Abstract
Autophagy is a conserved cellular process to degrade and recycle cytoplasmic components. During autophagy, lysosomes fuse with an autophagosome to form an autolysosome. Sequestered components are degraded by lysosomal hydrolases and presumably released into the cytosol by lysosomal efflux permeases. Following starvation-induced autophagy, lysosome homeostasis is restored by autophagic lysosome reformation (ALR) requiring activation of the “target of rapamycin” (TOR) kinase. Spinster (Spin) encodes a putative lysosomal efflux permease with the hallmarks of a sugar transporter. Drosophila spin mutants accumulate lysosomal carbohydrates and enlarged lysosomes. Here we show that defects in spin lead to the accumulation of enlarged autolysosomes. We find that spin is essential for mTOR reactivation and lysosome reformation following prolonged starvation. Further, we demonstrate that the sugar transporter activity of Spin is essential for ALR.
Keywords: lysosomal storage disease
During autophagy, lysosomes fuse with autophagosomes to form autolysosomes, where contents are degraded by lysosomal hydrolases and released by lysosomal efflux transporters (1). The autophagic/lysosomal pathway is critical to cellular homeostasis. Defects in autophagy lead to the accumulation of damaged organelles, misfolded proteins, and toxic metabolites, and are associated with neurodegeneration and other abnormalities (1, 2). Defects in specific lysosomal hydrolysis have been implicated in lysosomal storage disorders (LSDs; ref. 2). Loss of lysosomal protease activity can lead to the accumulation of undigested material, as well as neurodegenerative disease. In addition, defective efflux of lysosomal contents by lysosomal transporters can lead to accumulation of lysosomal substrates and defective lysosomal function (3).
Lysosomal efflux transporters are a family of lysosomal membrane proteins required for the export of lysosomal degradation products, such as amino acids and monosaccharides (4). A subset of lysosomal storage diseases has been linked to mutations found in lysosomal efflux transporters. For example, defects in Sialin, a sialic acid transporter, leads to sialic acid storage diseases (SASD), and defects in the lysosome Arginine transporter lead to Juvenile Batten Disease (5, 6). Spinster (Spin) (also known as benchwarmer) is a late endosomal/lysosomal membrane protein with the amino acid sequence of a lysosomal sugar carrier in the major facilitator superfamily (4, 7). Spin is a transmembrane protein containing 8–12 transmembrane domains (8). In Drosophila, hypomorphic mutations in spin lead to decreased adult life span, defects in courtship behavior, accumulation of autoflourescent pigments, and neurodegeneration (5, 8, 9). Drosophila spin mutants also exhibit neuromuscular synaptic overgrowth (8) and enhanced tau-mediated toxicity (4). In zebrafish, loss of the spin homolog not really started (nrs) leads to embryonic lethality characterized by the accumulation of opaque substances in the yolk (9). Interestingly, Drosophila spin mutants exhibit endocytic defects, as well as widespread accumulation of lysosomal carbohydrates and enlarged lysosomes (4). Little is known, however, about the mechanism leading to the accumulation of enlarged lysosomes in spin mutants.
ALR is an evolutionarily conserved lysosome regeneration cycle that governs nutrient sensing and lysosome homeostasis following starvation-induced autophagy (10). In response to starvation, mTOR is inhibited, leading to the induction of autophagy. After prolonged starvation, however, mTOR is reactivated. Upon mTOR reactivation, tubules extrude from autolysosomal membranes and give rise to vesicles that ultimately mature into functional lysosomes (10). The degradation of autophagic cargo is required for mTOR reactivation after starvation, and inhibiting mTOR reactivation leads to the accumulation of enlarged autolysosomes. In addition, ALR requires the dissociation of the small GTPase Rab7 from autolysosomes, and overexpression of constitutively active Rab7 results in the accumulation of enlarged autolysosomes (10).
Here we report that loss of spin leads to the accumulation of enlarged autolysosomes that fail to degrade their contents in both mammalian cells and Drosophila. We show that spin is required for mTOR reactivation and lysosome reformation following prolonged starvation. Interestingly, reactivation of mTOR signaling after starvation is sufficient to induce lysosome reformation even in the context of decreased spin function. Importantly, we find that the sugar transporter activity of Spin is essential for ALR. Our findings elucidate the role of this lysosomal efflux transporter in ALR and reveal its contribution to LSDs.
Results
Mammalian Spin Colocalizes with the Lysosomal Membrane Marker Lamp1.
We used RNAi to screen a collection of permeases, and identified Spinster (Spin) as a candidate regulator of autophagic lysosome reformation. spin encodes a protein with the hallmarks of a sugar transporter in the major facilitator superfamily related to the arabinose efflux permease (4, 7). Spin has been localized to the late endosome/lysosome in Drosophila and zebrafish (4, 9). In mammalian cells, Spin has been reported to localize to mitochondria (11). We expressed Spin-GFP in normal rat kidney (NRK) cells and found that Spin-GFP largely colocalizes with the acidic compartment dye Lysotracker, but not the mitochondrial marker Mitotracker (Fig. 1A). Furthermore, we found that Spin-GFP specifically colocalizes with the late endosomal/lysosomal membrane marker lysosomal-associated membrane protein 1 (Lamp1; Fig. 1B). These data suggest that mammalian Spin localizes to Lamp1-positive compartments, and like its fly and zebrafish counterparts, is a late endosomal/lysosomal protein.
