Abstract
Current models of stem cell biology assume that normal and neoplastic stem cells reside at the apices of hierarchies and differentiate into nonstem progeny in a unidirectional manner. Here we identify a subpopulation of basal-like human mammary epithelial cells that departs from that assumption, spontaneously dedifferentiating into stem-like cells. Moreover, oncogenic transformation enhances the spontaneous conversion, so that nonstem cancer cells give rise to cancer stem cell (CSC)-like cells in vitro and in vivo. We further show that the differentiation state of normal cells-of-origin is a strong determinant of posttransformation behavior. These findings demonstrate that normal and CSC-like cells can arise de novo from more differentiated cell types and that hierarchical models of mammary stem cell biology should encompass bidirectional interconversions between stem and nonstem compartments. The observed plasticity may allow derivation of patient-specific adult stem cells without genetic manipulation and holds important implications for therapeutic strategies to eradicate cancer.
Keywords: breast cancer, dedifferentiation
Tissue-specific stem cells exist in many adult tissues and can be identified and isolated using specific antigen profiles. Their potential utility in regenerative medicine holds great promise. However, our current ability to isolate and propagate adult stem cells for such purposes is limited (1). This limitation is due in part to the paucity of stem cells in most epithelial tissues and our fragmentary understanding of the survival, proliferation, and differentiation signals that these cells receive in highly specialized stem-cell niches (2, 3).
The discovery of stem-like cells in a number of human solid tumor types has suggested a central role for stem cells in tumorigenesis. Thus, stem-like cells, often termed cancer stem cells (CSCs), have been defined experimentally by their ability to seed new tumors and to spawn non-CSC populations lacking tumor-initiating ability. CSC subpopulations have now been identified in a variety of malignancies (4). Importantly, CSC-rich tumors are associated with aggressive disease and poor prognosis (5), indicating that an understanding of their biology is pertinent to developing effective therapies.
Both normal and neoplastic stem cells are thought to be self-renewing and to reside at the apex of a cellular hierarchy. Through asymmetric division, these stem cells generate more differentiated progeny that lack self-renewal capacity (6, 7). Intratumor heterogeneity may thus derive from neoplastic cells at various differentiation stages. In the case of human mammary epithelial cells (HMECs), the structure of the associated stem-cell hierarchy is yet to be definitively described. It is becoming apparent, however, that the differentiation states of cells-of-origin can influence the organization of derived neoplastic cell populations (8, 9).
These hypotheses have been difficult to validate because of the apparent critical role of the stem-cell niche and associated microenvironment in the survival and differentiation of both normal and neoplastic stem cells. However, evidence of normal and neoplastic cells with stem-cell properties residing naturally among populations of epithelial cells propagated in culture has been reported (10–12).
In light of the latter observations, we undertook to study the biology of subpopulations of HMECs that exist in culture and share certain properties with either stem-like cells or their more differentiated derivatives. We identified an unexpected degree of plasticity between stem-like and nonstem cell compartments, leading to the demonstration that differentiated cell types can convert to stem-like cells. Moreover, these observations hold true for the neoplastic counterparts of such cells.
Results
Enrichment of a Rare Floating Population of Cells from Cultured Human Mammary Epithelial Cells.
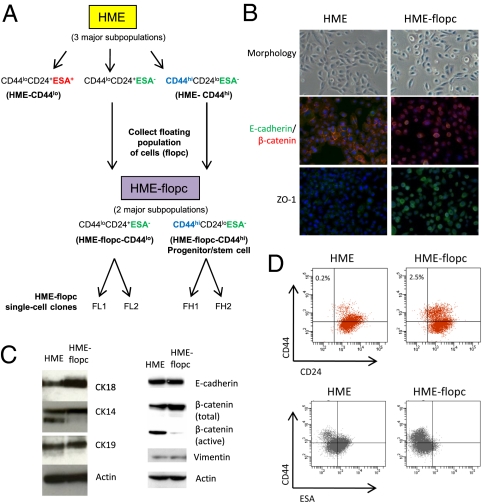
In the work described below we used primary HMECs as well as HMECs immortalized with human telomerase (hTERT) that are termed here HME cells (13, 14). We observed that populations of HME cells cultured in their normal mammary epithelial growth medium contained a small proportion of cells that grew as floating cells above the majority population of adherent cells. These populations, termed here HME–floating population of cells (HME-flopcs), were collected from the conditioned media of HME cells through centrifugation and introduced into new culture dishes, yielding fully viable, adherent cell populations.
The epithelial nature of HME and HME-flopc cells was confirmed by the expression of cytokeratins and low vimentin expression (Fig. 1C) and by formation of acinar structures when plated at clonal density on a layer of Matrigel (15). These polarized epithelial structures contained cells expressing vimentin, cytokeratins, and MUC1, the latter marking luminal epithelial cells in the mammary gland (Fig. S1C). However, under two-dimensional (2D) culture conditions, HME cells formed a typical epithelial monolayer with junctional E-cadherin, β-catenin, and ZO-1, whereas HME-flopc cells did not, despite similarly high levels of those proteins (Fig. 1 B and C). Thus, HME-flopc cells were stably morphologically distinct from the bulk HME cells.
