery of the transfer of male ACP transcripts to females during copulation raises new questions about the nature and complexity of reproductive tract interactions, paving the way for exciting avenues of future research.
Materials and Methods
Mating and Sample Preparation.
We used one D. mojavensis isofemale line collected from Anza Borrego Desert State Park, Borrego Springs, CA, in April of 2002, and one D. arizonae isofemale line collected from Guaymas, Sonora, Mexico, in March of 2007 for our experiments. We collected virgin males and females within 24 h of emergence (flies take at least 5 d to reach reproductive maturity) and aged them separately for 10–12 d. For mating experiments, one D. mojavensis female was introduced to a vial containing two D. mojavensis or D. arizonae males. Following copulation males were removed from vials and females were kept in isolation until lower reproductive tracts were removed at either 15 min, 2 h, or 6 h postmating; we also removed lower reproductive tracts from virgin females at this time. Tissues were placed immediately in TRIzol and frozen at −80 °C until total RNA was extracted using a standard TRIzol protocol. We performed two biological replicates for each time point by pooling 20–25 lower reproductive tracts for each sample (14 samples total). Following RNA extraction, mRNA was amplified using the MessageAmp II aRNA amplification kit (Applied Biosystems/Ambion).
Microarray Assay and Data Analysis.
The D. mojavensis microarray, which included 14,591 genes, was designed for the NimbleGen 4-Plex platform. Before analysis, hybridization intensities were normalized using the robust multichip average (RMA) method, which includes quantile normalization of intensities (41, 42). We analyzed the log2 of the RMA normalized intensities using a two-step mixed model ANOVA (43). More details on the microarray design and analysis can be found in (SI Materials and Methods). All expression data have been deposited in the Gene Expression Omnibus under series entry GSE27454.
We verified microarray results for three genes (Dmoj/Acp2, GI17858, and GI23890) using quantitative PCR. Working from the same mRNA pools used in the microarray, we synthesized cDNA using ABI’s high-capacity RNA-to-cDNA kit. Quantitative PCR reactions were performed on an ABI 7000 Sequence Detection system machine using ABI’s Power SYBR Green PCR kit. We ran each gene (including a control: Ribosomal subunit 18S) in triplicate using gene specific primers (Table S4). Statistical significance was calculated by performing 10,000 bootstraps using the REST 2008 software (44).
Analysis of the representation of gene ontology terms was performed using the online Database for Annotation, Visualization and Integrated Discovery (DAVID) at http://david.abcc.ncifcrf.gov/. The analysis was based on the level of gene ontology term representation of D. melanogaster orthologs of the differentially expressed D. mojavensis genes.
Verification of Male-Derived Transcripts.
To determine whether the increase in transcript abundance of previously identified male Acps following mating was due to female up-regulation of these genes or whether they were included in the male ejaculate, we created cDNA libraries from the original aRNA of two samples used in the microarray experiment (conspecifically mated/15 min and heterospecifically mated/15 min). Libraries were constructed using the Bio-Rad Iscript select cDNA synthesis kit with random primers. We used PCR to amplify 23 transcripts that included previously identified D. mojavensis/D. arizonae Acps (27, 45) and/or previously identified transcripts from D. melanogaster male reproductive tract (46, 47), in addition to the 18 genes that differed in transcript abundance between the conspecifically mated females and virgins (Table 1). We then sequenced the amplified products and used fixed differences between the species to determine whether transcripts from the heterospecific cross were from D. arizonae (i.e., of male origin) or D. mojavensis (i.e., of female origin). Sequences from this analysis that are longer than 200 bp are deposited under GenBank accession nos. JF512479–JF512494; all sequences, including those under 200 bp, are included in Dataset S1.
To independently verify the transfer of male transcripts, we repeated the heterospecific matings using the same D. mojavensis line and randomly chosen males from 12 D. arizonae lines from Guaymas, Sonora, Mexico (not including the original line used in the microarray). All procedures (matings, RNA extraction, cDNA synthesis) were identical to the original experiment except that we did not perform the mRNA amplification step.
Supplementary Material
Acknowledgments
We thank Miguel Escalona for help with laboratory work. This study was supported by the National Institute of General Medical Sciences 2K12 G000798-06 Postdoctoral Excellence in Research and Teaching Fellowship through the Center for Insect Science at the University of Arizona (to J.M.B.), the University of California at San Diego, National Science Foundation (NSF) Award DEB-0921514 (to T.A.M.), a grant from Science Foundation Arizona (to T.A.M. and L.M.M.), and NSF Award DEB-1020009 (to L.M.M.). E.S.K. was supported by a dissertation fellowship from the American Association of University Women.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus accession no. GSE27454, and the sequences reported in this paper have been deposited in the GenBank database (accession nos. JF512479–JF512494).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100388108/-/DCSupplemental.