Fig. 1.
Spin colocalizes with the lysosomal membrane marker Lamp1, and defects in spin lead to enlargement of Lamp1-positive compartments. (A) NRK cells were transfected with Spin-GFP (green) and stained for Mitotracker (red, Left) or Lysotracker (red, Right). (Scale bar, 5 μm.) (B) NRK cells were transfected with Spin-GFP and Lamp1-Cherry. (Scale bar 5 μm.) (C) NRK cells were transfected with nonspecific (NS)- or spin-RNAi. After 2 d, cells were again transfected with NS- or spin-RNAi and Lamp1-YFP; 16 h after transfection, cells were starved for 0 or 10 h and observed by confocal microscopy. (Scale bar, 5 μm.) (D) NRK cells were transfected with nonspecific (NS)-RNAi or RNAi against spin. Two days after transfection, cells were retransfected with RNAi and either hSpin-CFP (Rescue)/Lamp1-YFP or Vector/Lamp1-YFP. Twenty-four hours after the second transfection, cells were starved for 10 h and then observed by confocal microscopy. (E) Cells from D were assessed in a blind fashion for rescue of the enlarged lysosome phenotype after starvation for 10 h and quantified. One hundred cells were counted. Error bars represent s.d. from more than three independent experiments. (F) Control Drosophila expressing tub-Lamp1-GFP versus transheterozygous spin10403/spinK09905 (spin mutant) larvae expressing tub-Lamp1-GFP were either fed or starved for 12 h, and lysosomes in fatbody cells were observed by fluorescent microscopy. Blue: Höescht. Green: Lamp1-GFP. (Scale bars, 20 μm.)
Defects in spin Lead to Accumulation of Enlarged Lamp1-Positive Compartments.
We next generated Spin knockdown cells (Fig. S1). When cultured in nutrient-rich conditions, these cells exhibited grossly normal morphology of Lamp1-positive structures, although we noted that a fraction of these cells exhibit a slight enlargement and subtle increase in the perinuclear localization of Lamp1-positive structures (Fig. 1C). However, we found that serum withdrawal led to the dramatic enlargement of Lamp1-positive structures in spin knockdown cells which differed strikingly from control cells (Fig. 1C). To test the specificity of this phenotype, we knocked down rat spin by expressing an RNAi construct that fails to bind the human spin sequence due to a single nucleotide mismatch (Fig. S2), and overexpressed human Spin in these knockdown cells. We found that whereas 94% of spin knockdown cells exhibited enlarged Lamp1-positive compartments, only 12% of cells overexpressing human Spin exhibit this phenotype indicating that the human sequence rescues the lysosomal defect (Fig. 1 D and E). This result suggests that the phenotype is due to a specific Spin depletion effect and not an off-target effect.
Significantly, we observed similar effects in vivo in the fatbody of Drosophila spin mutants, which is a nutrient storage and mobilization organ akin to the mammalian liver. Upon starvation, the fatbody cells of spin mutants accumulate enlarged Lamp1-GFP-marked structures compared with fatbody cells of unstarved control animals (Fig. 1F). Together, these data indicate that defects in spin lead to the accumulation of enlarged Lamp1-positive structures in vivo.
Decreased spin Function Causes Enlarged Autolysosomes.
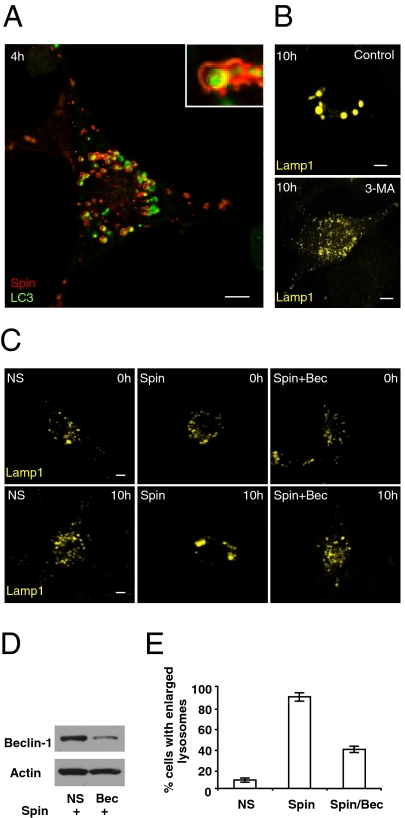
We found that the enlargement of Lamp1-positive structures upon loss of spin is starvation-dependent. We therefore tested whether this phenotype is dependent upon starvation-induced autophagy. We coexpressed Spin-YFP and cyan fluorescent protein (CFP)-tagged microtubule-associated light chain 3 (CFP-LC3), a marker of autophagosomes, in NRK cells. We found that, after 4 h of starvation, Spin-YFP (Red) localized to discrete ring-like structures surrounding CFP-LC3 marked autophagosomes (Fig. 2A). Cotreatment of cells with the autophagy inhibitor 3-methyl adenine (3-MA) completely inhibited the enlargement of Lamp1-positive structures upon starvation (Fig. 2B and Fig. S3 A–C). Moreover, concomitant knockdown of spin and beclin1, a gene required for autophagy and also known as Atg6, reduced the percentage of cells exhibiting enlarged Lamp1-positive compartments after starvation from 89% to 38% (Fig. 2 C–E). These data demonstrate that the starvation-induced enlargement of Lamp1-positive compartments upon spin knockdown is autophagy-dependent.