Fig. 1.
Enrichment of a rare population of floating cells from cultured mammary epithelial cells. (A) Schematic illustrating the derivation of HME-flopcs and single-cell clones (SCCs), as well as the antigen profiles of the major cell subpopulations described in this paper. (B) Phase contrast micrographs (20×) of HME and HME-flopc populations in 2D culture. Immunofluorescence of HME and HME-flopc cells is shown, for E-cadherin (green), β-catenin (red), and ZO-1 [nuclei stained blue with 4,6-diamidino-2-phenylindole (DAPI)]. (C) Immunoblot with antibodies to cytokeratins 14, 18 and 19, vimentin, E-cadherin and β-catenin (total and active forms). (D) Flow cytometry plots for CD44, CD24, and ESA on bulk HME and HME-flopc cells. Gating is set to unstained control cells.
HME-flopc Cultures Are Enriched for CD44loCD24+ESA− and CD44hiCD24loESA− Cells.
The human mammary gland contains at least three distinct epithelial cell types: luminal cells (CD44loCD49floCD24+ESA+), basal/myoepithelial cells (CD44loCD49fhiCD24+ESA−), and bipotent progenitor/stem cells (CD44hiCD24loESA−) (10, 16). We note that a definitive marker profile to distinguish mammary progenitor cells from stem cells does not currently exist and, thus, tentatively consider cell populations with the CD44hiCD24loESA− marker profile to be enriched for stem-like cells. By flow cytometry (FACS) for CD44hiCD24loESA− cells we determined that HME-flopc cells are enriched for a putative stem-like fraction over HME cells (2.5% vs. 0.2%, respectively) (Fig. 1D).
We also analyzed the CD44loCD24+ nonstem cell fractions of HME and HME-flopc cells and found HME CD44loCD24+ cells were ∼51% ESA+ and 49% ESA− cells, whereas HME-flopc CD44loCD24+ cells were 96% ESA− (Fig. 1D). Together these results indicated the presence of three major subtypes of cells coexisting within HME cultures, which we have designated as follows (Fig. 1A): (i) HME-CD44lo cells (CD44loCD24+ cells of ESA+ and ESA− phenotypes), (ii) HME-flopc-CD44lo cells (CD44loCD24+ cells of ESA− phenotype only), and (iii) HME-flopc-CD44hi cells (exhibiting the CD44hiCD24loESA− antigen profile).
The flopc isolation method enriched for the latter two cell populations. Importantly, HME-flopc-CD44hi cells bear a marker profile similar to mammary progenitor/stem cells and HME-CD44lo cells and HME-flopc-CD44lo cells bear marker profiles of more differentiated cell populations.
HME-flopc-CD44lo Cells Spontaneously Convert into CD44hi Cells.
We explored the biological properties of the two major cell types that constitute the HME-flopc population: the HME-flopc-CD44hi putative stem-like fraction and the nonstem HME-flopc-CD44lo fraction. We purified these cell types through single-cell cloning yielding seven single-cell clones (SCCs) that were stably CD44hi and 28 SCCs that were predominantly CD44lo. The latter CD44lo SCCs also contained a small component of CD44hi cells (ranging from 1 to 10% of the population) (Fig. S1 and data not shown). This result provided the first indication that one population of cells (HME-flopc-CD44lo) can give rise to the other (HME-flopc-CD44hi).
To further explore this notion, we fractionated two HME-flopc-CD44lo SCC populations (termed FL1 and FL2, where “L” designates that the clones are predominantly CD44lo) (Fig. 1A and Fig. S1E) into CD44hi and CD44lo subpopulations by FACS, introduced the purified populations into 2D culture, and monitored them by FACS over the subsequent 12 d. Purified HME-flopc-CD44hi cells did not regenerate the HME-flopc-CD44lo cell subpopulation and remained as pure CD44hi populations (data not shown). However, the HME-flopc-CD44lo cultures developed a progressively increasing CD44hi subpopulation (Fig. 2 A and C).
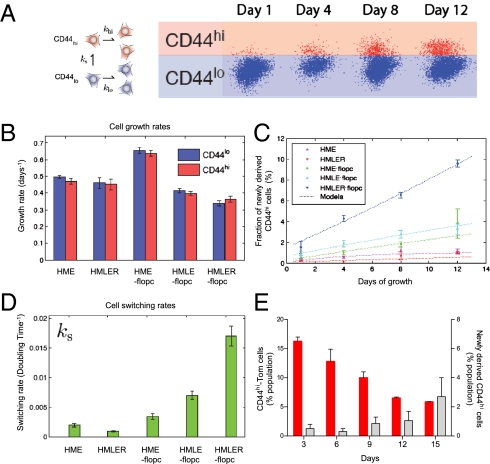
Fig. 2.