References
- 1.Markow TA, Reed LK, Kelleher ES. Sperm fate and function in reproductive isolation in Drosophila. Soc Reprod Fertil Suppl. 2007;65:155–173. [PubMed] [Google Scholar]
- 2.Pitnick S, Wolfner MW, Suarez SS. In: Sperm Biology: An Evolutionary Perspective. Birkhead TR, Hosken DJ, Pitnick S, editors. Burlington, MA: Academic; 2009. pp. 247–304. [Google Scholar]
- 3.Arnqvist G, Rowe L. Sexual Conflict. Princeton: Princeton Univ Press; 2005. [Google Scholar]
- 4.Birkhead TR, Pizzari T. Postcopulatory sexual selection. Nat Rev Genet. 2002;3:262–273. doi: 10.1038/nrg774. [DOI] [PubMed] [Google Scholar]
- 5.Howard DJ, Palumbi SR, Birge L, Manier MK. In: Sperm Biology: An Evolutionary Perspective. Birkhead TR, Hosken JD, Pitnick S, editors. Burlington, MA: Academic; 2009. pp. 367–395. [Google Scholar]
- 6.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 7.Innocenti P, Morrow EH. Immunogenic males: A genome-wide analysis of reproduction and the cost of mating in Drosophila melanogaster females. J Evol Biol. 2009;22:964–973. doi: 10.1111/j.1420-9101.2009.01708.x. [DOI] [PubMed] [Google Scholar]
- 8.Kapelnikov A, et al. Mating induces an immune response and developmental switch in the Drosophila oviduct. Proc Natl Acad Sci USA. 2008;105:13912–13917. doi: 10.1073/pnas.0710997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawniczak MK, Begun DJ. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome. 2004;47:900–910. doi: 10.1139/g04-050. [DOI] [PubMed] [Google Scholar]
- 10.Mack PD, Kapelnikov A, Heifetz Y, Bender M. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103:10358–10363. doi: 10.1073/pnas.0604046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGraw LA, Clark AG, Wolfner MF. Post-mating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics. 2008;179:1395–1408. doi: 10.1534/genetics.108.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGraw LA, Gibson G, Clark AG, Wolfner MF. Strain-dependent differences in several reproductive traits are not accompanied by early postmating transcriptome changes in female Drosophila melanogaster. Genetics. 2009;181:1273–1280. doi: 10.1534/genetics.108.099622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGraw LA, Gibson G, Clark AG, Wolfner MF. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr Biol. 2004;14:1509–1514. doi: 10.1016/j.cub.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Prokupek AM, Kachman SD, Ladunga I, Harshman LG. Transcriptional profiling of the sperm storage organs of Drosophila melanogaster. Insect Mol Biol. 2009;18:465–475. doi: 10.1111/j.1365-2583.2009.00887.x. [DOI] [PubMed] [Google Scholar]
- 15.Rogers DW, et al. Molecular and cellular components of the mating machinery in Anopheles gambiae females. Proc Natl Acad Sci USA. 2008;105:19390–19395. doi: 10.1073/pnas.0809723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kocher SD, Richard FJ, Tarpy DR, Grozinger CM. Genomic analysis of post-mating changes in the honey bee queen (Apis mellifera) BMC Genomics. 2008;9:232. doi: 10.1186/1471-2164-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kocher SD, Tarpy DR, Grozinger CM. The effects of mating and instrumental insemination on queen honey bee flight behaviour and gene expression. Insect Mol Biol. 2010;19:153–162. doi: 10.1111/j.1365-2583.2009.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfner MF. “S.P.E.R.M.” (seminal proteins (are) essential reproductive modulators): The view from Drosophila. Soc Reprod Fertil Suppl. 2007;65:183–199. [PubMed] [Google Scholar]
- 19.Matzkin LM. The molecular basis of host adaptation in cactophilic Drosophila: Molecular evolution of a glutathione S-transferase gene (GstD1) in Drosophila mojavensis. Genetics. 2008;178:1073–1083. doi: 10.1534/genetics.107.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markow TA. Evolutionary Biology. New York: Plenum; 1996. pp. 73–106. [Google Scholar]
- 21.Knowles LL, Markow TA. Sexually antagonistic coevolution of a postmating-prezygotic reproductive character in desert Drosophila. Proc Natl Acad Sci USA. 2001;98:8692–8696. doi: 10.1073/pnas.151123998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller GT, Starmer WT, Pitnick S. Quantitative genetic analysis of among-population variation in sperm and female sperm-storage organ length in Drosophila mojavensis. Genet Res. 2003;81:213–220. doi: 10.1017/s0016672303006190. [DOI] [PubMed] [Google Scholar]