Fig. 2.
Enlargement of lysosomes upon spin knockdown is autophagy dependent. (A) NRK cells were transfected with CFP-LC3 (pseudocolored as green) and Spin-YFP (pseudocolored as red), and 24 h after transfection, cells were starved for 4 h. (Inset) A close-up of the discrete ring structure of Spin-YFP surrounding the LC3 assembly. (Scale bars, 5 μm.) (B) NRK cells were transfected with nonspecific (NS)- or spin-RNAi. After 2 d, cells were again transfected with NS- or spin-RNAi and Lamp1-YFP; 16 h after transfection, cells were starved for 10 h with or without the addition of 1 mM 3-MA, and observed by confocal microscopy. (Scale bars, 5 μm.) (C) NRK cells were transfected with nonspecific (NS)-RNAi, spin-RNAi, or spin-RNAi plus Beclin 1 (Bec)-RNAi. After 2 d, cells were again transfected with RNAi and Lamp1-YFP; 16 h after transfection, cells were starved for 0 or 10 h and observed by confocal microscopy. (Scale bars, 5 μm.) (D) Western blot for the Beclin-1 protein and Actin control for a representative sample of cells transfected with NS-RNAi plus spin-RNAi, or spin-RNAi plus beclin 1-RNAi. (E) Cells expressing nonspecific (NS)-RNAi, spin-RNAi, or spin-RNAi plus Beclin 1 (Bec)-RNAi were quantified in a blind fashion for the presence of enlarged lysosomes after 10 h of starvation. One hundred cells were counted. Error bars indicate the SD from more than three independent experiments.
We next investigated whether the enlarged Lamp1-positive structures are autolysosomes. We knocked down spin in NRK cells stably expressing CFP-LC3 and Lamp1-YFP, and found that the accumulated Lamp1-positive structures are LC3 positive (Fig. S4 A and B). Consistent with the idea that autolysosomes accumulate upon starvation, TEM analysis revealed that single-membrane-bound vesicles containing undigested cytoplasmic contents accumulate upon starvation in control and spin-knockdown cells (Fig. S4C). However, spin knockdown led to the accumulation of larger, more distended single-membrane structures than those found in control cells, which is most evident 10 h after starvation (Fig. S4D). Similarly, TEM revealed that large autolysosomes with cargo accumulate in vivo in Drosophila fat body (Fig. S5). We have shown that, in NRK cells, the formation of autolysosomes peaks around 4 h after starvation, and at 10 h after starvation, most autolysosomes have disappeared (10). However, the TEM analyses indicate that, in spin-knockdown cells, autolysosomes are long lasting, and that 10 h after starvation, significant numbers of autolysosomes persist (Fig. S4 C and D). These data indicate that cells with decreased spin function accumulate enlarged autolysosomes upon starvation.
spin Is Required for ALR Following Starvation.
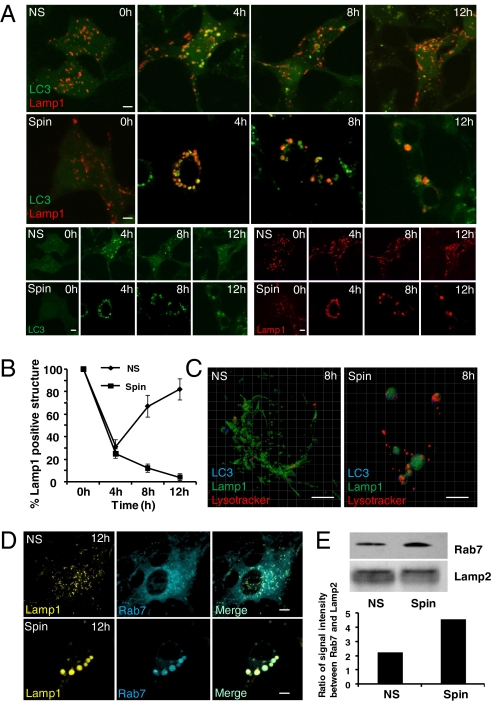
Lysosome numbers are restored by ALR following starvation-induced autophagy (10). To determine whether ALR is defective upon spin knockdown, we coexpressed LC3 and Lamp1 in NRK cells and analyzed the kinetics of autophagosome formation and lysosome number during starvation. Similar to our previous results (10), we found that after 4 h of starvation, LC3 and Lamp1 largely colocalize, indicating the formation of LC3 and Lamp1-positive autolysosomes and the consumption of most of the lysosomes in the cell into autolysosomes (Fig. 3A). By 12 h of starvation, however, LC3 is diffuse indicating that autophagy has ended and aggregated LC3 has been degraded (Fig. 3A). In addition, after 12 h of starvation in control cells, Lamp1-positive, LC3-negative lysosomes recover in number to 80% of the levels found in nonstarved control cells (Fig. 3B). We found that, upon spin knockdown, LC3 and Lamp1 colocalize after 4 h of starvation (Fig. 3B). By contrast, in spin knockdown cells LC3 and Lamp1 colocalize even after 12 h of starvation, and lysosome number fails to recover in these cells (Fig. 3 A and B).