Normal and transformed HME-flopc-CD44lo cells spontaneously convert into CD44hi cells. (A) Population dynamics modeled by a simple growth model in which CD44lo cells either divide (klo) or spontaneously switch (ks) to a CD44hi state. CD44hi cells divide (khi) but cannot switch to the CD44lo state. Cell populations are quantified by flow cytometry. (B) Growth rates of purified CD44lo and CD44hi cell populations analyzed in C, n = 6, Promega CellTiter 96 AQueous Assay. (C) Quantification of the spontaneous dedifferentiation of purified CD44lo cells isolated from bulk HME cells, HME-flopcs, and HME-flopcs expressing SV40 early region only (HMLE-flopc), as well as fully transformed HME cells (HMLER) and HME-flopc cells (HMLER-flopcs) expressing the SV40 early region and Ras oncoproteins (n = 3–6, results are mean ± SEM). (D) Switching rate (ks) of various CD44lo cell types per population doubling time. (E) HME-flopc-CD44lo cells purified from two independent single-cell clones (no color) were mixed with fluorescent (pLV-Tomato) HME-flopc-CD44hi (CD44hi-Tom) cells to recreate a heterogeneous population. The percentages of CD44hi-Tom cells (red bars) and of de novo-derived CD44hi cells arising from HME-flopc-CD44lo cells expressing no fluorescent protein (gray bars) were measured over a time course of 12 d. Results are mean ± SEM.
The observed increase in CD44hi cells in these cultures was compatible with either of two mechanisms: the initial CD44lo cells spontaneously dedifferentiated into CD44hi cells or the emerging population of CD44hi cells grew out from CD44hi cells that had contaminated the purified CD44lo population following the initial FACS. In fact, the proliferation rates of the two cell populations (and of second-generation CD44hi cells repurified from HME-flopc-CD44lo cells) were not significantly different (Fig. 2B). Moreover, in a reconstructed heterogeneous population, in which we mixed purified CD44hi cells expressing the tomato fluorescent protein (CD44hi-Tom) with purified, unlabeled HME-flopc-CD44lo cells, the percentage of CD44hi-Tom cells decreased progressively from 15 to 5% over 12 d (Fig. 2E). This result demonstrated directly that the CD44hi cells actually proliferated more slowly than HME-flopc-CD44lo cells in a mixed cell population.
Together these results indicated that the emerging CD44hi cells could not have arisen from a contaminating CD44hi subpopulation that outgrew the majority CD44lo cell population. We thus concluded that HME-flopc-CD44lo cells spontaneously generated CD44hi cells in 2D cultures.
Interestingly, the presence of admixed CD44hi-Tom cells (Fig. 2E) did not inhibit the de novo generation of CD44hi cells arising from the unlabeled HME-flopc-CD44lo cells, arguing against some type of homeostatic control of CD44hi cell number under these culture conditions.
We repeated the same time-course experiment with purified populations of HME-CD44hi and HME-CD44lo cells fractionated by FACS directly from parental HME cells. Unlike HME-flopc-CD44lo cells, HME-CD44lo cells were poorly able to generate CD44hi cells in vitro (Fig. 2C).
Display of Mammary Progenitor Traits by Preexisting and de Novo-Generated CD44hi Cells.
We used the in vitro mammosphere-forming assay (17) to acquire functional evidence that de novo-derived and preexisting HME-flopc-CD44hi cells were enriched for cells with mammary stem-like traits. Cells were plated as single cells at a density of 300 cells/well (96-well plate). Bulk HME cells did not form any detectable mammospheres, demonstrating that the stem-like fraction represented <1 in 2,000 cells in this population. In contrast, bulk HME-flopc cells and HME-flopc-CD44lo SCCs (n = 3) formed mammospheres at a frequency of 1.2/300 cells and 0.4/300 cells, respectively (Fig. 3A). Stable HME-flopc-CD44hi SCCs formed mammospheres at the highest rate (2.3/300 cells) (Fig. 3A), indicating that mammosphere formation correlates with CD44hi expression.
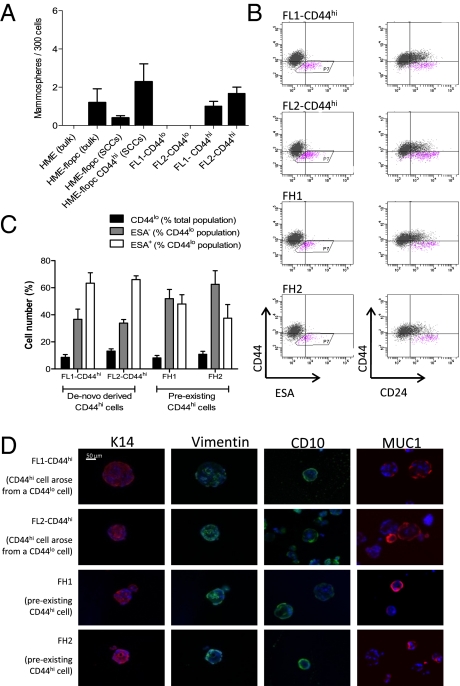
Fig. 3.