- 23.Pitnick S, Miller GT, Schneider K, Markow TA. Ejaculate-female coevolution in Drosophila mojavensis. Proc Biol Sci. 2003;270:1507–1512. doi: 10.1098/rspb.2003.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almeida FC, Desalle R. Evidence of adaptive evolution of accessory gland proteins in closely related species of the Drosophila repleta group. Mol Biol Evol. 2008;25:2043–2053. doi: 10.1093/molbev/msn155. [DOI] [PubMed] [Google Scholar]
- 25.Kelleher ES, Swanson WJ, Markow TA. Gene duplication and adaptive evolution of digestive proteases in Drosophila arizonae female reproductive tracts. PLoS Genet. 2007;3:e148. doi: 10.1371/journal.pgen.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelleher ES, Markow TA. Duplication, selection and gene conversion in a Drosophila mojavensis female reproductive protein family. Genetics. 2009;181:1451–1465. doi: 10.1534/genetics.108.099044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagstaff BJ, Begun DJ. Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics. 2005;171:1083–1101. doi: 10.1534/genetics.105.043372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagstaff BJ, Begun DJ. Adaptive evolution of recently duplicated accessory gland protein genes in desert Drosophila. Genetics. 2007;177:1023–1030. doi: 10.1534/genetics.107.077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markow TA. Courtship behavior and control of reproductive isolation between Drosophila mojavensis and Drosophila arizonensis. Evolution. 1981;35:1022–1026. doi: 10.1111/j.1558-5646.1981.tb04968.x. [DOI] [PubMed] [Google Scholar]
- 30.Massie KR, Markow TA. Sympatry, allopatry and sexual isolation between Drosophila mojavensis and D. arizonae. Hereditas. 2005;142:51–55. doi: 10.1111/j.1601-5223.2005.01911.x. [DOI] [PubMed] [Google Scholar]
- 31.Reed LK, Markow TA. Early events in speciation: Polymorphism for hybrid male sterility in Drosophila. Proc Natl Acad Sci USA. 2004;101:9009–9012. doi: 10.1073/pnas.0403106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domanitskaya EV, Liu H, Chen S, Kubli E. The hydroxyproline motif of male sex peptide elicits the innate immune response in Drosophila females. FEBS J. 2007;274:5659–5668. doi: 10.1111/j.1742-4658.2007.06088.x. [DOI] [PubMed] [Google Scholar]
- 33.Peng J, Zipperlen P, Kubli E. Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr Biol. 2005;15:1690–1694. doi: 10.1016/j.cub.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 34.Zhao C, et al. Role of translation by mitochondrial-type ribosomes during sperm capacitation: An analysis based on a proteomic approach. Proteomics. 2009;9:1385–1399. doi: 10.1002/pmic.200800353. [DOI] [PubMed] [Google Scholar]
- 35.Gur Y, Breitbart H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006;20:411–416. doi: 10.1101/gad.367606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lalancette C, Miller D, Li Y, Krawetz SA. Paternal contributions: New functional insights for spermatozoal RNA. J Cell Biochem. 2008;104:1570–1579. doi: 10.1002/jcb.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markow TA, Ankney PF. Drosophila males contribute to oogenesis in a multiple mating species. Science. 1984;224:302–303. doi: 10.1126/science.224.4646.302. [DOI] [PubMed] [Google Scholar]
- 38.Kannemeier C, et al. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc Natl Acad Sci USA. 2007;104:6388–6393. doi: 10.1073/pnas.0608647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altincicek B, Stötzel S, Wygrecka M, Preissner KT, Vilcinskas A. Host-derived extracellular nucleic acids enhance innate immune responses, induce coagulation, and prolong survival upon infection in insects. J Immunol. 2008;181:2705–2712. doi: 10.4049/jimmunol.181.4.2705. [DOI] [PubMed] [Google Scholar]
- 40.Gowaty PA. Reproductive compensation. J Evol Biol. 2008;21:1189–1200. doi: 10.1111/j.1420-9101.2008.01559.x. [DOI] [PubMed] [Google Scholar]
- 41.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 42.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 43.Wolfinger RD, et al. Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- 44.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelleher ES, Watts TD, LaFlamme BA, Haynes PA, Markow TA. Proteomic analysis of Drosophila mojavensis male accessory glands suggests novel classes of seminal fluid proteins. Insect Biochem Mol Biol. 2009;39:366–371. doi: 10.1016/j.ibmb.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Findlay GD, Yi X, Maccoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 2008;6:e178. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takemori N, Yamamoto MT. Proteome mapping of the Drosophila melanogaster male reproductive system. Proteomics. 2009;9:2484–2493. doi: 10.1002/pmic.200800795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.