Fig. 3.
spin is required for autophagic lysosome reformation. (A) NRK-CFP-LC3 (pseudocolored as green)-Lamp1-YFP (pseudocolored as red) stable cells were transfected with nonspecific (NS)- or spin-RNAi and starved for 0, 4, 8, or 12 h. (Scale bar, 5 μm.) (B) Cells from (A) were assessed in a blind fashion for change in number of Lamp1-positive structures relative to nonstarved cells. Fifty cells were counted per timepoint. Error bars represent S.D. derived from multiple experiments. (C) Nonspecific (NS)- or spin-RNAi-transfected NRK-CFP-LC3-Lamp1-YFP cells were starved for 8 h. Cells were then Z-stack scanned at 0.5-μm intervals, and images were collapsed to construct 3D models using IMRIS. Blue, LC3; Green, Lamp1; Red, Lysotracker. (Scale bars, 5 μm.) (D) NRK cells were transfected with nonspecific (NS) or spin-RNAi and after 2 d, cells were transfected with NS or spin-RNAi, CFP-Rab7, and Lamp1-YFP. Twenty-four hours after transfection, cells were starved for 0 or 12 h before imaging. (Scale bars, 5 μm.) (E) NS or spin-RNAi-transfected NRK cells were starved for 12 h, autolysosome/lysosome were purified, and the protein levels of Rab7 and Lamp2 were analyzed by Western blot. The signal intensity of Rab7 and Lamp2 were measured by IPP and the histogram shows the ratio between Rab7 and Lamp2.
During ALR, Lamp1-positive tubules extrude from autolysosomal membranes after 8 h of starvation, and give rise to Lamp1-positive and LC3-negative vesicles that ultimately mature into functional lysosomes (10). We expressed CFP-LC3 and Lamp1-YFP in NRK cells and performed 3D reconstructions of Z-stacked scanning confocal images of control and spin knockdown cells after 8 h of starvation. We found that whereas Lamp1-YFP tubules were abundant in control cells, they were reduced in spin knockdown cells (Fig. 3C). Thus, spin knockdown prevents ALR.
We previously reported that ALR requires the dissociation of the small GTPase Rab7 from autolysosomes, and that overexpression of constitutively active Rab7 prevents ALR and results in the accumulation of enlarged autolysosomes (10). We investigated the localization of CFP-Rab7 and Lamp1-YFP after 12 h of starvation in control and spin knockdown cells. We found that, unlike in control cells, CFP-Rab7 colocalizes with Lamp1-YFP in spin knockdown cells even after 12 h of starvation, and the percentage of cells with large persistent CFP-Rab7 vesicles is dramatically increased in spin knockdown cells (Fig. 3D). Furthermore, the amount of endogenous Rab7 on lysosomal/autolysosomal membranes is increased in starved spin knockdown cells (Fig. 3E) Together, these data indicate that spin knockdown leads to defects in several markers of ALR.
Sugar Transporter Activity of Spin Is Essential for ALR.