CD44hi cells display stem-like properties in vitro. (A) Mammosphere-forming ability of bulk HME or HME-flopc cells, HME-flopc-CD44lo single-cell clones (SCCs, n = 5), HME-flopc-CD44hi SCCs (n = 4), and CD44hi and CD44lo cells purified by FACS from two independent HME-flopc SCCs (FL1 and FL2). Results are mean ± SEM. (B) Representative flow cytometry plots for the markers CD44, CD24, and ESA of cells isolated from dissociated mammospheres from de novo-derived CD44hi cells (FL1-CD44hi and FL2-CD44hi) and two stable HME-flopc-CD44hi SCCs (FH1 and FH2). (C) Quantification of total CD44lo cells, CD44loESA− and CD44loESA+ populations [from (B) box P7; n = 3]. Results are mean ± SEM. (D) Frozen sections of mammospheres (5 μm) stained by immunofluorescence for myoepithelial [CD10, vimentin, and cytokeratin-14 (K14)] and luminal (K14 and MUC1) differentiation markers.
Because HME-flopc-CD44lo SCCs inevitably contained subpopulations of CD44hi cells (Fig. S1E), we purified CD44lo and CD44hi fractions by FACS and demonstrated that CD44lo cells did not form mammospheres, whereas CD44hi cells formed mammospheres quite efficiently (∼1.5/300 cells) (Fig. 3A). These results indicated that (i) mammosphere formation is largely if not entirely restricted to CD44hi cells and (ii) both preexisting CD44hi cells (HME-flopc-CD44hi SCCs) and de novo arising CD44hi cells (arising from HME-flopc-CD44lo cells) display mammosphere-forming ability.
CD44hi Cells Can Differentiate into CD44loCD24+ESA− and CD44loCD24+ESA+ Progeny.
Whereas HME-flopc-CD44hi stem-like cells did not give rise to more differentiated progeny in 2D culture (Fig. S1), we reasoned that the 3D mammosphere culture environment may be more conducive for differentiation to occur. Mammospheres that had been initially seeded by de novo-derived CD44hi cells (purified CD44hi cells from the FL1 and FL2 CD44lo SCCs, designated FL1-CD44hi and FL2-CD44hi) and two stable CD44hi SCCs (FH1 and FH2, where “H” designates that the clones are pure CD44hi) (Fig. 1A) were dissociated by trypsinization and analyzed for CD44, CD24, and ESA expression. All CD44hi-initiated mammospheres contained both CD44loCD24+ESA+ and CD44loCD24+ESA− cells (Fig. 3 B and C). Immunofluorescence revealed that each mammosphere contained vimentin+, CK14+, and CD10+ cells (markers of myoepithelial differentiation) and MUC1+ cells (a marker of luminal differentiation) (Fig. 3D). Hence, 3D but not 2D culture conditions permitted the conversion of the various CD44hi cells into at least two types of more differentiated derivatives. Moreover, in the case of de novo-derived CD44hi cells, 3D culture conditions were able to reverse the process that led to their recent de novo formation in 2D culture.
By gene set enrichment analysis (18) we compared the gene expression profiles of cultured HME-flopc-CD44hi mammary stem-like cells to those of primary human mammary stem-like cells (19). HME-flopc-CD44hi cells were enriched in both bipotent progenitor cell and luminal progenitor cell gene expression signatures compared with bulk HME-flopc cells (Fig. S1D), indicating that HME-flopc-CD44hi cells are enriched in a gene signature that resembles that of primary human mammary progenitor cells.
CD44hi Cells Reconstitute the Humanized Mouse Mammary Fat Pad.
We used the humanized mouse mammary fat pad reconstitution assay (20) to test for human stem cell activity by assessing a cell's ability to generate mammary structures in a humanized murine fat pad of NOD/SCID mice. We implanted a variety of cell populations (2 × 105 cell aliquots): (i) parental unfractionated HME cells, (ii) preexisting pure HME-flopc-CD44hi cells (clone FH1, Fig. 1A), (iii) spontaneously arising CD44hi cells purified by FACS from HME-flopc SCC FL1, and (iv) uncultured, single-cell suspensions of freshly isolated HMECs (which served as a positive control).
At 12 wk, ductal structures arose from the uncultured HMECs as well as from both FH1 and FL1-CD44hi cells, whereas HME cells gave rise only to cell aggregates and simple cystic structures. The ductal structures arising from FH1, FL1-CD44hi, and uncultured HMECs were composed of stratified epithelium that stained for keratin-14 in the basal layer and pan-cytokeratin in the luminal layer (Fig. 4A). The human origin of the ductal structures was confirmed by the expression of pLV-Tomato using fluorescent microscopy of whole-mount mammary fat pads (Fig. S2A). By this stringent measure, de novo-derived and preexisting CD44hi cells contain bona fide HMEC stem-like cells. We note additionally that the CD44hi cells could maintain their stem-like state under 2D culture conditions that did not recreate a specialized niche microenvironment.