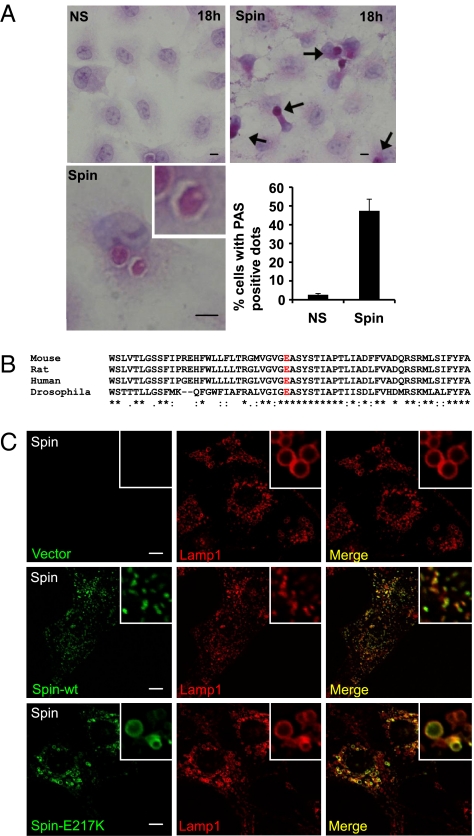
Spin encodes a sugar transporter, and an E to K mutation in the 217 position leads to a loss of function in the sugar transporter activity in flies (4). Consistent with this observation, we found that carbohydrates accumulate in the autolysosomes of spin knockdown NRK cells following prolonged starvation as indicated by the accumulation of periodic acid Schiff staining material (Fig. 4A). We next studied the E217 residue, which is conserved in spin across species and presumed to be functionally required (Fig. 4B). Hence, we tested whether the sugar transporter activity is required for autophagic lysosome reformation by comparing the rescue efficiency of wild-type spin plasmid with the E217K mutant human spin plasmid. Whereas expression of wild-type Spin rescued ALR, expression of mutant Spin E217K failed to rescue the accumulation of giant Lamp1-positive structures and ALR, indicating the critical role of this residue in the human ortholog (Fig. 4C).
Fig. 4.
Sugar transporter activity is essential for Spin's role in the regulation of autophagic lysosome reformation. (A) Carbohydrates accumulate in spin knockdown cells under prolonged starvation. NRK (CFP-LC3) cells were transfected with NS- or spin-RNAi. After 2 d, cells were starved for 18 h and subjected to periodic acid-Schiff (PAS) staining. (Scale bars, 5 μm.) Arrows indicate PAS-positive structures. Histogram shows the percentage of PAS-positive structures from multiple experiments counting a total of 100 cells, with mean and SD shown. (B) The alignment of the spin amino acid sequence from representative species shown in standard single letter code. The conserved glutamic acid residue (E) which is essential for sugar transporter activity is highlighted in red. (C) NRK cells were transfected with RNAi against spin. Two days after transfection, cells were retransfected with RNAi and vector/Lamp1-YFP, hSpin-CFP-wt/Lamp1-YFP or hspin-CFP-mutant (spinE217K)/Lamp1-YFP. Twenty-four hours after the second transfection, cells were starved for 10 h and then observed by confocal microscopy. (Scale bar, 5 μm.)
Degradative Capacity of Autolysosomes, but Not Lysosomes, Is Impaired by spin Knockdown.
LC3 that is internalized upon formation of autolysosomes is subject to degradation. We noticed that, after 10 h starvation, LC3 is retained in the autolysosomes of spin knockdown cells (Fig. S4A). In addition, electron microscopy revealed that compared with control cells, spin knockdown cells accumulate what appear to be undigested material in autolysosomes after 10 h of starvation, and this is also observed in starved Drosophila spin mutants (Figs. S4C and S5). To test the degradation capacity of lysosomes and autolysosomes, we loaded control or spin knockdown cells expressing Lamp1-cherry red with the lysosomal substrate DQ-BSA. If grown in normal growth medium, lysosomes of both control and spin knockdown cells exhibit normal degradation capacity. However, after 12 h of starvation, whereas the Lamp1-positive structures of control cells stain positive for DQ- BSA, the majority of giant, distended Lamp1-positive structures in spin knockdown cells lack DQ-BSA staining (Fig. S6). Thus, only after prolonged starvation does loss of spin lead to defects in the degradation capacity of autolysosomes. This result indicates that the degradation capacity of autolysosomes, but not lysosomes, is impaired upon spin knockdown, and suggests a possible role for ALR in maintaining the degradation capacity of autolysosomes. To further test the degradation capacity of autolysosomes, we stained cells with BODIPY FL-conjugated Pepstatin A (BODIPY FL-Pep), a cathepsin D substrate that stains only the active form of cathepsin D. Consistent with what we observed with DQ-BSA, we found that in nutrient- rich conditions, spin knockdown only slightly decreases BODIPY FL-Pep staining in lysosomes. However, after 12 h of starvation, most Lamp1-positive structures in spin knockdown cells are BODIPY FL-Pep negative (Fig. S7), indicating that cathepsin D activity is impaired in starved spin knockdown cells.
We next examined the mechanism of degradation impairment in spin knockdown cells by staining cells with lysosensor green DND-189, a pH indicator that exhibits a pH-dependent increase in fluorescence intensity upon acidification. We found that, after 12 h of starvation, autolysosomes from spin knockdown cells have much higher fluorescence intensity than nonspecific RNAi-transfected cells (Fig. S8), suggesting that spin knockdown augments lysosome acidification.
spin Is Required for mTOR Reactivation Following Starvation.
We recently reported that mTOR activity is inhibited during the initiation of autophagy, but is reactivated after prolonged starvation. We found that the degradation of autophagic cargo is required for mTOR reactivation after starvation, and that mTOR reactivation attenuates autophagy and triggers ALR (10). Because mTOR directly regulates ALR, we next tested whether mTOR reactivation is impaired upon spin knockdown. We used Phospho-p70 S6 Kinase (P-S6K) as a marker for mTOR activity. We found that, whereas in control cells P-S6K is lost after 2 h of starvation but recovers after 8 h of starvation, spin knockdown markedly inhibits the recovery of S6K phosphorylation (Fig. 5A). Thus, mTOR reactivation is impaired in spin knockdown cells.
Fig. 5.
spin is required for mTOR reactivation following starvation. (A) NRK-LC3 cells were transfected with nonspecific RNAi (NS) or RNAi against spin. Sixty hours after transfection, cells were starved 0, 2, 6, or 8 h, harvested, and analyzed by Western blot using antibodies against Phospho-p70 S6 Kinase (P-S6K), Total S6 Kinase (S6K), and Actin. (B and C) NRK cells were transfected with spin-RNAi as in A, and starved for 10 h. FCS was then added back for 0–120 min; cells were harvested and analyzed by Western blot using antibodies against Phospho-p70 S6 Kinase (P-S6K) (B), or observed by confocal microscopy for the Lamp1 intracellular pattern (C). (Scale bar, 5 μm.) (Inset) Enlargement of tubules extruding from lysosomes.
The addition of serum is sufficient to rapidly activate mTOR (12). To test whether mTOR reactivation after prolonged starvation suffices to trigger autophagic lysosome reformation in spin knockdown cells, we induced giant autolysosomes by starving spin knockdown cells for 10 h, and then added 10% FCS. We found that phosphorylated S6K levels were rapidly boosted by the addition of FCS (Fig. 5B). Interestingly, shortly after the addition of FCS, tubules extended from giant autolysosomes in spin knockdown cells (Fig. 5C) similar to the tubules that have been associated with ALR and establishment of lysosome homeostasis (12). Within 2 h of the addition of FCS, most of the giant autolysosomes had disassembled into small Lamp1-positive vesicles (Fig. 5C). These data demonstrate that reactivation of mTOR signaling, which can be achieved by replacing nutrients exogenously, is sufficient to induce ALR even in spin knockdown cells. This finding supports our conclusion that the stagnation of autolysosome progression to ALR in starved spin-deficient cells is due to a failure in mTOR activation, which likely results from defective sugar efflux, which has a deleterious effect on autolysosomal degradation of a variety of macromolecules, leading to a failure of mTOR activation in these cells.
Discussion
Drosophila spin mutants have been shown to exhibit progressive neurodegeneration, and lysosomal abnormalities have been shown to contribute to neurodegeneration in this context (4, 7). Nonetheless, how defects in spin lead to lysosomal abnormalities has been unclear. Here, we demonstrate that the structures that accumulate in spin mutants are autolysosomes, and that spin is required for ALR following prolonged starvation.
It is interesting that abnormalities in lysosome function and morphology become apparent only under starvation conditions in cultured mammalian cells. This might be explained by the fact that under nutrient-rich conditions, the influx of lysosome cargo is limited, but when cells undergo autophagy, lysosome cargo influx increases, magnifying the severity of the defect. If this hypothesis is correct, then one must wonder why spin mutant flies exhibit an accumulation of slightly enlarged Lamp1-positive structures even when fed (4). One possibility is that the metabolism of flies in this respect is greater than the metabolism of mammals. Another possible explanation is that the fluctuation of nutrients in vivo is much greater than in cells grown in culture medium. Thus, a nonstarved animal could have higher basal levels of autophagy compared with cells maintained in culture. This might be particularly important in the brain because selective deletion of Atg5 in neuronal cells leads to neurodegeneration even in unstarved mice (13).
One question is how Spin, a lysosomal efflux sugar transporter, alters the protein degradation capacity of lysosomes. Lysosome protein degradation capacity is dependent on the lysosomal internal environment. Lysosome pH is one of the major factors regulating lysosomal degradation ability, as the optimal pH for many lysosomal proteases is around 4.5; in either lower or higher pH, lysosome protease activity is compromised. Interestingly, we found that upon starvation, spin knock-down cells exhibit a dramatically decreased lysosomal pH. However, it is unknown how spin knock-down leads to an increase in lysosomal acidity. One possibility is that Spin is a H+/sugar symporter. H+/amino acid symporters have been indentified in lysosomes, for example, LYAAT1, a lysosome amino acid efflux transporter, is a H+/amino acid symporter, and the efflux transport of amino acids by LYAAT1 is driven by the H+ gradient (14). Similarly, cystinosin, a lysosomal cystine efflux transporter which also contains seven transmembrane domains, has been reported to be a H+/cystine symporter which uses H+ to drive cystine efflux transport (14). A recent structural study for FucP, a H+/Fucose symporter which also belongs to the major facilitator superfamily (MFS) of transporters further supports this hypothesis. A structural study showed that a conserved E residue must be protonated for proper Fucose transport (15). Interestingly, in our rescue experiment, the E217K mutation failed to rescue ALR in spin knockdown cells. If this hypothesis is correct, we would expect that spin knockdown would block both sugar and H+ efflux and lead to lysosomal acidification, and autolysosomal degradation defects. Further investigation will be required to explore this possibility.