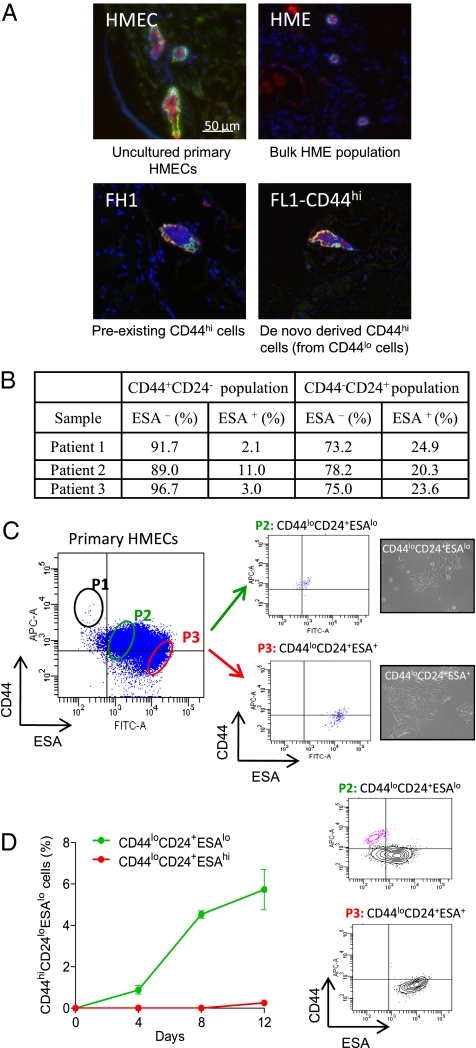
Fig. 4.
CD44loCD24+ESAlo cells isolated from primary HMECs convert into the CD44hiCD24loESAlo mammary stem-like state. (A) Single-cell suspensions of (i) uncultured primary human mammary epithelial cells (HMECs), (ii) bulk HME cells (expressing hTERT), (iii) FH1 [preexisting HME-flopc CD44hi single-cell clone (SCC)], and (iv) spontaneously arising CD44hi cells (purified from HME-flopc CD44lo SCC-FL1) were injected into the humanized mouse mammary fat pad. After 12 wk, mammary glands were sectioned and stained by immunofluorescence for basal (keratin 14, green) and luminal (pan-cytokeratin, red) markers. (B) Quantification of subpopulations of uncultured HMECs following direct isolation from primary tissue (SI Materials and Methods). (C) Flow cytometry plot illustrating the gating strategy used to isolate subpopulations of HMECs. P1, proposed mammary stem cell-enriched fraction; P2, CD44loCD24+ESAlo basal cells; P3, CD44loCD24+ESA+ luminal fraction with phase contrast micrographs (20×) of purified P2 and P3 cell populations. (D) Purified (via flow cytometry) primary CD44loCD24+ESAlo and CD44loCD24+ESA+ HMECs were monitored for 12 d in 2D culture for their ability to spontaneously convert into the CD44hiCD24+ESAlo cell state. Results are mean ± SEM. Flow cytometry plots of the purified CD44loCD24+ESAlo and CD44loCD24+ESA+ cell populations at day 12 are shown.
Conversion of Primary HMECs to the CD44hi24loESA− Stem-Like State.
To demonstrate that cells similar to the three subpopulations of mammary epithelial cells identified in cultured HME cells (Fig. 1A) mirrored cell types that exist in vivo in the normal human mammary gland, we analyzed uncultured primary HMECs for CD44, CD24, and ESA expression (Fig. S6 and SI Materials and Methods). We also analyzed other commonly used markers (9, 21) on cultured HMECS to further identify HMEC subpopulations (Fig. S2B). We detected a CD44hiCD24loESA− stem-like fraction, a CD44loCD24+ESA+ luminal cell population (present in HME-CD44lo cells), and a CD44loCD24+ESA− basal cell population (equivalent to HME-flopc-CD44lo cells). In uncultured HMECs, the basal cell population was on average 3.3 times more abundant than the luminal fraction (Fig. 4B). The ploidy of all cells was checked (Fig. S3). Hence HME and HME-flopc populations likely have biological equivalents in primary normal human mammary breast tissue.
We next sought to demonstrate that freshly isolated primary basal HMECs (equivalent to HME-flopc-CD44lo cells) could spontaneously convert to the CD44hiCD24loESA− stem-like state. To this end, basal cells (P2) and luminal cells (P3) were purified from 2D-cultured bulk primary HMECs by FACS (Fig. 4C) and monitored for 12 d in 2D culture. The percentages of P1 (the putative CD44hiCD24loESA− mammary stem cell fraction), P2 (basal population), and P3 (luminal population) in cultured HMECs were on average 0.3%, 62%, and 38%, respectively.