There is a bidirectional regulation between autolysosomal degradation and mTOR reactivation/ALR. On one hand, degradation of autolysosomal content is required for mTOR reactivation and ALR; on the other hand, defective mTOR reactivation/ALR also causes the impairment of degradation (10). For example, if mTOR reactivation/ALR is blocked during starvation by either adding the mTOR inhibitor rapamycin or knocking down mTOR, the degradation of autophagy substrate LC3 is impaired. Interestingly, we found that adding FCS rapidly, albeit partially, restores the pH and degradation capacity of spin knockdown cells (Fig. S9), indicating that mTOR may play a role in regulating the pH, and thus the degradation capacity, of autolysosomes. These data raise the interesting possibility that mTOR may regulate ALR by affecting the degradation capacity of autolysosomes.
One important implication of our findings is that ALR may play an important role in disease progression in LSDs. The phenotype of spin mutant flies has been long considered to be similar to certain LSDs (4). In this study, we demonstrate that a spin knockdown also leads to defects in autolysosomal degradation and ALR. These data clearly demonstrate that ALR is important to the maintenance of lysosome-based cellular degradation capacity. As we discuss above, the basal level of autophagy may be higher in vivo than cultured cells due to the greater fluctuation of nutrients during the feeding cycle. Thus, it is conceivable that a defect in ALR could cause LSD phenotypes in fed animals.
A long-lasting question in LSDs is why a mutation in a single lysosomal enzyme can cause overall lysosome degradation failure. Because the regulation between lysosomal degradation and ALR is bidirectional, our data suggest the interesting possibility that a minor defect in lysosomal degradation capacity could cause a minor defect in ALR, which could in turn amplify the lysosomal/autolysosomal degradation defect. The positive feedback nature of this regulation loop then may eventually cause the progressive pathological features of LSDs.
Materials and Methods
Reagents and Antibodies.
Lysotracker-red, dextran-red (MW 10000), DQ-BSA-red, and DQ-ovalbumin-green were from Invitrogen (Carlsbad, CA). Anti-Lamp1 antibody was from Sigma-Aldrich (St. Louis, MO).
Cell Culture and Transfection.
NRK cells were obtained from American Type Culture Condition (ATCC) and cultured in DMEM (Life Technologies) medium supplemented with 10% FBS (5% CO2). Cells were starved by removal of serum and glutamine. Cells were transfected via Amaxa nucleofection using solution T and program X-001, using 200 pmol of RNAi or total 2 μg of DNA. Cells were then cultured in growth medium for further analysis. For two rounds of transfection, cells were transfected with 200 pmol RNAi, and 72 h after transfection, cells were transfected again with 100 pmol of RNAi and up to 2 μg of DNA.
Constructs.
The human Spinster construct was kindly provided by D. Yamamoto (Waseda University). Lamp1 and LC3 constructs were provided by J. Lippincott-Schwartz (National Institutes of Health). GTP-Rab7 and Rab7 constructs were provided by J Bonifacino (National Institutes of Health). spinster mutant flies were generously provided by G. Davis (University of California), and tub-Lamp1-GFP flies were provided by H. Kramer (University of Texas Southwestern Medical Center).
Live Cell Imaging.
Transfected cells were replated in Lab Tek Chambered coverglass (Nunc) the night before imaging, and cells were maintained at 37 °C with 5% CO2 in a PeCon open chamber (PeCon, Erbach, Germany). Images were acquired by a Leica sp5 or Olympus FV1000 confocal microscope. Three-dimensional models were constructed by collecting images by Z-stack scanning at 0.5-μm intervals, and images were collapsed to construct 3D models by IMRIS. For fly experiments, Canton-S third instar larvae expressing tub-Lamp1-GFP (control), or transheterozygous spinster mutant larvae expressing Lamp1-GFP (tub-Lamp1-GFP; spinster10403/spinsterK09905) were starved in moist Petri dishes at 25 °C. The fatbody was dissected and imaged immediately without fixation on a Zeiss AxiovisionZ.1 microscope with fluorescence.
Staining.
Cells were washed with PBS (PBS), fixed in 2% paraformaldehyde for 10 min, and permeabilized in 0.2% Triton X-100 for 5 min. Cells were blocked with 10% FBS in PBS for 30 min, stained with 10 μg/mL of rabbit anti-Lamp1 (Sigma) in blocking buffer for 1 h, and washed with PBS three times. Cells were then stained with Fluorescein isothiocyanate conjugated (FITC)-anti rabbit secondary antibody (BD, San Jose, CA) in PBS for 1 h and washed with PBS three times. For PAS staining, cells were fixed with 10% formalin and embedded in paraffin, and 5-μm sections were deparaffinized hydrated with water. Sections were oxidized in 0.5% periodic acid solution for 5 min, rinsed in distilled water, and stained in Schiff reagent for 15 min. Sections were washed in lukewarm tap water for 5 min before dehydration and mounting. For lysosensor staining, cells were labeled with 1 μM LysoSensor Green DND-153 for 30 min, and then the probe-containing medium was replaced with fresh medium. For BODIPY FL pepstatin A staining, cells were labeled with 1 μM BODIPY FL pepstatin A for 30 min, and then the probe-containing medium was replaced with fresh medium.
Electron Microscopy.
Cells were fixed in 3% glutaraldehyde in 0.1 M Mops buffer (pH 7.0) for 8 h at room temperature, then 3% glutaraldehyde/1% paraformaldehyde in 0.1 M Mops buffer (pH 7.0) for 16 h at 4 °C. They were then postfixed in 1% osmium tetroxide for 1 h, and embedded in Spurr's resin, sectioned, doubly stained with uranyl acetate and lead citrate, and analyzed using a Zeiss EM 10 transmission electron microscope. For fly EM analyses, spinster mutant larvae (spinster10403/spinsterK09905) were compared with control progeny of spinster mutant lines (either spinster10403/Cyo-TM6B or spinsterK09905/Cyo-TM6B) crossed to wild-type Canton-S. third instar larvae were starved for 12 h on moist plates and fat was dissected and fixed in 2% glutaraldehyde-4% paraformaldehyde in PBS. Fat was then fixed for EM analysis as described above.
Supplementary Material
Acknowledgments
We are grateful to Olympus China and the Tsinghua cell biology core facility for providing technical support and O. Schwartz, Juraj Kabat, Lily Koo, Meggan Czapiga (Biological Imaging Facility, National Institute for Allergy and Infectious Diseases, National Institutes of Health (NIH), Qi Dong, and Ying Li for assistance with confocal microscopy, TEM, and imaging processing. We thank J. Lippincott-Schwartz and J. Bonifacino for helpful discussions and D. Yamamoto, G. Davis, and H. Kramer for constructs and fly strains. This research was supported by National Science Foundation Grants 31030043 and 30971484, 973 Program 2010CB833704, 2011CB910100, and Tsinghua University Grants 2010THZ0 and 2009THZ03071 (to L.Y.), NIH Grant GM079431 (to E.H.B.), and the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.M.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013800108/-/DCSupplemental.
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Walkley SU. Pathogenic cascades in lysosomal disease-Why so complex? J Inherit Metab Dis. 2009;32:181–189. doi: 10.1007/s10545-008-1040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 4.Dermaut B, et al. Aberrant lysosomal carbohydrate storage accompanies endocytic defects and neurodegeneration in Drosophila benchwarmer. J Cell Biol. 2005;170:127–139. doi: 10.1083/jcb.200412001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruivo R, et al. Molecular pathogenesis of sialic acid storage diseases: insight gained from four missense mutations and a putative polymorphism of human sialin. Biol Cell. 2008;100:551–559. doi: 10.1042/BC20070166. [DOI] [PubMed] [Google Scholar]
- 6.Vesa J, Peltonen L. Mutated genes in juvenile and variant late infantile neuronal ceroid lipofuscinoses encode lysosomal proteins. Curr Mol Med. 2002;2:439–444. doi: 10.2174/1566524023362311. [DOI] [PubMed] [Google Scholar]
- 7.Nakano Y, et al. Mutations in the novel membrane protein spinster interfere with programmed cell death and cause neural degeneration in Drosophila melanogaster. Mol Cell Biol. 2001;21:3775–3788. doi: 10.1128/MCB.21.11.3775-3788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweeney ST, Davis GW. Unrestricted synaptic growth in spinster-a late endosomal protein implicated in TGF-beta-mediated synaptic growth regulation. Neuron. 2002;36:403–416. doi: 10.1016/s0896-6273(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 9.Young RM, et al. Zebrafish yolk-specific not really started (nrs) gene is a vertebrate homolog of the Drosophila spinster gene and is essential for embryogenesis. Dev Dyn. 2002;223:298–305. doi: 10.1002/dvdy.10060. [DOI] [PubMed] [Google Scholar]
- 10.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanagisawa H, Miyashita T, Nakano Y, Yamamoto D. HSpin1, a transmembrane protein interacting with Bcl-2/Bcl-xL, induces a caspase-independent autophagic cell death. Cell Death Differ. 2003;10:798–807. doi: 10.1038/sj.cdd.4401246. [DOI] [PubMed] [Google Scholar]
- 12.Dennis PB, et al. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 13.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 14.Boll M, Daniel H, Gasnier B. The SLC36 family: proton-coupled transporters for the absorption of selected amino acids from extracellular and intracellular proteolysis. Pflugers Arch. 2004;447:776–779. doi: 10.1007/s00424-003-1073-4. [DOI] [PubMed] [Google Scholar]
- 15.Dang S, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467:734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.