Indeed, primary basal cells converted to the CD44hiCD24loESA− progenitor/stem-like state (which represented 6% of the population by day 12). As also predicted from our earlier studies, primary luminal cells poorly converted into the progenitor/stem-like state (Fig. 4D). These results demonstrate that primary mammary basal epithelial cells can spontaneously generate progenitor, if not stem-like, cells in 2D culture.
Transformed Epithelial Cells Spontaneously Generate Cancer Stem-Like Cells.
We next wanted to determine whether the oncogenic counterparts of HME-flopc-CD44lo cells could also undergo spontaneous conversion. If so, we reasoned that such newly arising cells would have the properties of CSCs. We therefore transformed HME and HME-flopc cells, by the sequential introduction of the SV40 early region (SV40-ER) and the H-ras oncogenes according to an established protocol (13). In the text that follows, cells expressing hTERT and SV40-ER are denoted with the suffix LE (e.g., “HMLE-flopc”), and those expressing all three introduced genes—hTERT, SV40-ER, and Ras—are denoted with the suffix LER (e.g., “HMLER-flopc”).
We determined that HMLE-flopc-CD44lo cells converted into the CD44hi state more efficiently than the nontransformed HME-flopc population. Moreover, HMLER-flopc-CD44lo cells converted into the CD44hi state at an even higher rate (Fig. 2C). Like their nontransformed HME-CD44lo counterparts, HMLER-CD44lo cells displayed minimal interconversion ability (Fig. 2C).
We developed a simple proliferation-switching model (SI Materials and Methods) to determine the rate of conversions of CD44lo cells into the CD44hi state (shown schematically in Fig. 2A), by using the proliferation rates (ki) of purified CD44lo and CD44hi cells and the percentage of spontaneously arising CD44hi cells (ks) from each population over a 12-d period (Fig. 2 B and C).
HME-flopc-CD44lo, HMLE-flopc-CD44lo, and HMLER-flopc-CD44lo cells convert into the CD44hi state at rates of 0.0034, 0.0070, and 0.0170 per cell division, respectively (Fig. 2D). As such, conversion was increased twofold by the addition of SV40-ER alone and fivefold by the addition of SV40-ER plus Ras, relative to nontransformed HME-flopc cells. Hence, the sequential addition of SV40-ER and Ras progressively lowered the barriers to spontaneous conversion.
Spontaneous Generation of Cancer Stem-Like Cells in Vivo.
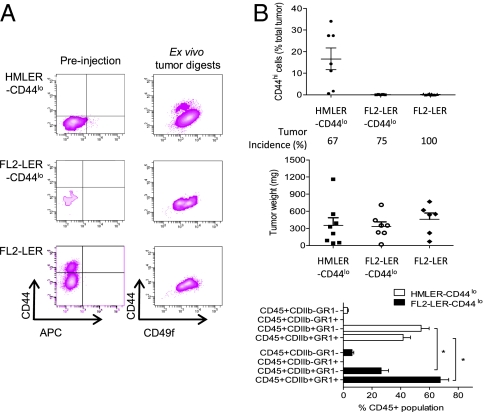
To determine whether transformed CD44lo cells could spontaneously dedifferentiate into CD44hi-CSCs in vivo, we injected FACS-purified cells expressing pLV-Tomato s.c. into NOD/SCID mice; the injected cells were HMLER-CD44lo (purified CD44lo cells from the oncogenic counterparts of HME cells, which convert poorly to the CD44hi state in 2D culture), FL2-LER-CD44lo cells (purified CD44lo cells from the oncogenic counterpart of clone FL2, which convert efficiently to the CD44hi state in 2D culture), or the bulk FL2-LER population (the bulk oncogenic counterpart of clone FL2 containing both CD44hi and CD44lo cells). After 8–10 wk, tumors were removed, digested, and analyzed by FACS for CD44 status in the tomato-positive tumor cell population (Fig. 5A). We found that HMLER-CD44lo cells had readily converted to the CD44hi-CSC state in vivo, where CD44hi cells constituted ∼16% of the total tumor cell population.
Fig. 5.
CSCs are created de novo in vivo. (A) Representative FACS profiles of tumor cell populations before injection into NOD/SCID mice and of digested tumor cell populations following 8–10 wk in vivo. (B) Quantitation of in vivo de novo-derived CD44hi cells isolated from various tumor populations, including tumor incidence, tumor weight, and a FACS analysis of immune cells recruited to the different tumor types (n = 6–9 animals/group, P < 0.001, two-way ANOVA).
In contrast to their efficient ability to convert to the CD44hi state in 2D culture, FL2-LER-CD44lo–derived tumors contained <0.2% of CD44hi cells. FL2-LER-CD44lo cells induced a substantial inflammatory response resulting in highly cystic tumors. Relative to HMLER-CD44lo tumors, FL2-LER-CD44lo–initiated tumors contained a significantly decreased proportion of differentiated myeloid cells (CDllb+GR1−) with a concomitant significant increase in CDllb+GR1+ myeloid-derived suppressor cells (22) (Fig. 5B). We do not yet know how this inflammatory response affects the in vivo conversion of CD44lo to the CD44hi-CSC state. Nonetheless, the observed conversion of HMLER-CD44lo cells to the CD44hi state in vivo demonstrated that CD44hi cells can indeed arise de novo in vivo and highlights the key role of the tumor microenvironment in affecting this process.
Characteristics of Tumors Generated by Transformed Progenitor and Differentiated Epithelial Cells.
Breast carcinoma is a heterogeneous collection of diseases whose diversity may be explained by the biology of the respective cells-of-origin from which these tumors arise (8, 9). We thus compared the tumorigenicity of transformed derivatives of the HME bulk population (HMLER), of the HME-flopc bulk population (HMLER-flopc), of two HME-flopc-CD44lo SCCs (FL1-LER and FL2-LER), and of two stable HME-flopc-CD44hi SCCs (FH1-LER and FH2-LER). Importantly, the morphological differences that distinguished the untransformed precursors of these various cell populations were maintained in the respective transformed derivative populations and the in vitro proliferation rates of these various transformed cell populations were not significantly different (Fig. S4).
We implanted 5 × 105 cell aliquots of the various RAS-transformed cell populations orthotopically into NOD/SCID mice. Cell populations with more differentiated cell phenotypes (HMLER, HMLER-flopc, F1-LER, and F2-LER) were on average poorly tumorigenic (25%, 50%, 80%, and 30% incidences, respectively), with well-differentiated tumor morphologies, whereas cell lines with a normal mammary stem-like phenotype (FH1-LER and FH2-LER) were highly tumorigenic (100% incidence) with poorly differentiated tumor morphologies (Fig. S5). Limiting dilution analysis of the highly tumorigenic FH1-LER and FH2-LER cell populations determined the frequencies of tumor-initiating cells to be 1:1,420 and 1:1,804, respectively (determined by extreme limiting dilution analysis) (23) (Fig. S5E). We estimate these frequencies to be at least 1,000 times greater than the tumor-initiating frequency of the other RAS-transformed cell populations tested here.
The CSC phenotype of the different CD44hi cell populations was assessed in vitro by assessing tumorsphere-forming ability, where only CSC-derived tumorspheres have the ability to be serially passaged (11, 24). HMLER cells, containing a very low CD44hi component, formed low numbers of primary tumorspheres and could not be serially passaged, whereas de novo-derived and preexisting CD44hi cells efficiently formed primary, secondary, and tertiary tumorspheres (Fig. S5D).
Together these observations demonstrate that oncogenic transformation of mammary stem-like cells yields more aggressive tumors than does oncogenic transformation of differentiated mammary epithelial cells and provide further evidence that the biological state of normal cells-of-origin before transformation strongly influences the behavior of their descendants following transformation (8, 25).
Discussion
The most unanticipated discovery that has emerged from this study is the plasticity that we can now ascribe to human mammary epithelial cells. We have shown that differentiated mammary epithelial cells can convert to a stem-like state, doing so in an apparent stochastic manner in vitro. This conversion occurs in transformed and nontransformed HMECs isolated from cell lines and primary tissue. In each case, the conversion proceeded without genetic manipulation.
These findings represent a profound divergence from the currently accepted unidirectional hierarchical model of mammary epithelial cells and have widespread implications for the use of cultured cells. In mammalian cells, the idea that nonstem cells dedifferentiate to form functional stem cells has been restricted to the notion that progenitor cells can reacquire stem cell activity in mouse differentiating spermatogonia (26). As such, our work demonstrates in mammalian cells that differentiated epithelial cells can revert to a stem-like state.
Our findings also hold implications for the development of anticancer therapeutics. As we previously reported, cells that have been forced experimentally into a mesenchymal/stem-like state can be used to screen for candidate therapeutic agents that specifically target CSCs (27); the intent here was to eliminate these cells and thereby deprive tumors of their ability to regenerate and thrive following initial therapy. However, if non-CSCs can spontaneously dedifferentiate into CSCs, then targeting CSC populations will, on its own, be unlikely to yield durable clinical responses, because the therapeutic eradication of existing CSC populations might be followed by their regeneration from non-CSCs within the tumor under treatment.
Given the present findings, the known ability of microenvironmental signals to provoke epithelial–mesenchymal transitions (EMTs) and the close connection between passage through an EMT and entrance into a stem-cell state, we suspect that the presently observed spontaneous conversion in vitro may be augmented in vivo by contextual signals in the tumor microenvironment, such as those that drive the EMT (28, 29). Relevant here are studies demonstrating that hypoxia-inducible factors (HIFs) can induce the EMT phenotype and promote metastasis and the CSC phenotype (30, 31). Spontaneous dedifferentiation in vivo may involve the reactivation of one or more of the described pluripotency factors (Oct4, Klf4, c-myc, and Sox-2) (32). Hence, the representation of CSCs within tumor cell populations is likely to be influenced both by contextual signals and by the intrinsic phenotypic plasticity of these cells, as observed here.
The ability of non-CSCs to convert into CSCs in vivo might resolve many of the current inconsistencies of the CSC model. In particular, the observed plasticity that was once reserved for CSCs alone can now be associated with nonstem cells. As such, CSC populations may differ profoundly between various tumor types according to the inherent plasticity of cells in their respective nonstem fractions and their ability to spawn CSCs de novo.
The present observations lend further support to the emerging view that the biological state of cells-of-origin is an important determinant of the phenotype of their transformed derivatives (8, 9), where experimental transformation of cells that have a phenotype related to that of mammary stem cells generates cell populations with a high frequency of tumor-initiating cells (∼1:1,420–1:1,804 cells) and metastasis, which contrasts with the low tumor-initiating ability and nonmetastatic nature of tumors derived via transformation of more differentiated cell types.
The present findings hold the implication that patient- and tissue-specific stem-like cells may one day be created in vitro via spontaneous conversion of a patient's own terminally differentiated epithelial cells, a process that would not require any genetic alteration of these cells. Such stem-like cells could be important for regenerative therapies. Our results further emphasize the pathological implications of cellular plasticity in cancer development, progression, and recurrence. Further research needs to be undertaken to determine the mechanism underlying the de novo generation of CSCs from non-CSCs in vivo, with the promise of potential novel targets for future cancer therapies aimed at eradicating CSCs.
Materials and Methods
Detailed materials and methods are provided in SI Materials and Methods.
Cell Culture.
HME cells and all derivatives were cultured in MEGM media as previously described (13). HMECs were isolated from primary tissue as previously described (33) and cultured in M87A+X (34). A list of antibodies is provided in Table S1.
Mammosphere Culture.
Mammosphere culture was performed as previously described (17).
Flow Cytometry.
Cells were prepared according to standard protocols.
Animal Studies.
Athymic female nude mice were 2–4 mo of age at time of injections. Tumor cells were resuspended in 10% Matrigel/MEGM (20 μL) for mammary fat pad injections. GFP-positive lung metastases were counted from individual lobes by fluorescent microscopy.
Statistical Analysis.
Data are presented as mean ± SEM. Student's t test (two-tailed) was used to compare two groups (P < 0.05 was considered significant) unless otherwise indicated.
Supplementary Material
Acknowledgments
We thank the Core Facilities at Whitehead Institute for Biomedical Research at Massachusetts Institute of Technology, Koch Institute for Cancer Research, and Annie Gifford for technical assistance at Whitehead Institute for Biomedical Research. This work was supported by the National Health and Medical Research Council of Australia (C.L.C.), National Institutes of Health Grant U54 CA12515, Massachusetts Institute of Technology's Ludwig Center for Molecular Oncology, the Breast Cancer Research Foundation, the Advanced Medical Research Foundation, and a Department of Defense Breast Cancer Research Program Idea Award. R.A.W. is an American Cancer Society Research Professor and a Daniel K. Ludwig Foundation Cancer Research Professor.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102454108/-/DCSupplemental.
References
- 1.Teo AK, Vallier L. Emerging use of stem cells in regenerative medicine. Biochem J. 2010;428:11–23. doi: 10.1042/BJ20100102. [DOI] [PubMed] [Google Scholar]
- 2.Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tavazoie M, et al. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alison MR, Islam S, Wright NA. Stem cells in cancer: Instigators and propagators? J Cell Sci. 2010;123:2357–2368. doi: 10.1242/jcs.054296. [DOI] [PubMed] [Google Scholar]
- 5.Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18:460–466. doi: 10.1016/j.copbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Alison MR, Islam S. Attributes of adult stem cells. J Pathol. 2009;217:144–160. doi: 10.1002/path.2498. [DOI] [PubMed] [Google Scholar]
- 7.Tan BT, Park CY, Ailles LE, Weissman IL. The cancer stem cell hypothesis: A work in progress. Lab Invest. 2006;86:1203–1207. doi: 10.1038/labinvest.3700488. [DOI] [PubMed] [Google Scholar]
- 8.Ince TA, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 10.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charafe-Jauffret E, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn WC, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 16.Stingl J. Detection and analysis of mammary gland stem cells. J Pathol. 2009;217:229–241. doi: 10.1002/path.2457. [DOI] [PubMed] [Google Scholar]
- 17.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 19.Raouf A, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 20.Kuperwasser C, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci USA. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eirew P, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14:1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- 22.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Ponti D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 25.Gupta PB, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barroca V, et al. Mouse differentiating spermatogonia can generate germinal stem cells in vivo. Nat Cell Biol. 2009;11:190–196. doi: 10.1038/ncb1826. [DOI] [PubMed] [Google Scholar]
- 27.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 29.Brabletz T, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang MH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67:93–109. doi: 10.1023/a:1010615124301. [DOI] [PubMed] [Google Scholar]
- 34.Garbe JC, et al. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 2009;69:7557–7568. doi: 10.1158/0008-5472.CAN-09-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.